An adipokine feedback regulating diurnal food intake rhythms in mice
Figures
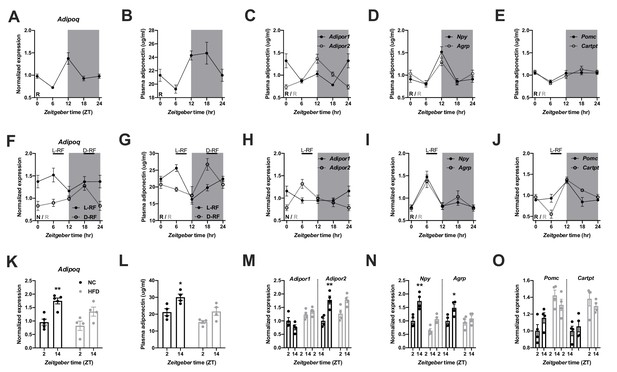
Adiponectin signaling integrates energy metabolic state in male mice.
(A) Daily profile of Adipoq mRNA expression in epididymal white adipose tissue (eWAT; n = 5 per time point). (B) Daily profile of ADIPOQ peptide in plasma (n = 3 per time point). (C) Daily profiles of Adipor1 (filled circles) and −2 (open circles) mRNA expression in the mediobasal hypothalamus (MBH; n = 5 per time point). (D, E) Daily profiles of Npy, Agrp (D), Pomc and Cartpt (E) mRNA expression in the MBH (n = 5 per time point). (F, G) Daily profiles of eWAT Adipoq mRNA expression (F) and plasma ADIPOQ levels (G) under light phase (L-RF; filled circles) or dark phase restricted feeding conditions (D-RF; open circles; n = 3 per time point). (H–J) Daily profiles of MBH Adipor1/2 (H), Npy/Agrp (I) and Pomc/Cartpt (J) mRNA expression under L-RF conditions (n = 3 per time point). (K, L) Time of day differences in eWAT Adipoq mRNA expression (K) and plasma ADIPOQ levels (L) under normal chow (NC; black) and after 1 week of high-fat diet (HFD; grey) conditions (n = 4). (M–O) Time of day differences in MBH mRNA expression of Adipor1/2 (M), Npy/Agrp (N) and Pomc/Cartpt (O) under NC and HFD conditions (n = 4). All data are means ± SEM. R – rhythmic expression (p<0.05; CircWave); N – non-rhythmic expression (p>0.05); */** - p<0.05/0.01; 2-way ANOVA with Sidak’s multiple comparisons. p values: (K) 0.005 (NC; ZT2 vs. ZT14), ANOVA dF = F(1, 14)=0.90 (interaction)/22.63 (time)/3.73 (diet); (L) 0.029 (NC; ZT2 vs. ZT14), ANOVA dF = F(1, 12)=0.45 (interaction)/15.66 (time)/14.18 (diet); (M) 0.001 (Adipor2; NC; ZT2 vs. ZT14), ANOVA dF = F(3, 24)=7.98 (interaction)/13.74 (time)/12.35 (diet); (N) 0.002 (Npy; NC; ZT2 vs. ZT14) and 0.044 (Agrp; NC; ZT2 vs. ZT14), ANOVA dF = F(3, 24)=2.51 (interaction)/32.44 (time)/8.70 (diet).
-
Figure 1—source data 1
Raw data of experiments shown in Figure 1.
- https://cdn.elifesciences.org/articles/55388/elife-55388-fig1-data1-v2.xlsx
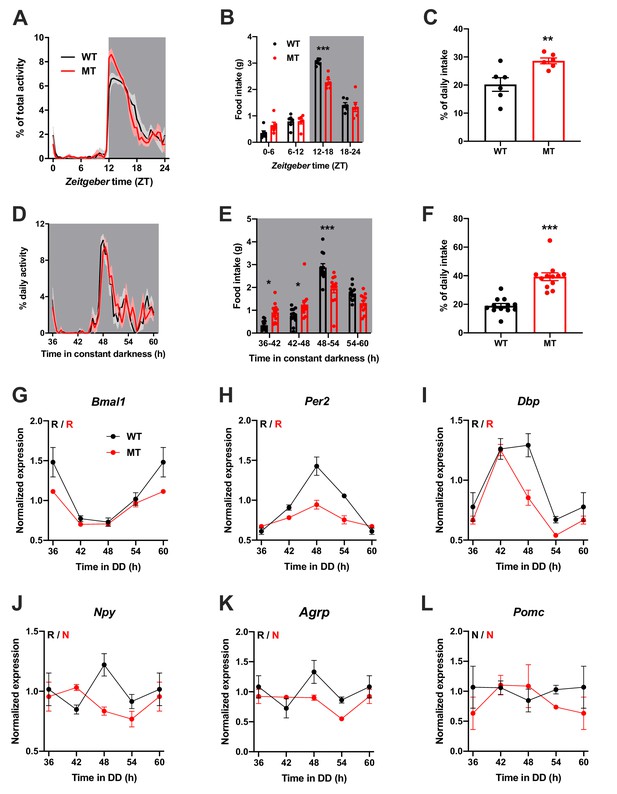
Dampened circadian rhythms of food intake and hypothalamic clock gene expression in Adipoq-deficient male mice.
(A) Daily profiles of running-wheel activity of wild-type (WT; black) and Adipoq-deficient mice (MT; red) under 12 hr light: 12 hr dark (LD) conditions (n = 6 per group; grey shading indicates dark phase). (B) Daily food intake profiles of WT and MT mice in LD in 6 hr bins (n = 6 per group). (C) Relative light phase food intake of WT and MT mice in LD (n = 6). (D–F) circadian profiles of running-wheel activity (D; n = 12 per group), food intake (E; n = 12 per group) and relative rest phase food intake (F) of WT and MT mice in constant darkness (DD; n = 12 per group). (G–L) Circadian profiles of MBH clock gene (G–I) and metabolic gene (J–L) mRNA expression in WT and MT mice in DD (n = 3 per time point). All data are means ± SEM. R – rhythmic expression (p<0.05; CircWave); N – non-rhythmic expression (p>0.05); */**/*** - p<0.05/0.01/0.001; 2-way ANOVA with Sidak’s multiple comparisons for B, E; unpaired Student’s t-test for C, F. p-values: (B) < 0.0001 (MT vs. WT ZT12-18), ANOVA dF = F(3, 40)=8.32 (interaction)/155.4 (time)/3.03 (genotype); (C) 0.0097 (MT vs. WT); (E) 0.015 (MT vs. WT 36–42), 0.040 (42-48),<0.0001 (48-54), ANOVA dF = F(3, 88)=14.79 (interaction)/66.95 (time)/0.61 (genotype); (F) < 0.0001 (MT vs. WT).
-
Figure 2—source data 1
Raw data of experiments shown in Figure 2.
- https://cdn.elifesciences.org/articles/55388/elife-55388-fig2-data1-v2.xlsx
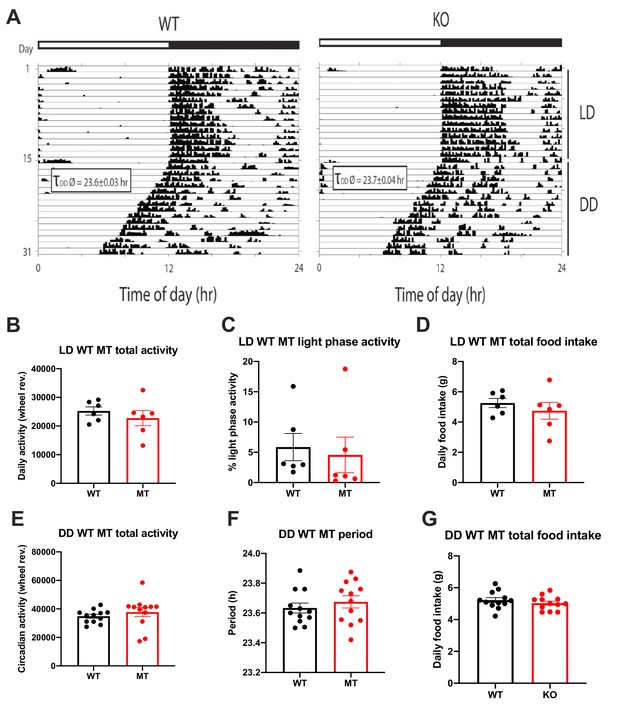
Normal rest-activity rhythms in Adipoq-deficient male mice.
(A) Actograms of representative male wild-type (WT; left) and Adipoq-deficient (MT; right) mice under 12 hr light: 12 hr dark (LD) and constant darkness (DD) conditions. Average DD free-running periods are depicted in the inset boxes. White-black bar on top indicates light regimen in LD. (B–D) Total running-wheel activity (B), relative light phase activity (C) and daily total food intake (D) of WT (black) and MT mice (red) in LD (n = 6 per group). (E–G) Total running-wheel activity (E), free-running period length (F) and daily total food intake (G) of WT and MT mice in DD (n = 12). All data are means ± SEM.
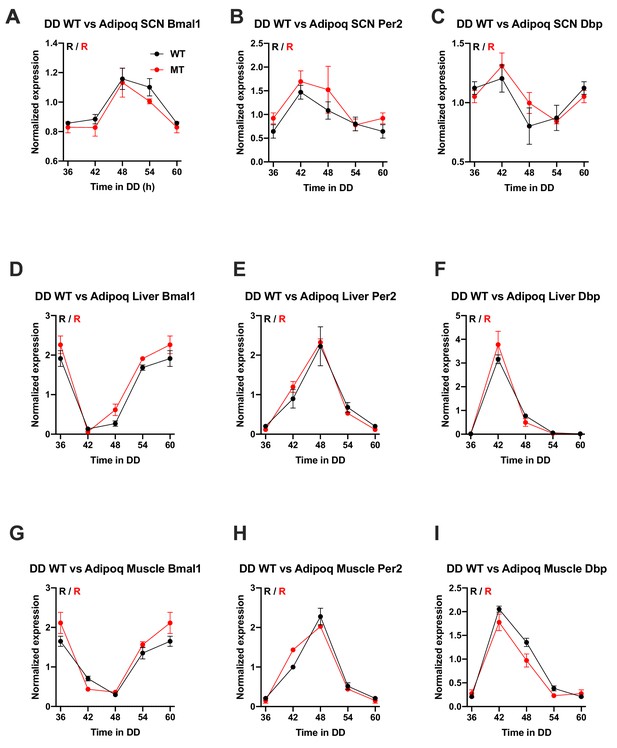
Preserved circadian clock gene expression rhythms in the SCN and peripheral tissues of Adipoq-deficient male mice.
(A–I) Circadian rhythms of clock gene mRNA expression in the SCN (A–C), liver (D–F) and skeletal muscle (G–I) of male wild-type (WT; black) and Adipoq-deficient mice (MT; red) in constant darkness (DD). All data are means ± SEM (n = 3 per timepoint and genotype). R – rhythmic expression (p<0.05; CircWave); N – non-rhythmic expression (p>0.05).
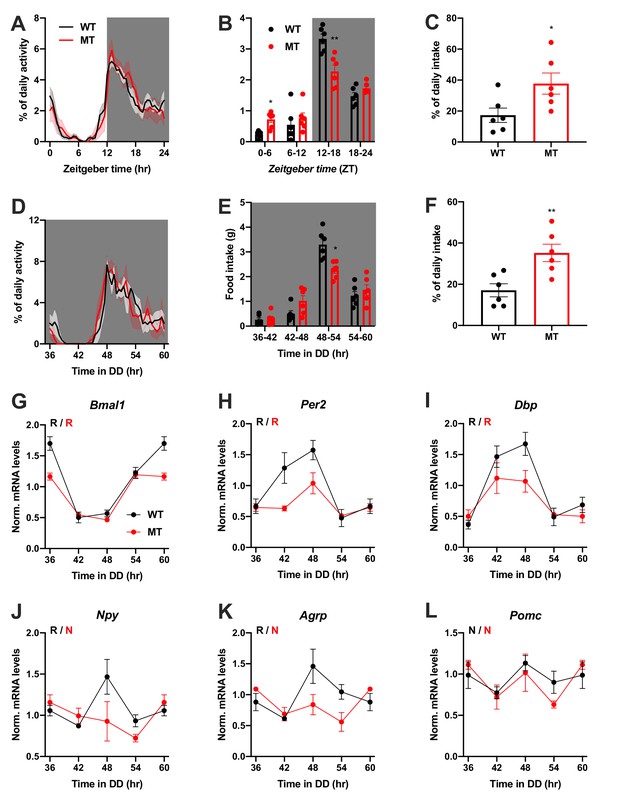
Dampened circadian rhythms of food intake and hypothalamic clock gene expression in Adipoq-deficient female mice.
(A) Daily profiles of running-wheel activity of wild-type (WT; black) and Adipoq-deficient mice (MT; red) under 12 hr light: 12 hr dark (LD) conditions (n = 6 per group; grey shading indicates dark phase). (B) Daily food intake profiles of WT and MT mice in LD in 6 hr bins (n = 6 per group). (C) Relative light phase food intake of WT and MT mice in LD (n = 6 per group). (D–F) Circadian profiles of running-wheel activity (D; n = 6 per group), food intake (E; n = 6 per group) and relative rest phase food intake (F) of WT and MT mice in constant darkness (DD; n = 6 per group). (G–L) Circadian profiles of MBH clock gene (G–I) and metabolic gene (J–L) mRNA expression in WT and MT mice in DD (n = 3 per time point). All data are means ± SEM. R – rhythmic expression (p<0.05; CircWave); N – non-rhythmic expression (p>0.05); */**/*** - p<0.05/0.01/0.001; 2-way ANOVA with Sidak’s multiple comparisons for B, E; unpaired Student’s t-test for C, F. p-values: (B) 0.0145 (MT vs. WT ZT0-6) and 0.0097 (MT vs. WT ZT12-18), ANOVA dF = F(3, 30)=10.21 (interaction)/95.89 (time)/0.10 (genotype); (C) 0.0325 (MT vs. WT); (E) 0.0206 (MT vs. WT 48–54), ANOVA dF = F(3, 30)=7.63 (interaction)/79.32 (time)/0.30 (genotype); (F) 0.0062 (MT vs. WT).
-
Figure 3—source data 1
Raw data of experiments shown in Figure 3.
- https://cdn.elifesciences.org/articles/55388/elife-55388-fig3-data1-v2.xlsx
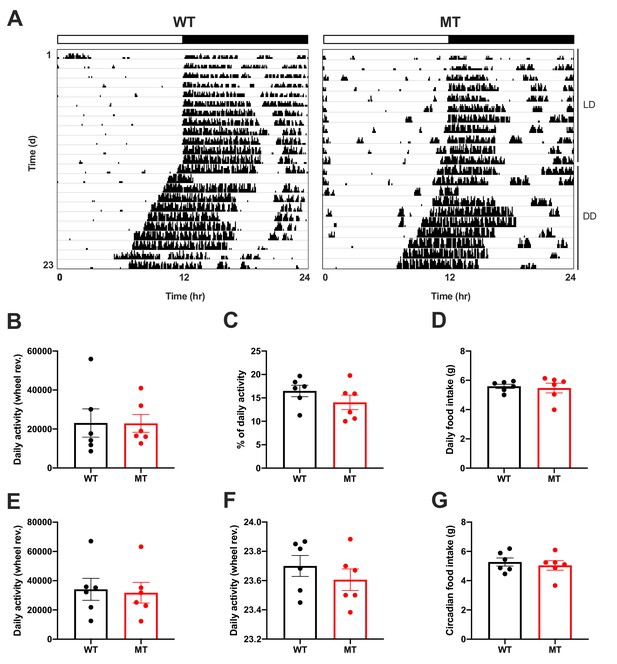
Normal rest-activity rhythms in Adipoq-deficient female mice.
(A) Actograms of representative female wild-type (WT; left) and Adipoq-deficient (MT; right) mice under 12 hr light: 12 hr dark (LD) and constant darkness (DD) conditions. White-black bar on top indicates light regimen in LD. (B–D) Total running-wheel activity (B), relative light phase activity (C) and daily total food intake (D) of WT (black) and MT mice (red) in LD (n = 6). (E–G) Total running-wheel activity (E), free-running period length (F) and daily total food intake (G) of WT and MT mice in DD (n = 6). All data are means ± SEM.
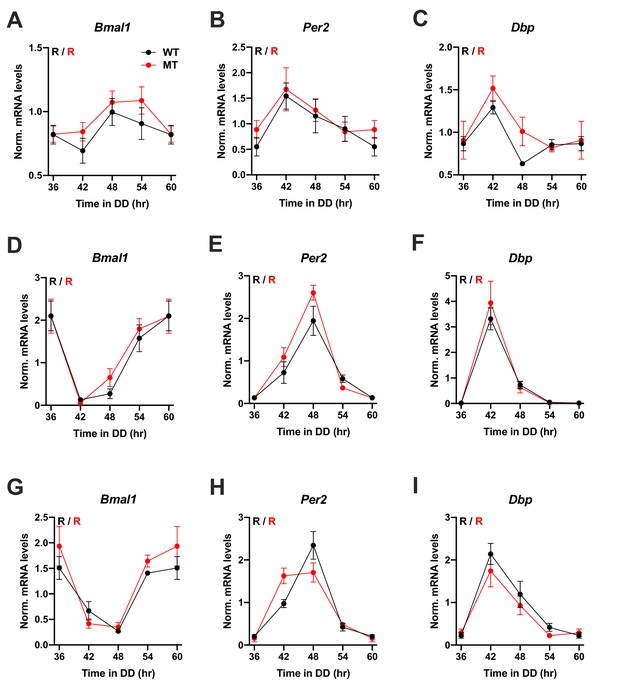
Preserved circadian clock gene expression rhythms in the SCN and peripheral tissues of Adipoq-deficient female mice.
(A–I) Circadian rhythms of clock gene mRNA expression in the SCN (A–C), liver (D–F) and skeletal muscle (G–I) of female wild-type (WT; black) and Adipoq-deficient mice (MT; red) in constant darkness (DD). All data are means ± SEM (n = 3 per timepoint and genotype). R – rhythmic expression (p<0.05; CircWave); N – non-rhythmic expression (p>0.05).
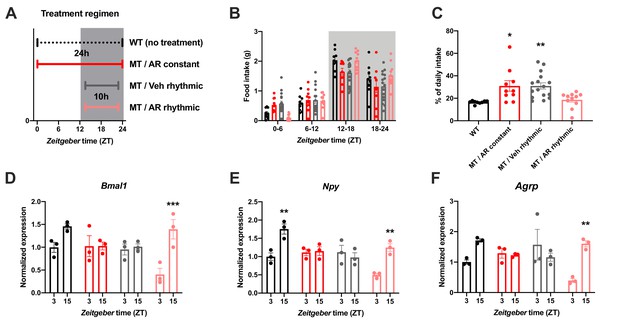
Restoration of food intake rhythms by rhythmic activation of central adiponectin signaling in male mice.
(A) Treatment regimen sketch. Grey shading indicates dark phase. (B) Daily food intake profiles of untreated wild-type mice (WT; black; n = 9), Adipoq-deficient mice with constant AdipoRon i.c.v. infusion (MT/AR constant; red; n = 10), MT mice with rhythmic (10 hr per day) vehicle infusion (MT/Veh rhythmic; grey; n = 16), and MT mice with rhythmic AdipoRon infusion (MT/AR rhythmic; pink; n = 11). (C) Relative light phase food intake of WT mice and MT mice with constant or rhythmic AdipoRon or vehicle i.c.v infusion. (D–F) Time of day differences in MBH mRNA expression of Bmal1 (D), Npy (E) and Agrp (F) in WT and MT mice with constant (red) or rhythmic i.c.v. infusion of AdipoRon (pink) or vehicle (grey) (n = 3 per time point). All data are means ± SEM. */**/*** - p<0.05/0.01/0.001; 1-way ANOVA (C) or 2-way ANOVA with Sidak’s multiple comparisons (D–F). p-values: (C) 0.017 (MT/AR constant vs. WT), 0.007 (MT/Veh rhythmic vs. WT), ANOVA dF = F(3, 42)=6.56; (D) 0.0005 (AR rhythmic, ZT3 vs. ZT15), ANOVA dF = F(3, 16)=30.24 (interaction)/27.8 (time)/11.37 (treatment); (E) 0.0019 (WT, ZT3 vs. ZT15) and 0.0017 (AR rhythmic, ZT3 vs. ZT15), ANOVA dF = F(3, 16)=30.98 (interaction)/22.76 (time)/24.39 (treatment); (F) 0.0024 (AR rhythmic, ZT3 vs. ZT15), ANOVA dF = F(3, 16)=42.73 (interaction)/13.77 (time)/9.25 (treatment).
-
Figure 4—source data 1
Raw data of experiments shown in Figure 4.
- https://cdn.elifesciences.org/articles/55388/elife-55388-fig4-data1-v2.xlsx
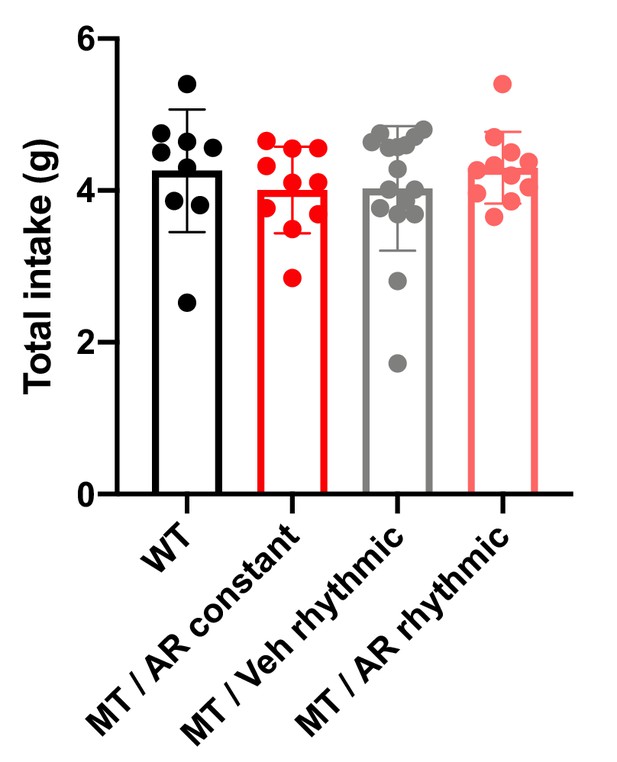
Central AdipoRon treatment has no effect on total food intake in Adipoq-deficient male mice.
Total daily normal chow food intake in wild-type mice (WT; black; n = 9), Adipoq-deficient mice with constant i.c.v. infusion of AdipoRon (MT/AR constant; red; n = 10) and MT mice with rhythmic infusion of AdipoRon (MT/AR rhythmic; pink; n = 11) or vehicle (MT/Veh rhythmic; grey; n = 16) in LD. All data are means ± SEM.
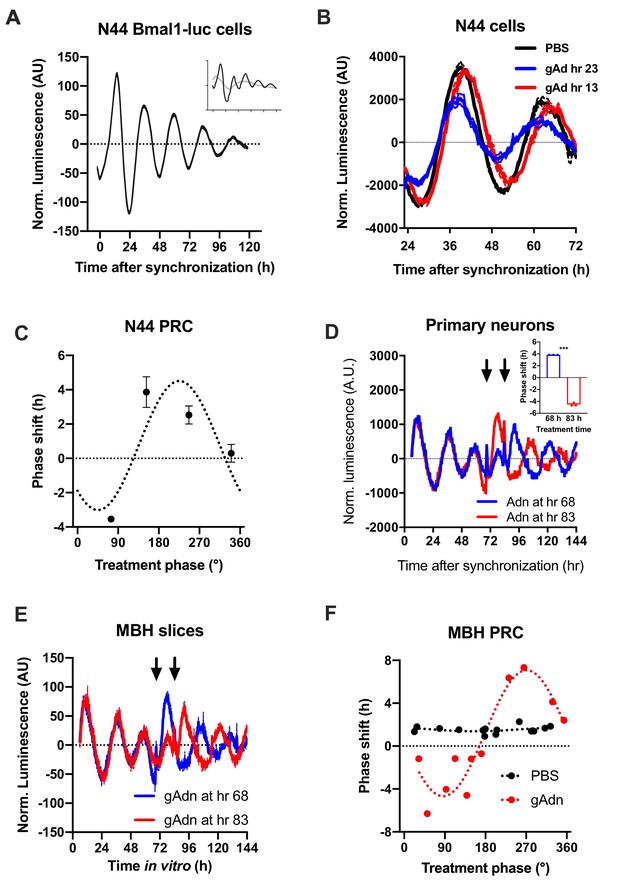
Resetting of hypothalamic clocks by ADIPOQ.
(A) Representative normalized Bmal1-luc luminescence rhythms of dexamethasone (DEX) synchronized hypothalamus-derived mHypo-N44 (N44) cells. Inset depicts raw luminescence data of the same set. (B) Normalized Bmal1-luc luminescence rhythms of N44 cells after treatment with ADIPOQ (Adn; red and blue) peptide or PBS (black) at the depicted time points after synchronization. Shown are averages ± SEM of 3 traces each. (C) Phase response curve for ADIPOQ-mediated resetting of N44/Bmal1-luc cells (n = 3 per time point; treatment time given in degrees with 90 °=maximal luminescence and 270 °=minimal luminescence – see (A) for reference). (D) Normalized PER2::LUC luminescence rhythms of primary hypothalamic neurons after treatment with ADIPOQ (Adn; red and blue) peptide at the depicted time points after synchronization. Shown are averages ± SEM of 3 traces each. Inset shows quantification of phase shifts (p<0.0001; unpaired Student’s t-test). (E) Normalized PER2::LUC luminescence rhythms of organotypic MBH slices after treatment with ADIPOQ (Adn; red and blue) peptide at the depicted time points. Shown are averages ± SEM of 3 traces each. (F) Phase response curve for ADIPOQ-mediated resetting of PER2::LUC MBH slices (treatment time given in degrees with 90 °=maximal luminescence and 270 °=minimal luminescence – see (E) for reference.
-
Figure 5—source data 1
Raw data of experiments shown in Figure 5.
- https://cdn.elifesciences.org/articles/55388/elife-55388-fig5-data1-v2.xlsx

ADIPOQ resets hypothalamic clocks in a dose-dependent manner.
(A, B) Sample traces (A) and quantification (B) of resetting of N44/Bmal1-luc cells by increasing doses of ADIPOQ (Adn) peptide added at 23 hr after synchronization. (C, D) Sample traces (C) and quantification (D) of resetting of N44/Bmal1-luc cells by increasing doses of ADIPOQ (Adn) peptide added at 13 hr after synchronization. All data are means ± SEM (n = 3 per condition). Small letters depict significantly different groups (p<0.05; 1-way ANOVA). p-values: (B) 0.0121 (1 µg/ml vs. PBS), 0.0005 (10 µg/ml vs. PBS), ANOVA dF = F(2, 6)=33.44; (D) 0.0095 (10 µg/ml vs. PBS), ANOVA dF = F(2, 6)=11.43.

Hormonal resetting of clocks in N44/Bmal1-luc cells.
(A–F) Normalized Bmal1-luc luminescence rhythms of N44 cells after treatment with leptin (Lep; A), visfatin (Vis; B), insulin (Ins; C), ghrelin (Ghr; D), glucagon (Gcg; E), resistin (Res; F) or PBS at the depicted time points after synchronization. Shown are averages ± SEM of 3 traces each.
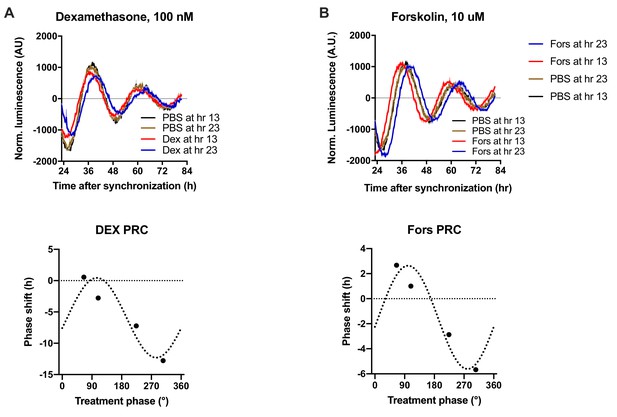
Resetting of clocks in N44/Bmal1-luc cells by dexamethasone and forskolin.
(A, B) Normalized Bmal1-luc luminescence rhythms of N44 cells after treatment with dexamethasone (Dex; A) or forskolin (Fors; B) or PBS at the depicted time points after synchronization. Shown are averages ± SEM of 3 traces each. Bottom panels: phase response curves for dexamethasone (left) and forskolin (right). Shown are averages of three experiments per time point each with sine wave regressions (treatment time given in degrees with 90 °=maximal luminescence and 270 °=minimal luminescence – see (Figure 5A) for reference.
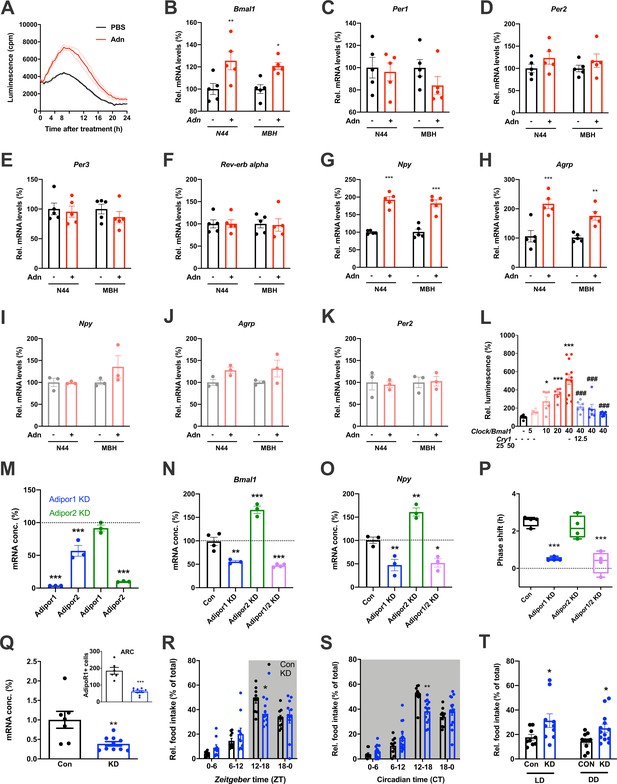
Adiponectin signaling modulates hypothalamic appetite regulation in male mice.
(A) Transient upregulation of Bmal1-luc activity in non-synchronized N44 cells after adiponectin (Adn) treatment. Shown are averages ± SEM of 3 traces each. (B–H) Induction of mRNA levels in N44 cells after Adn treatment and in the mediobasal hypothalamus (MBH) after i.v. Adn injection in wild-type mice; (B) Bmal1, (C) Per1, (D) Per2, (E) Per2, (F) Rev-erb alpha, (G) Npy, (H) Agrp (n = 5 per condition). (I–K) Induction of Npy (I), Agrp (J) and Per2 (K) mRNA levels in Bmal1-deficient N44 cells after Adn treatment and in the mediobasal hypothalamus (MBH) after i.v. Adn injection in Bmal1 knockout mice (n = 3 per condition). */**/***: p<0.05/0.01/0.001, unpaired Student’s t-test. (L) Luciferase promotor assay for Npy in HEK293T cells after transient expression of increasing doses of Clock/Bmal1 or Cry1 (n = 6–10). */***: p<0.05/0.001 vs. -/-, ###: p<0.001 vs. 40/-, 1-way ANOVA. (M) Expression of Adipor1/2 mRNA levels after shRNA-mediated knockdown of Adipor1 (blue) or Adipor2 (green) in N44 cells (n = 3 per condition). (N, O) Expression of Bmal1 (N) and Npy (O) mRNA after Adipor1/2 knockdown in N44 cells (n = 3–4 per condition). (P) Adiponectin-induced phase shift of N44/Bmal1-luc rhythms after Adipor1/2 knockdown in N44 cells (n = 4 per condition). */**/***: p<0.05/0.01/0.001 vs. Con, 1-way ANOVA. (Q) Expression of Adipor1 mRNA in the mediobasal hypothalamus (MBH; n = 7–10) and of ADIPOR1 protein in the arcuate nucleus (ARC; inset; n = 6–9) after shRNA-mediated viral knockdown of Adipor1. (R, S) Daily (LD; R) and circadian (DD; S) food intake profiles of mice after shRNA-mediated viral knockdown of Adipor1 in the MBH (KD) and of scramble-AAV treated control animals (Con) (n = 9–13 per group). (T) Relative light phase food intake in LD (left) and rest phase food intake in DD (right) in Adipor1-KD animals (KD) and controls (Con) (n = 9–13 per group). */**/***: p<0.05/0.01/0.001 vs. Con, unpaired Student’s t-test (Q, T) or 2-way ANOVA with Sidak’s multiple comparisons (R, S). All data are averages ± SEM. Box plots show medians, quartiles and min/max. p-values: (B) 0.0084 (N44, +/- Adn), 0.0309 (MBH, +/- Adn); (G) < 0.001 (N44, +/- Adn and MBH, +/- Adn); (H) 0.0002 (N44, +/- Adn), 0.0059 (MBH, +/- Adn); (L) 0.019 (10 /- vs. -/-),<0.001 (20 /- and 40 /- vs. -/-),<0.001 (40/12, 40/25 and 40/50 vs. 40/-), ANOVA dF = F(7, 51)=15.63; (M) < 0.001 (Adipor1 KD, Adipor1 and Adipor2 vs. control and Adipor2 KD, Adipor2), ANOVA dF = F(4, 10)=68.39; (N) 0.002 (Adipor1 KD vs. control),<0.001 (Adipor2 KD and Adipor1/2 KD vs. control), ANOVA dF = F(3, 10)=68.50; (O) 0.010 (Adipor1 KD vs. control), 0.005 (Adipor2 KD vs. control), 0.016 (Adipor1/2 KD vs. control), ANOVA dF = F(3, 8)=31.95; (P) < 0.001 (Adipor1 KD and Adipor1/2 KD vs. Con), ANOVA dF = F(3, 12)=25.72; (Q) 0.006 (mRNA) and <0.001 (protein), unpaired Student’s t-test; (R) 0.037 (KD vs. Con, ZT12-18), ANOVA dF = F(3, 54)=2.359 (interaction)/37.82 (time)/0.56 (knockdown); (S) 0.002 (KD vs. Con, ZT12-18), ANOVA dF = F(3, 66)=5.337 (interaction)/87.32 (time)/1.43 (knockdown); (T) 0.0454 (LD, Con vs. KD) and 0.0199 (DD, Con vs. KD), unpaired Student’s t-test.
-
Figure 6—source data 1
Raw data of experiments shown in Figure 6.
- https://cdn.elifesciences.org/articles/55388/elife-55388-fig6-data1-v2.xlsx
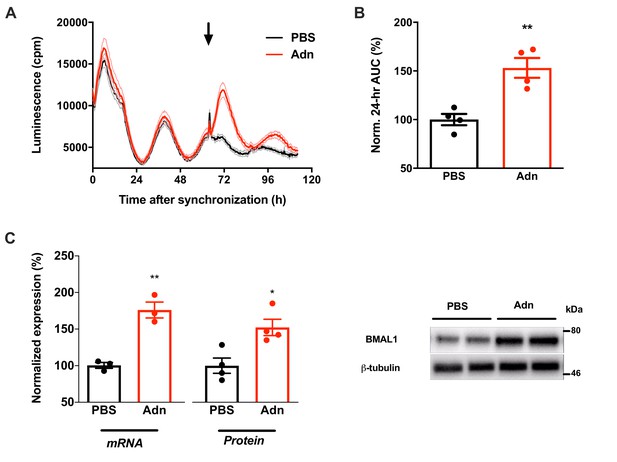
Adiponectin induces Bmal1 expression in synchronized N44 cells.
(A) Raw luminescence data of N44/Bmal1-luc cells treated with ADIPOQ (Adn) or PBS at hr 63 after synchronization (black arrow). Shown are means ± SEM of 3 traces. (B) Normalized luminescence changes during 24 hr after Adn treatment at hr 63 (n = 3). (C) Bmal1 mRNA (left) and BMAL1 protein expression at 3 hr (mRNA) or 6 hr (protein) after Adn treatment at hr 63 (n = 3 for mRNA and four for protein). Right panel: representative Western blot for BMAL1 and ß-tubulin as loading control. All data are means ± SEM; */**: p<0.05/0.01, unpaired Student’s t-test. p-values: (B) 0.004 (Adn vs. PBS); (C) 0.003 (mRNA, Adn vs. PBS) and 0.014 (protein, Adn vs. PBS).
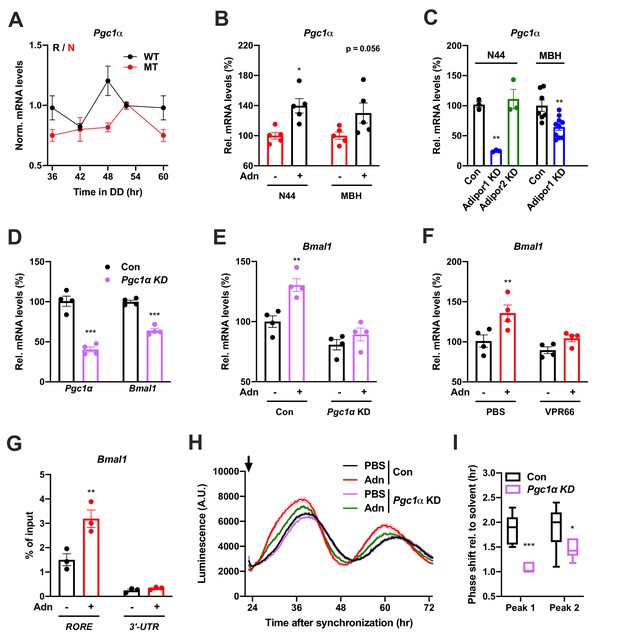
PGC1a-mediated induction of Bmal1 by ADIPOQ.
(A) Circadian profile of Pgc1a mRNA levels in the MBH of WT (black) and Adipoq deficient (MT) mice (red; n = 3 per time point). (B) Induction of Pgc1a mRNA levels in N44 cells after Adn treatment and in the mediobasal hypothalamus (MBH) after i.v. Adn injection in wild-type mice (n = 5). (C) Expression of Pgc1a in N44 cells and in the MBH after shRNA-mediated knockdown of Adipor1 or Adipor2 (n = 3 for N44 cells and 7 (Con)/10 (KD) in MBH). (D) mRNA levels of Pgc1a (left) and Bmal1 (right) in non-synchronized N44 cells after shRNA-mediated knockdown of Pgc1a (n = 4). (E) Bmal1 mRNA levels in unsynchronized N44 cells in response to Adn with (right) or without (left) prior knockdown of Pgc1a (n = 4). (F) Bmal1 mRNA levels in unsynchronized N44 cells in response to Adn with (right) or without (left) prior treatment with 5 µM VPR66 (n = 4). (G) ChIP-qPCR for PGC1a binding to Bmal1 RORE (and Bmal1 3’-UTR as negative control) in unsynchronized N44 cells in response to Adn treatment (n = 3). (H, I) Response of Bmal1-luc luminescence rhythms in synchronized N44 cells to Adn treatment at 23 hr after synchronization (arrow) with or without prior knockdown of Pgc1a. (H) Normalized luminescence data (n = 3 per condition). (I) Phasing of the first and second peak after Adn treatment relative to solvent (n = 8). All data are means ± SEM. Box plots show medians, quartiles and min/max. p-values: (B) 0.0116 (N44, +Adn vs. -Adn), ANOVA dF = F(1, 16)=0.299 (interaction)/0.294 (tissue)/15.60 (treatment); (C) 0.0001 (N44, Adipor1 KD vs. Con), 0.0020 (MBH, Adipor1 KD vs. Con), ANOVA dF = F(1, 19)=0.028 (interaction)/0.044 (tissue)/40.90 (treatment); (D) < 0.0001 (Pgc1a, Pgc1a KD vs. Con),<0.0001 (Bmal1, Pgc1a KD vs. Con), ANOVA dF = F(1, 12)=9.693 (interaction)/8.566 (gene)/151.5 (treatment). (E) 0.0019 (Con, +Adn vs. -Adn), ANOVA dF = F(1, 12)=4.894 (interaction)/37.05 (genotype)/15.39 (treatment); (F) 0.0087 (PBS, +Adn vs. -Adn), ANOVA dF = F(1, 12)=2.044 (interaction)/9.281 (inhibition)/12.45 (treatment); (G) 0.0013 (RORE, +Adn vs. -Adn), ANOVA dF = F(1, 8)=13.17 (interaction)/87.25 (site)/16.20 (treatment); (I) < 0.0001 (peak 1, Con vs. KD), 0.0258 (peak 2, Con vs. KD), ANOVA dF = F(1, 24)=2.809 (interaction)/3.192 (peak)/29.93 (genotype).
-
Figure 7—source data 1
Raw data of experiments shown in Figure 7.
- https://cdn.elifesciences.org/articles/55388/elife-55388-fig7-data1-v2.xlsx
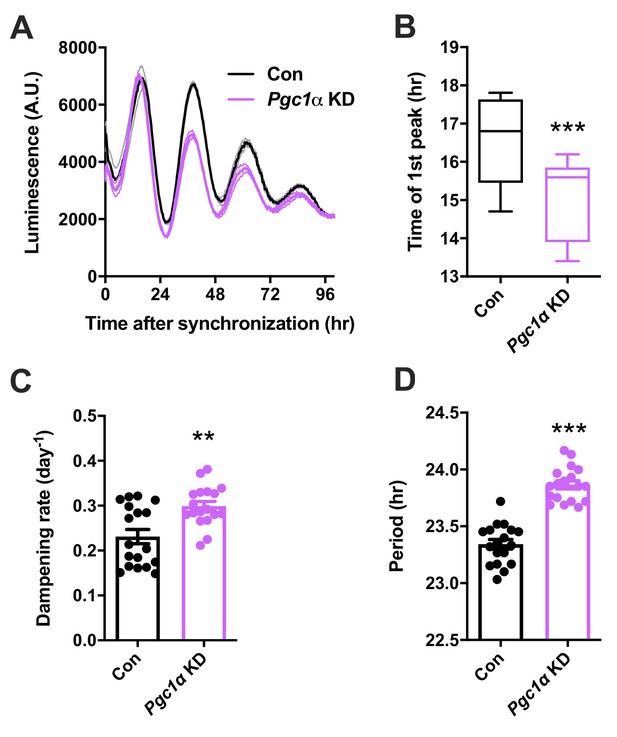
Knockdown of Pgc1a affects circadian rhythms in N44 cells.
(A) Representative Bmal1-luc luminescence traces of N44 cells after transfection with Pgc1a shRNA (purple) or scramble controls (black; n = 3). (B) Timing of the first peak (n = 18), (C) dampening rates (n = 18) and (D) period length (n = 18). All data are means ± SEM; **/***: p<0.01/0.001, unpaired Student’s t-test. p-values: (B) 0.0002; (C) 0.0011; (D) < 0.0001.
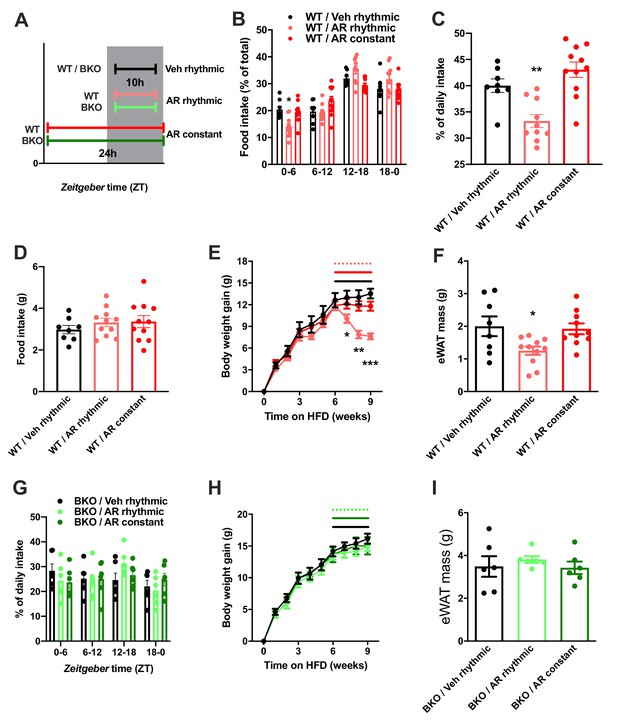
Rhythmic AdipoRon administration rescues food intake rhythms and body weight in obese male mice.
(A) Treatment regimen and groups. (B–D) Daily food intake profiles (B), relative light phase food intake (C) and total daily food intake (D) of HFD-fed male WT mice treated 10 hr or 24 hr per day i.c.v. with AdipoRon or vehicle. (E) Body weight development of HFD-fed WT mice before and during constant or rhythmic i.c.v. AdipoRon treatment (lines indicate treatment phases). (F) Epididymal adipose tissue (eWAT) mass of HFD-fed WT mice after 3 weeks of rhythmic or constant i.c.v. treatment with AdipoRon (A-F: n = 8–11 per group). (G) Daily food intake profiles of HFD-fed male Bmal1 deficient (BKO) mice treated 10 hr or 24 hr per day i.c.v. with AdipoRon or vehicle. (H) Body weight development of HFD-fed BKO mice before and during constant or rhythmic i.c.v. AdipoRon treatment (n = 6 per group; lines indicate treatment phases). (I) eWAT mass of HFD-fed BKO mice after 3 weeks of rhythmic or constant i.c.v. treatment with AdipoRon (G-I: n = 6 per group). Shown are averages ± SEM. */**/***: p<0.05/0.01/0.001, 1-way ANOVA (C, D, F) or 2-way ANOVA with Sidak’s multiple comparisons (B, E). p-values: (B) 0.002 (WT/AR rhythmic vs. WT/Veh rhythmic; ZT0-6), ANOVA dF = F(6, 78)=4.933 (interaction)/ (61.05 (time)/0.03 (treatment); (C) 0.007 (WT/AR rhythmic vs. WT/Veh rhythmic), ANOVA dF = F(2, 26)=14.47; (E) 0.04 (WT/AR rhythmic vs. WT/Veh rhythmic; week 7), 0.001 (WT/AR rhythmic vs. WT/Veh rhythmic; week 8),<0.001 (WT/AR rhythmic vs. WT/Veh rhythmic; week 9), ANOVA dF = F(18, 234)=13.51 (interaction)/550.1 (time)/3.68 (treatment); (F) 0.033 (WT/AR rhythmic vs. WT/Veh rhythmic), ANOVA dF = F(2, 26)=4.709.
-
Figure 8—source data 1
Raw data of experiments shown in Figure 8.
- https://cdn.elifesciences.org/articles/55388/elife-55388-fig8-data1-v2.xlsx





