Smchd1 is a maternal effect gene required for genomic imprinting
Figures
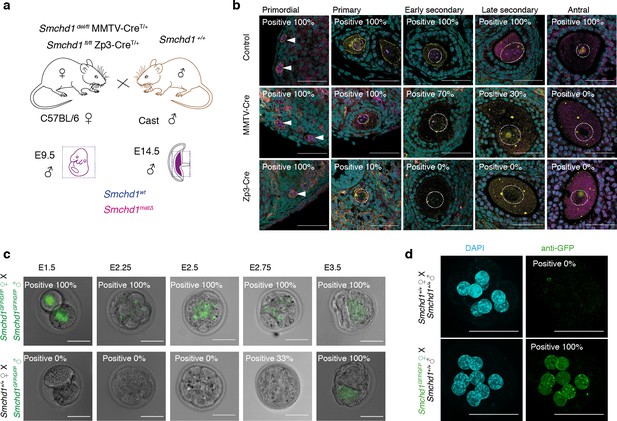
Maternal deletion of Smchd1 during oocyte development depletes SMCHD1 until the 16-cell embryonic stage.
(a) Schematic for maternal deletion of Smchd1. (b) Deletion of Smchd1 in oocyte development with MMTV-Cre and Zp3-Cre. Arrowheads indicate primordial follicle oocyte nuclei, white dotted lines surround primary-antral follicle oocytes. Smchd1 (magenta), c-KIT (yellow), DAPI (cyan). n = 15–27 sections for two ovaries per cohort. A total of 5–20 follicles were observed for primordial – late secondary stages and 2–3 antral follicles for each genotype. (c) Detection of paternal SMCHD1-GFP from day 1.5 to 3.5 in pre-implantation embryos. Smchd1GFP/GFP embryos were used as positive controls. (d) Detection of maternal SMCHD1-GFP in the nuclei of E2.5 (8 cell) embryos, with Smchd1+/+ used as negative controls. Nuclei marked with DAPI (cyan). Scale bar – 50 µm.
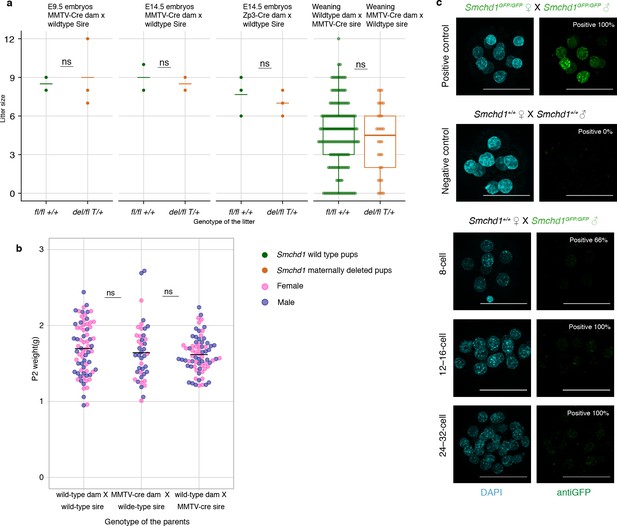
Maternal deletion of Smchd1 does not affect pup viability or weight.
Zygotic SMCHD1 has a limited contribution to the embryonic SMCHD1 pool in the early embryo. (a) Embryo lethality data for Smchd1 maternal null embryos and animals at weaning, compared to wild-type control litters generated from Smchd1fl/fl non-transgenic dams, or heterozygous litters generated from a reciprocal cross. The p-value was calculated using Student's two-tailed t-test. (b) Smchd1 mutations in the dam do not influence pup weights at P2. Student's two-tailed t-test. (c) Immunofluorescence detection of maternal SMCHD1-GFP in the nuclei of 8-cell embryos, with Smchd1+/+ used as negative controls and Smchd1GFP/GFP embryos were used as positive controls. Zygotic SMCHD1 was detectable in some cells from the 8-cell stage onward but comparatively less signal intensity to the GFP control when embryos were imaged with identical settings, in experiments performed on the same day. n ≥ 2 embryos.
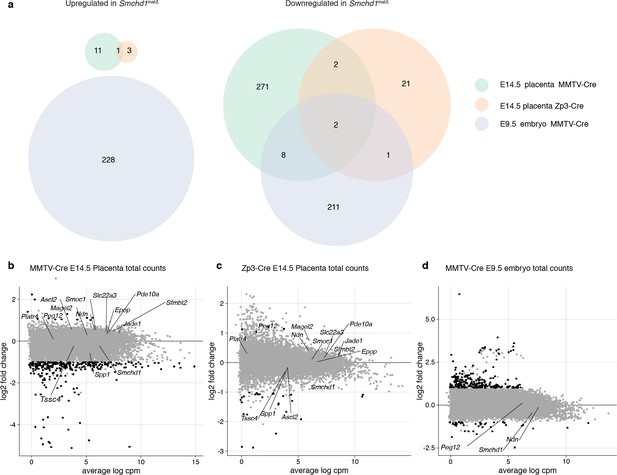
Minor global gene expression changes in Smchd1matΔ placentae and embryos.
(a) Intersection of the differentially expressed gene lists in MMTV-Cre and Zp3-Cre maternal deletion experiments, E14.5 placental samples. (b–d) MA-plots of total gene expression in MMTV-Cre (b) and Zp3-Cre (c) maternal deletion experiments, E14.5 placental samples, and MMTV-Cre E9.5 embryos (d). Genes below the 5% FDR and differentially expressed by at least two-fold are plotted in black. Smchd1 and genes with a partial loss of imprinting are labeled.
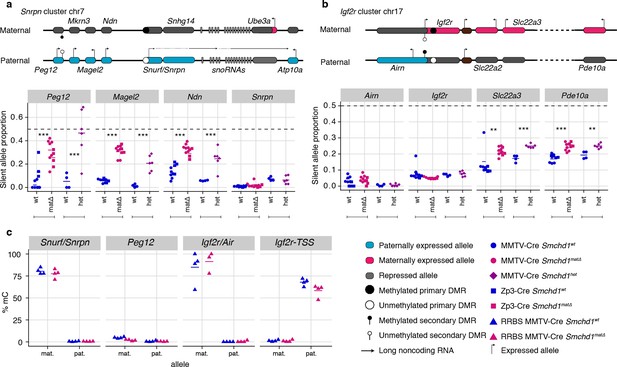
Heterozygous deletion of Smchd1 results in partial loss of imprinting at known SMCHD1-sensitive clusters.
(a-b) Expression of the silent allele as a proportion of total expression of the gene, obtained by allele-specific RNA-seq from the embryonic portion of the placenta of Smchd1 wild-type (wt), heterozygous (het) and maternal null (matΔ) conceptuses. Expression data for (a) Snrpn cluster genes on chromosome seven and (b) Igf2r cluster genes on chromosome 17. (c) Percentage methylation (% mC) on the maternal and paternal alleles at primary and secondary DMRs at Snrpn and Igf2r clusters in Smchd1 maternal null and wild-type placental samples. *p<0.05, **p<0.01, ***p<0.001, when the difference in mean silent allele proportions between genotypes is of at least 5%. RNA-seq sample sizes: for maternal deletion experiment, MMTV-Cre 6 wt and seven matΔ, Zp3-Cre 4 wt and four matΔ; for the heterozygous deletion experiment, 4 wt and six het E14.5 placental samples. RRBS: n = 4 MMTV-Cre for both matΔ and wt E14.5 placental samples.
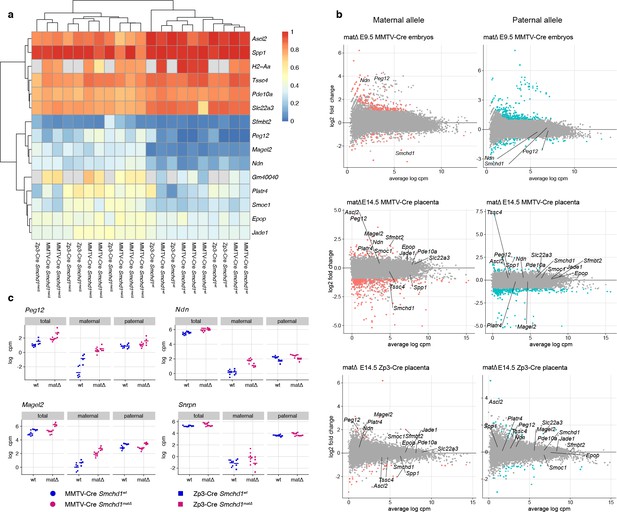
Allele-specific gene expression changes in Smchd1matΔ placentae and embryos.
(a) Heatmap of maternal allele expression proportion for genes that are differentially imprinted in at least one experimental set (MMTV-Cre and Zp3-Cre E14.5 placenta). (b) MA-plots of allelic gene expression in MMTV-Cre E9.5 embryos and E14.5 placentae, and Zp3-Cre E14.5 placentae. Genes below the 5% FDR and differentially expressed by at least two-fold are plotted in color. Smchd1 and genes with partial loss of imprinting are labeled. (c) Absolute expression levels (normalized log counts per million) for genes of the Snrpn cluster, showing non-split (total) and allele-split (maternal and paternal) counts. This shows the increase in silent-allele proportion for Peg12, Ndn, and Magel2 is explained by an increase in silent allele levels rather than a decrease in expressed allele levels.
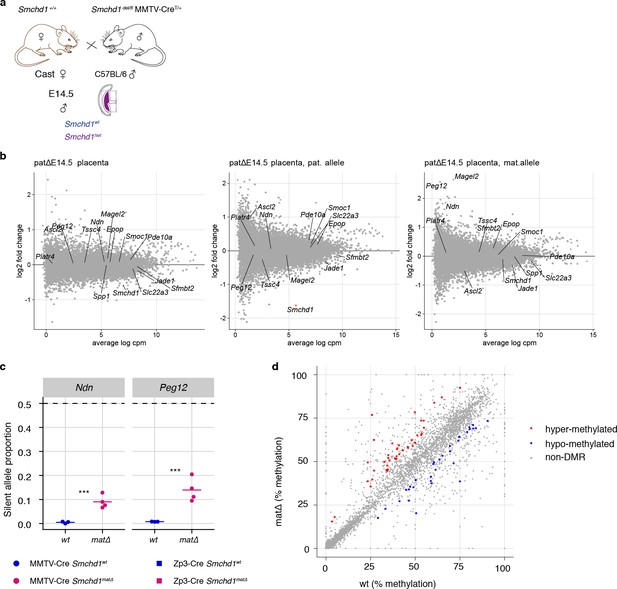
Global differential gene expression in Smchd1 heterozygous (paternally deleted) placentae, Snrpn cluster allele-specific expression in Smchd1matΔ embryos, and CpG island differential methylation in Smchd1matΔ placentae.
(a) Schematic for breeding scheme to generate heterozygous (het) deletion of Smchd1. Strain background is shown underneath each parent. Genotype is shown above each parent. (b) MA-plot of the total gene expression in the heterozygous deletion experiment. (c) Expression of the silent allele as a proportion of total expression of the gene at the Snprn cluster genes Ndn and Peg12, in E9.5 Smchd1 maternal null embryos. *p<0.05, **p<0.01, ***p<0.001, when the difference in silent allele proportions is at least 5%. (d) Scatter plot of CGI methylation in wt and maternal null placentae. 33 and 42 DMRs were found to be hypo- and hypermethylated in the matΔ placentae, respectively (logistic regression, 10 observations minimum, 5% FDR and at least 10% absolute difference in methylation levels between the wt and matΔ).
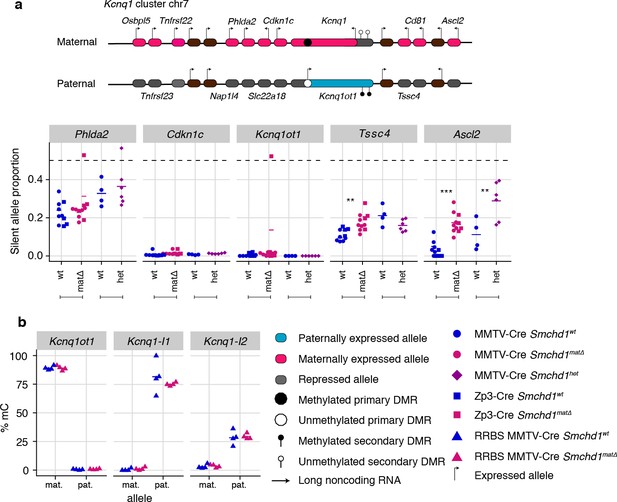
Maternal and heterozygous deletions of Smchd1 result in loss of imprinting at the Kcnq1 imprinted cluster without changes to primary or secondary DMR methylation.
(a) Expression of the silent allele as a proportion of total expression of the gene, obtained by allele-specific RNA-seq from the embryonic portion of the placenta of Smchd1 wild-type (wt), heterozygous (het), and maternal null (matΔ) conceptuses. Expression data for (a) Kcnq1 cluster genes on chromosome 7, and (b) Percentage methylation (% mC) for each parental allele at the DMRs for the Kcnq1 cluster. Kcnq1-I1: Kcnq1-Intergenic1; Kcnq1-I2: Kcnq1-Intergenic2 *p<0.05, **p<0.01, ***p<0.001, when the difference in silent allele proportions is of at least 5%. RNA-seq sample sizes: for maternal deletion experiment, MMTV-Cre 6 wt and seven matΔ, Zp3-Cre 4 wt and four matΔ; for the heterozygous deletion experiment, 4 wt and six het E14.5 placental samples. RRBS: n = 4 MMTV-Cre for both matΔ and wt E14.5 placental samples.
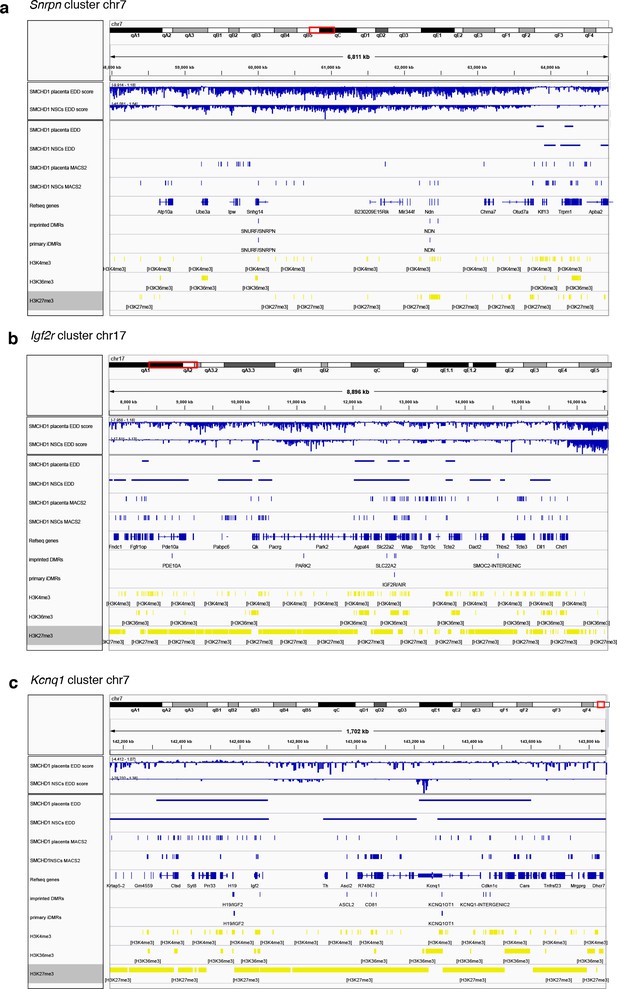
ChIP-seq for SMCHD1-GFP over the Kcnq1 (a), Snrpn (b), and Igf2r-Airn (c) imprinted clusters in E14.5 placenta and neural stem cells.
Enriched Domain Detector (EDD) enrichment scores over 3 kb bins and called enriched regions are shown for the placenta (one sample) and neural stem cells (pooled triplicate). MACS2-broad SMCHD1-GFP ChIP-seq peaks are also shown. H3K4me3, H3K36me3, and H3K27me3-marked regions are retrieved from Hanna et al., 2019 for E7.5 extraembryonic tissue.
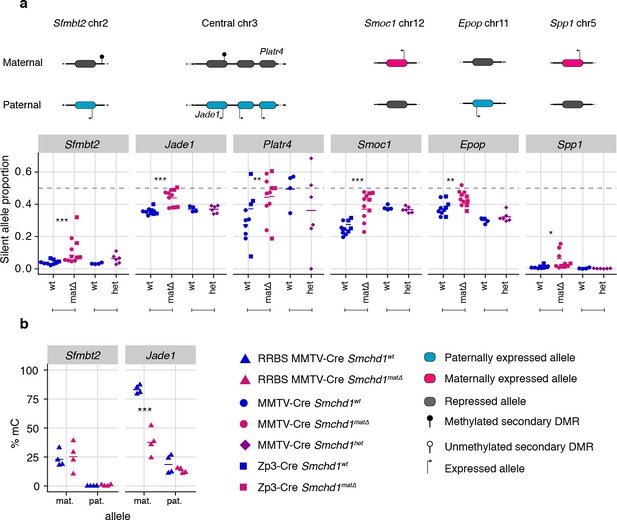
Only maternal deletion of Smchd1 results in loss of imprinting at lone imprinted genes.
(a) Expression of the silent allele as a proportion of total expression of the gene, obtained by allele-specific RNA-seq from the embryonic portion of the placenta of Smchd1 wild-type (wt), heterozygous (het), and maternal null (matΔ) conceptuses, at lone imprinted genes: Sfmbt2, Jade1, Platr4, Smoc1, Epop and Spp1. (b) Percentage methylation (% mC) for each parental allele at the DMRs for Sfmbt2 and Jade1. *p<0.05, **p<0.01, ***p<0.001, when the difference in silent allele proportions is of at least 5%. RNA-seq sample sizes: for maternal deletion experiment, MMTV-Cre 6 wt and seven matΔ, Zp3-Cre 4 wt and four matΔ; for the heterozygous deletion experiment, 4 wt and six het E14.5 placental samples. RRBS: n = 4 MMTV-Cre for both matΔ and wt E14.5 placental samples.
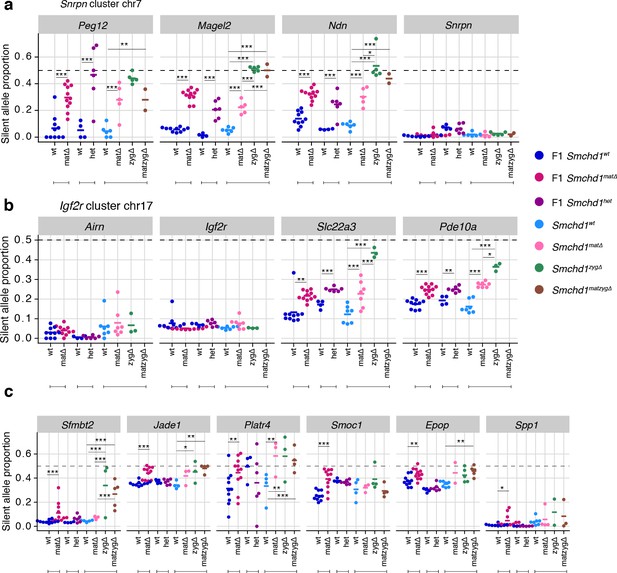
Homozygous zygotic deletion of Smchd1 generally results in more severe loss of imprinting at genes sensitive to maternal or heterozygous deletions.
MMTV-Cre Smchd1 maternal deletion data and heterozygous deletion data (F1 wt, F1 matΔ, F1 het) from Figures 1 and 2, along with samples produced to compare Smchd1 wild-type (wt), oocyte-deleted (matΔ), zygote-deleted (zygΔ), and oocyte-and-zygote-deleted (matzygΔ) genotypes. Samples from the embryonic portion of the placenta and expression of the silent allele is shown as a proportion of total expression of the gene, obtained by allele-specific RNA-seq. (a) Snrpn cluster genes. (b) Igf2r-Airn cluster genes. (c) Sfmbt2, Jade1, Platr4, Smoc1, Epop, and Spp1 genes. *p<0.05, **p<0.01, ***p<0.001, when the difference in silent allele proportions is at least 5%. RNA-seq sample sizes: for maternal deletion experiment, 10 wt and 11 matΔ; for the heterozygous deletion experiment, 4 wt and six het; for the maternal and zygotic deletion experiment, 13 wt, seven matΔ, eight zygΔ, and six matzygΔ E14.5 MMTV-Cre placentae.
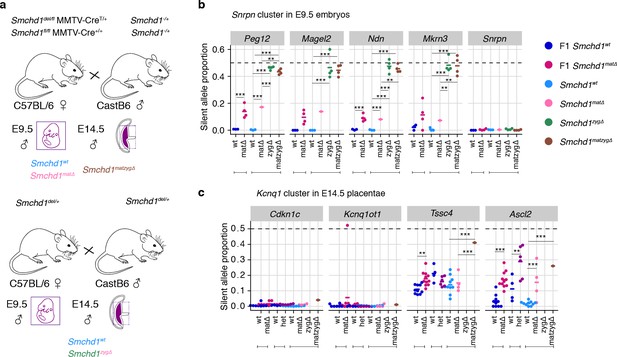
Allele-specific expression in maternal, zygotic, and maternal-and-zygotic Smchd1-deleted samples.
(a) Schematic for the breeding scheme to generate oocyte (matΔ), zygotic (zygΔ), oocyte-and-zygotic deletion (matzygΔ) of Smchd1. Strain background is shown underneath each parent. Genotype is shown above each parent. Allele-specific RNA-seq expression profiles of the silent allele are shown as a proportion of total expression at the (b) Snrpn cluster in E9.5 embryo (c) Kcnq1 cluster in the embryonic portion of the placenta at E14.5. *p<0.05, **p<0.01, ***p<0.001, when the difference in silent allele proportions is at least 5%.
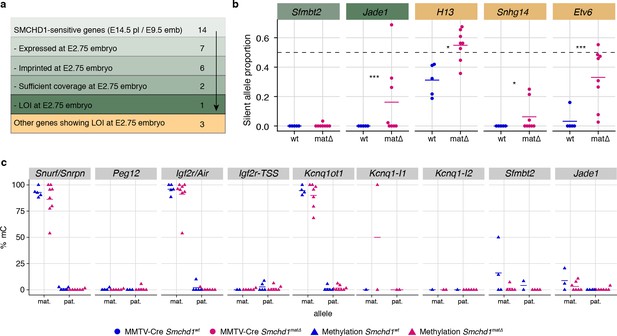
Maternal SMCHD1 establishes an epigenetic memory required for imprinted gene expression.
(a) Summarised analysis of SMCHD1-sensitive imprinted genes from E14.5 placentae and E9.5 embryos in E2.75 embryo transcriptome sequencing. (b) Expression of the silent allele as a proportion of total expression of the gene, obtained by allele-specific RNA-seq from whole E2.75 Smchd1 maternal null (matΔ) and wild-type (wt) embryos. (c) Percentage methylation (% mC) on each parental allele for the DMRs of SMCHD1-sensitive imprinted clusters and genes in Smchd1 maternal null (matΔ) compared with control (wt) E2.75 embryos. Kcnq1-I1: Kcnq1-Intergenic1; Kcnq1-I2: Kcnq1-Intergenic2 *p<0.05, **p<0.01, ***p<0.001, when the difference in silent allele proportions is at least 5%. n = 5 wt and n = 8 matΔ E2.75 embryos.
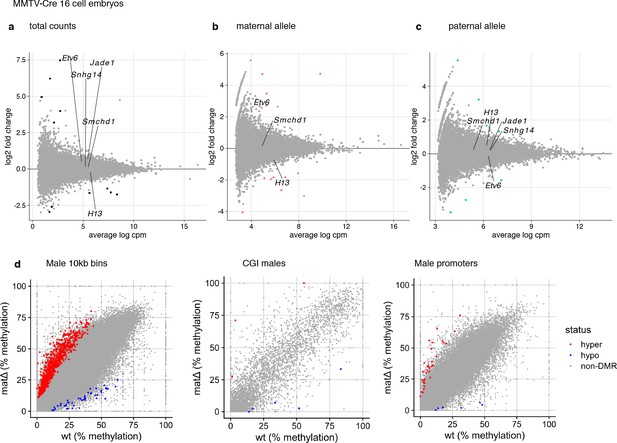
MA-plots of total (a) and allelic (b,c) gene expression in MMTV-Cre maternal deletion E2.75 embryos experiments.
Genes below the 5% FDR and differentially expressed by at least 2-fold are plotted in black. Smchd1 and genes with partial loss of imprinting are labeled. (d-f) Genome-wide differential DNA methylation analysis in matΔ E2.75 embryos at CGIs (d), promoters (e) and in 10 kb windows tiling the genome (f). Differentially methylated regions (edgeR, 5% FDR and 10% absolute difference in DNA methylation level): three hyper- and- five hypomethylated CpG islands, 37 hyper- and eight hypomethylated promoters, 826 hyper- and 46 hypomethylated 10 kb windows.
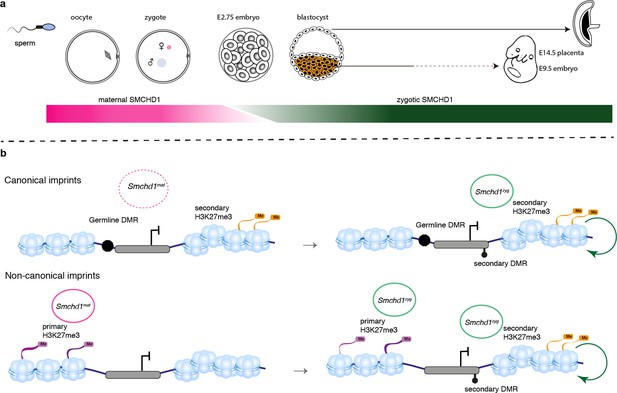
SMCHD1 translates the imprints to establish a heritable chromatin state required for imprinted expression later in development.
(a) Developmental windows of activity of maternal and zygotic SMCHD1. (b) Proposed model illustrating the regulation of imprinted genes by SMCHD1. Both oocyte and zygotic SMCHD1 contribute to an epigenetic memory downstream of polycomb repressive histone marks.
Tables
Summary of SMCHD1-sensitive imprinted genes.
| gene | cluster | type | sensitive tissue | Smchd1 sensitivity | haploinsufficiency |
|---|---|---|---|---|---|
| Peg12 | Snrpn | canonical | E9.5 embryo, E14.5 placenta | zygotic | yes |
| Magel2 | Snrpn | canonical | E9.5 embryo, E14.5 placenta | zygotic | yes |
| Ndn | Snrpn | canonical | E9.5 embryo, E14.5 placenta | zygotic | yes |
| Mkrn3 | Snrpn | canonical | E9.5 embryo, E14.5 placenta | zygotic | not significant |
| Snhg14 | Snrpn | canonical | E2.75 embryo | maternal | no |
| Slc22a3 | Airn/Igf2r | canonical | E14.5 placenta | zygotic | yes |
| Pde10a | Airn/Igf2r | canonical | E14.5 placenta | zygotic | yes |
| Tssc4 | Kcnq1 | canonical | E14.5 placenta | maternal and zygotic | no |
| Ascl2 | Kcnq1 | canonical | E14.5 placenta | zygotic | yes |
| Jade1 | Jade1 | non-canonical | E2.75 embryo, E14.5 placenta | maternal | no |
| Platr4 | Jade1 | non-canonical | E14.5 placenta | maternal | no |
| Sfmbt2 | lone | non-canonical | E14.5 placenta | maternal | no |
| Smoc1 | lone | non-canonical | E14.5 placenta | maternal | no |
| Epop | lone | ? | E14.5 placenta | maternal | no |
| Spp1 | lone | ? | E14.5 placenta | maternal | no |
| H13 | lone | canonical | E2.75 embryo | maternal | no |
| Etv6 | lone | non-canonical | E2.75 embryo | maternal | no |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (M. musculus) | Structural maintenance of chromosomes hinge domain containing 1 (Smchd1) | PMID:15890782 PMID:18425126 | NM_028887 | |
| Strain, strain background (M. musculus, males and females) | Castaneus EiJ | The Jackson Laboratory | 000928 | |
| Strain, strain background (M. musculus, females and males) | C57BL/6J | The Jackson Laboratory | 000664 | |
| Genetic reagent (M. musculus, females and males) | MMTV-Cre | PMID:9336464 | Strain back-crossed from FVB/N to C57BL/6J background for this study | |
| Genetic reagent (M. musculus, females and males) | Zp3-Cre | PMID:9016703 | C57BL/6J background | |
| Genetic reagent (M. musculus, females and males) | Smchd1 conditional knockout (Smchd1fl) | PMID:29281018 | C57BL/6J background | |
| Genetic reagent (M. musculus, females and males) | Smchd1fl/fl MMTV-Cre | This study | C57BL/6J background. MMTV-Cre transgene on the Smchd1fl background. | |
| Genetic reagent (M. musculus, females and males) | Smchd1fl/fl Zp3-Cre | This report | C57BL/6J background, Zp3-Cre transgene on the Smchd1fl/fl background. | |
| Genetic reagent (M. musculus, males and females) | Smchd1GFP | PMID:30127357 | C57BL/6J background | |
| Biological sample (M. musculus, male) | Smchd1GFP/GFP, primary Neural Stem Cells (NSCs) | This study | Produced fresh from Smchd1GFP/GFP E14.5 embryos in Blewitt lab. | |
| Antibody | anti-SMCHD1 ATPase domain (rat monoclonal) | In house. Source - PMID:30127357 | #5 or Clone 5H4 | (1:100) |
| Antibody | anti-SMCHD1 ATPase domain (rat monoclonal) | In house. Source - PMID:30428357 | #8 or Clone 2B8 | (1:100) |
| Antibody | anti-c-Kit (goat polyclonal) | Novus | Cat. #: af1356 | (1:500) |
| Antibody | anti-GFP (rabbit polyclonal) | ThermoFisher Scientific | Cat. #: A11122 | (1:100) |
| Antibody | anti-goat Alexa 488 (donkey polyclonal) | Thermo Fisher Scientific | Cat. #: A-11055 | (1:500) |
| Antibody | anti-rat IgG DyLight 550 (donkey polyclonal) | Invitrogen | Cat. #:SA5-10027 | (1:500) |
| Antibody | anti-rabbit Alexa 488 (donkey polyclonal) | Thermo Fisher Scientific | Cat. #: A21206 | (1:500) |
| Chemical compound, drug | ProLong Diamond Antifade Mountant with DAPI | Thermo Fisher Scientific | Cat. #: P36931 | |
| Chemical compound, drug | Vectashield | Vector Labs | H-1000 | |
| Commercial assay or kit | Qiagen All prep kit | Qiagen | Cat. #:80204 | |
| Commercial assay or kit | Zymo Quick DNA/RNA miniprep plus kit | Zymo research | Cat. #:D7003 | |
| Commercial assay or kit | TruSeq V1 or V2 RNA sample preparation kit | Illumina | Cat. #:RS-122–2001 Cat. #:RS-122–2002 | |
| Commercial assay or kit | Zymo research DNA Clean and concentrator-5 kit | Zymo research | Cat. #:D4103 | |
| Commercial assay or kit | Qubit dsDNA assay kit | ThermoFisher Scientific | Cat. #:Q32853 | |
| Commercial assay or kit | NuGEN Ovation RRBS methyl-seq system | Integrated sciences | Cat. #:0553–32 | |
| Commercial assay or kit | QIAGEN EpiTect Fast DNA Bisulfite Kit | Qiagen | Cat. #:59824 | |
| Commercial assay or kit | Illumina TruSeq DNA Sample Preparation Kit | Illumina | Cat. #:FC-121–2001 Cat. #:FC-121–2002 | |
| Commercial assay or kit | Nextera XT kit | Illumina | Cat. #:FC-131–1002 | |
| Commercial assay or kit | NEBNext Ultra II DNA Library Prep Kit for Illumina | NEB | Cat. #:E7645 | |
| Software, algorithm | FIJI | PMID:3855844 | ||
| Software, algorithm | SNPsplit | PMID:21493656 | v0.3.2 | |
| Software, algorithm | HISAT2 | PMID:25751142 | v2.0.5 | |
| Software, algorithm | R | R Core Team | 3.5.1 | Available: https://www.R-project.org/ |
| Software, algorithm | featureCounts | PMID:24227677 | ||
| Software, algorithm | Rsubread | PMID:30783653 | 1.32.1 | |
| Software, algorithm | rpart | R package version 4.1–15 | Available: https:// CRAN.R-project.org/package=rpart | |
| Software, algorithm | edgeR | PMID:19910308 PMID:22287627 | 3.24.0 | |
| Software, algorithm | Glimma | PMID:28203714 | 1.10.0 | |
| Software, algorithm | ggplot2 | Available: https://ggplot2.tidyverse.org | ||
| Software, algorithm | cowplot | R package version 1.0.0 | Available: https://CRAN.R-project.org/package=cowplot | |
| Software, algorithm | ggbeeswarm | R package version 0.6.0 | Available: https://CRAN.R-project.org/package=ggbeeswarm | |
| Software, algorithm | ggrepel | R package version 0.8.1 | Available: https://CRAN.R-project.org/package=ggrepel | |
| Software, algorithm | ggrastr | R package version 0.1.7 | ||
| Software, algorithm | pheatmap | R package version 1.0.12 | Available: https://CRAN.R-project.org/package=pheatmap | |
| Software, algorithm | TrimGalore! | v0.4.4 | ||
| Software, algorithm | Bismark | PMID:21493656 | v0.20.0 | |
| Software, algorithm | FastQC | v0.11.8 | Available: http://www.bioinformatics.babraham.ac.uk/projects/fastqc | |
| Software, algorithm | Bowtie2 | PMID:22388286 | v2.3.4.1 | |
| Software, algorithm | SeqMonk | v1.45.1 | Available: https://www.bioinformatics.babraham.ac.uk/projects/seqmonk/ | |
| Software, algorithm | TMM method | PMID:20196867 | ||
| Software, algorithm | quasi-likelihood F-tests | PMID:27008025 | ||
| Software, algorithm | Benjamini-Hochberg method | 10.1111/j.2517–6161.1995.tb02031.x | ||
| Software, algorithm | Enriched Domain Detector | PMID:24782521 | v1.1.19 |
Additional files
-
Supplementary file 1
All tables of statistical data for allele-specific RNA-seq analyses at imprinted genes.
(a) Differential imprinted expression in male E14.5 B6Cast placentae, Smchd1 matΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (b) Differential imprinted expression in male E14.5 CastB6 placentae, Smchd1 het versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (c) Differential imprinted expression in male E9.5 B6Cast embryos, Smchd1 matΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05. Gnas is likely a false positive because in the second dataset of E9.5 embryos (maternal and zygotic deletions) it did not display imprinted expression in the wt samples (d) Differential imprinted expression in male E2.75 B6Cast embryos, Smchd1 matΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (e) Differential imprinted expression in male E14.5 B6(CastB6) placentae, Smchd1 matΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (f) Differential imprinted expression in male E14.5 B6(CastB6) placentae, Smchd1 zygΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (g) Differential imprinted expression in male E14.5 B6(CastB6) placentae, Smchd1 matzygΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (h) Differential imprinted expression in male E14.5 B6(CastB6) placentae, Smchd1 zygΔ versus matΔ. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (i) Differential imprinted expression in male E14.5 B6(CastB6) placentae, Smchd1 matzygΔ versus matΔ. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (j) Differential imprinted expression in male E14.5 B6(CastB6) placentae, Smchd1 matzygΔ versus zygΔ. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (k) Differential imprinted expression in male E9.5 B6(CastB6) embryos, Smchd1 matΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (l) Differential imprinted expression in male E9.5 B6(CastB6) embryos, Smchd1 zygΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (m) Differential imprinted expression in male E9.5 B6(CastB6) embryos, Smchd1 matzygΔ versus wt. A gene is called differentially imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (n) Differential imprinted expression in male E9.5 B6(CastB6) embryos, Smchd1 zygΔ versus matΔ. A gene is called differentiallly imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (o) Differential imprinted expression in male E9.5 B6(CastB6) embryos, Smchd1 matzygΔ versus matΔ. A gene is called differentiallly imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05 (p) Differential imprinted expression in male E9.5 B6(CastB6) embryos, Smchd1 matzygΔ versus zygΔ. A gene is called differentiallly imprinted if the multiple-testing corrected p-value is below 0.05 and the absolute difference in silent allele proportion greater than 0.05.
- https://cdn.elifesciences.org/articles/55529/elife-55529-supp1-v1.xlsx
-
Supplementary file 2
Tables of statistical data for whole-genome RNA-seq analyses.
(a) Differential expression (non-split counts) in male E9.5 B6Cast embryos, Smchd1 matΔ versus wt. A gene is called differentially expressed if the q-value is below 0.05 and the fold change is greater than 2. (b) Differential expression (non-split counts) in male E14.5 B6Cast embryonic placentae, Smchd1 matΔ versus wt (MMTV-Cre). A gene is called differentially expressed if the q-value is below 0.05 and the fold change is greater than 2. (c) Differential expression (non-split counts) in male E14.5 B6Cast embryonic placentae, Smchd1 matΔ versus wt (Zp3-Cre). A gene is called differentially expressed if the q-value is below 0.05 and the fold change is greater than 2. (d) Differential expression (non-split counts) in male E14.5 CastB6 embryonic placentae, Smchd1 het versus wt. A gene is called differentially expressed if the q-value is below 0.05 and the fold change is greater than 2. (e) Differential expression (non-split counts) in male E2.75 B6Cast embryos, Smchd1 matΔ versus wt. A gene is called differentially expressed if the q-value is below 0.05 and the fold change is greater than 2.
- https://cdn.elifesciences.org/articles/55529/elife-55529-supp2-v1.xlsx
-
Supplementary file 3
Tables of statistical data for allele-specific DNA methylation analyses at imprinted DMRs.
(a) Differential imprinted methylation in E14.5 B6Cast male placentae, Smchd1 matΔ versus wt. A methylated allele is called differentially methylated if the multiple-testing corrected p-value is below 0.05. (b) Differential imprinted methylation in E2.75 B6Cast male embryos, Smchd1 matΔ versus wt. A methylated allele is called differentially methylated if the multiple-testing corrected p-value is below 0.05. (c) Differential imprinted methylation in E2.75 B6Cast male embryos, Smchd1 matΔ versus wt. A methylated allele is called differentially methylated if the multiple-testing corrected p-value is below 0.05.
- https://cdn.elifesciences.org/articles/55529/elife-55529-supp3-v1.xlsx
-
Supplementary file 4
Tables of statistical data for genome-wide DNA methylation analyses.
(a) Differentially methylated CGIs in E14.5 B6Cast male placentae, Smchd1 matΔ versus wt. A CGI is called differentially methylated if the multiple-testing corrected p-value is below 0.05 and the absolute difference in methylation level between the two conditions is at least 10%. (b) Differentially methylated CGIs in E2.75 B6Cast male embryos, Smchd1 matΔ versus wt. A CGI is called differentially methylated if the multiple-testing corrected p-value is below 0.05 and the absolute difference in methylation level between the two conditions is at least 10%. (c) Differentially methylated promoters (−3 to +1 kb) in E2.75 B6Cast male embryos, Smchd1 matΔ versus wt. A CGI is called differentially methylated if the multiple-testing corrected p-value is below 0.05 and the absolute difference in methylation level between the two conditions is at least 10%. (d) Differentially methylated bins (10 kb, tiling the genome without overlaps) in E2.75 B6Cast male embryos, Smchd1 matΔ versus wt. A CGI is called differentially methylated if the multiple-testing corrected p-value is below 0.05 and the absolute difference in methylation level between the two conditions is at least 10%.
- https://cdn.elifesciences.org/articles/55529/elife-55529-supp4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55529/elife-55529-transrepform-v1.docx





