A trans-eQTL network regulates osteoclast multinucleation and bone mass
Figures
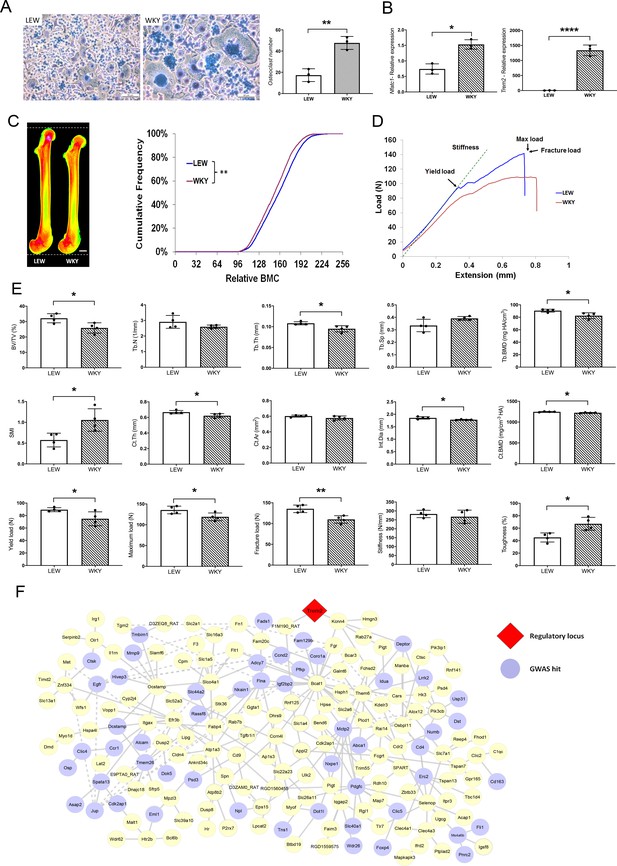
MMnet regulates adult bone homeostasis through osteoclast multinucleation.
(A) Left: Representative images of osteoclasts from Lewis (LEW) and Wistar Kyoto (WKY) rats stained with Giemsa stain. Right: number of osteoclasts with >10 nuclei from LEW and WKY rats. (B) Relative mRNA expression of Nfatc1 and Trem2 determined by qRT-PCR following osteoclast culture (n = 3/group). (C) (Left) Pseudocoloured X-ray microradiography images of femurs from male Lewis (LEW) and Wistar-Kyoto (WKY) rats (low BMC blue and green and high BMC red and pink) showing decreased femur length in WKY animals (scale bar = 2 mm). (Right) Cumulative frequency histogram showing reduced BMC in WKY rats; (n = 4 **p<0.01 vs. WT; Kolmogorov–Smirnov test). (D) Representative load displacement curves from femur three-point bend testing. (E) Micro-CT analysis of distal femur trabecular bone (trabecular bone volume/tissue volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), trabecular bone mineral density (Tb.BMD) and structure model index (SMI)) (Upper panel), micro-CT analysis from mid-diaphysis cortical bone (cortical thickness (Ct.Th), bone area (Ct.Ar), internal diameter (Int.Dia) and cortical bone mineral density (Ct.BMD)) (middle panel) and femur three-point-bend testing (lower panel) (Mean ± SD; n = 4 *p<0.05, **p<0.01 LEW vs WKY Student’s t-test). (F) MMnet comprising 190 trans-eQTLs under the master regulatory locus (Trem2, highlighted in red). Each gene in the network is represented as a circle (node), and the genes that are GWAS hits (See also Table 1 and Figure 1—source data 1) are highlighted in blue. Connecting lines indicate co-expression between the two transcripts.
-
Figure 1—source data 1
MMnet GWAS hits and their association with heel bone mineral density.
- https://cdn.elifesciences.org/articles/55549/elife-55549-fig1-data1-v2.xlsx
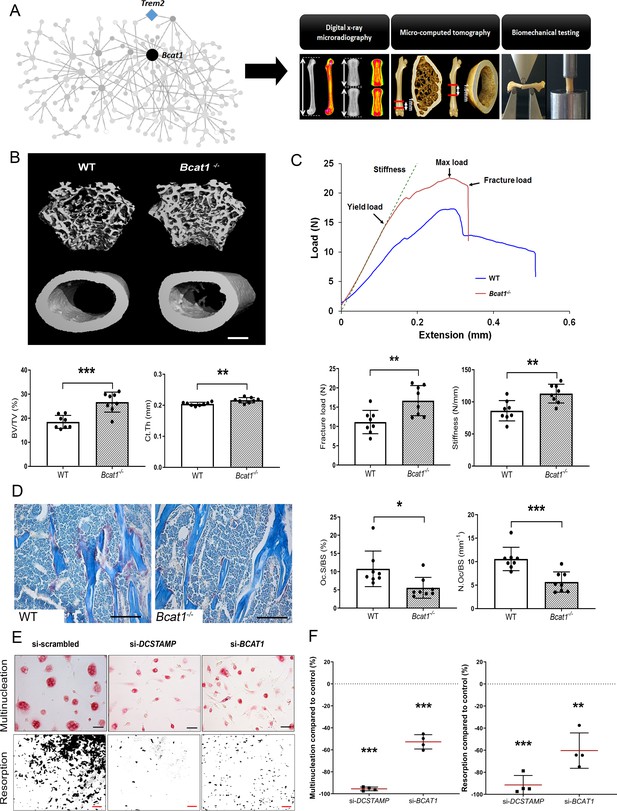
Bcat1 deficiency results in high bone mass due to decreased osteoclast fusion.
(A) Location of Bcat1 as the MMnet hub (left panel). Skeletal phenotyping of Bcat1-/- mice using X-ray microradiography, micro computed tomography (micro-CT) and three point bending testing (right panel). (B) Micro-CT images of distal femur trabecular bone and mid-diaphysis cortical bone in wild-type (WT) and Bcat1-/- mice (upper panel) (scale bar = 200 μm). Trabecular bone volume/tissue volume (BV/TV) and cortical thickness (Ct.Th) are shown in the lower panel; (Mean ± SD; n = 8 per genotype, **p<0.01, ***p<0.001 vs WT Student’s t-test). (C) Representative load displacement curves from femur three-point bend testing (upper panel). Fracture load and stiffness are shown in the lower panel; (Mean ± SD; n = 8 mice per genotype, **p<0.01 vs WT Student’s t-test). (D) Representative tartrate-resistant acid phosphatase (TRAP) stained sections from the distal femur of WT and Bcat1-/- mice showing osteoclasts in red (left panel). Quantification of the osteoclast surface/bone surface (Oc.S/BS) and osteoclast number/bone surface (N.Oc/BS) (right panel) (scale bar = 100 µm); (Mean ± SD; n = 8 per genotype, *p<0.05, ***p<0.001 vs WT Student’s t-test). (E) Representative images of TRAP+ multinucleated osteoclasts (upper panel) (scale bar = 40 µm) and hydroxyapatite resorption (lower panel) (scale bar = 1 mm) 2 days after siRNA transfection. (F) Percentage inhibition of multinucleation compared to scrambled siRNA (left panel) and percentage inhibition of hydroxyapatite resorption compared to scrambled siRNA (right panel); (Mean ± SD from n = 4 donors, **p<0.01, ***p<0.001 vs scrambled siRNA, one-sample-t test).
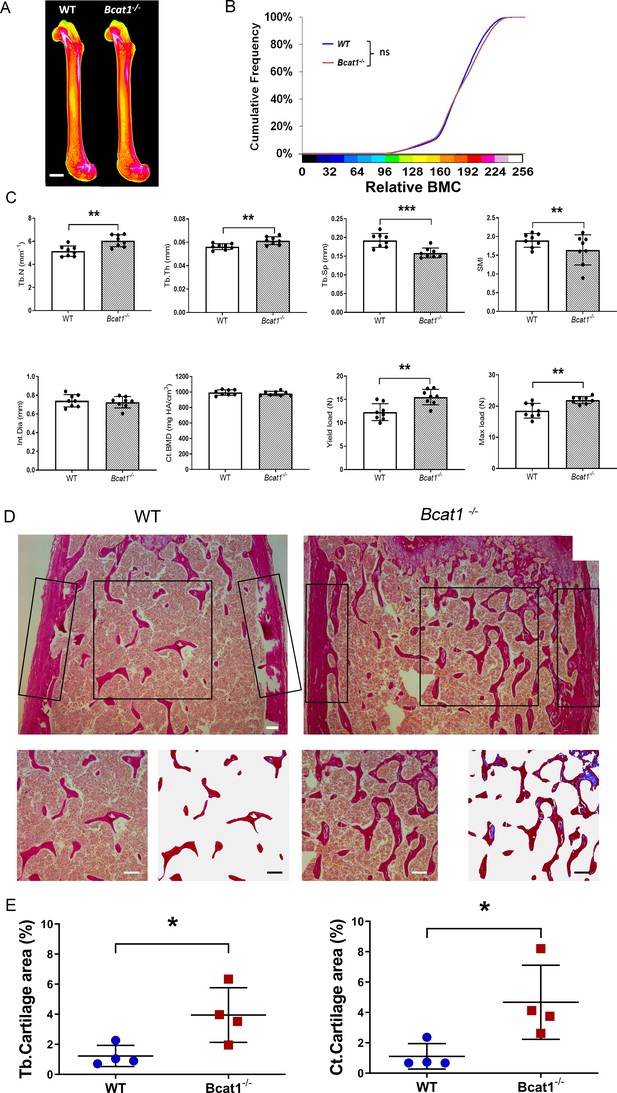
Deletion of Bcat1 results in increased bone mass and strength due to impaired osteoclast multinucleation and function.
(A) Representative X-ray microradiography images of femurs from male WT and Bcat1-/- mice. In pseudocoloured images low BMC is indicated by blue and green and high BMC by red and pink (scale bar = 1 mm). (B) Cumulative frequency histogram of relative bone mineral content (BMC) (n = 8 per genotype, by Kolmogorov–Smirnov test). (C) Micro-CT analysis of distal femur trabecular bone, micro-CT analysis of mid-diaphysis cortical bone and femur three-point-bend testing (trabecular number (Tb.N), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), structure model index (SMI), internal diameter (Int.Dia) and cortical bone mineral density (Ct.BMD)); (Mean ± SD; n = 8 per genotype, **p<0.01, ***p<0.001 vs WT Student’s t-test). (D) Representative images of retained cartilage stained with Alcian Blue (blue-green) in trabecular bone stained with van Gieson (red) from the distal femurs of WT and Bcat1-/- mice. Top panel shows images of the distal femoral metaphyses. Black boxes indicate the region of interest (ROI) used to determine trabecular and cortical cartilage areas. Lower panel shows trabecular ROI before and after image analysis to highlight retained trabecular cartilage in blue (scale bars = 100 μm). (E) Quantification of the retained cartilage area as a proportion of total trabecular bone area (Tb.Cartilage area) and as a proportion of total cortical bone area (Ct.Cartilage area); (Mean ± SD; n = 4–6 per genotype *p<0.05 vs WT Student’s t-test).
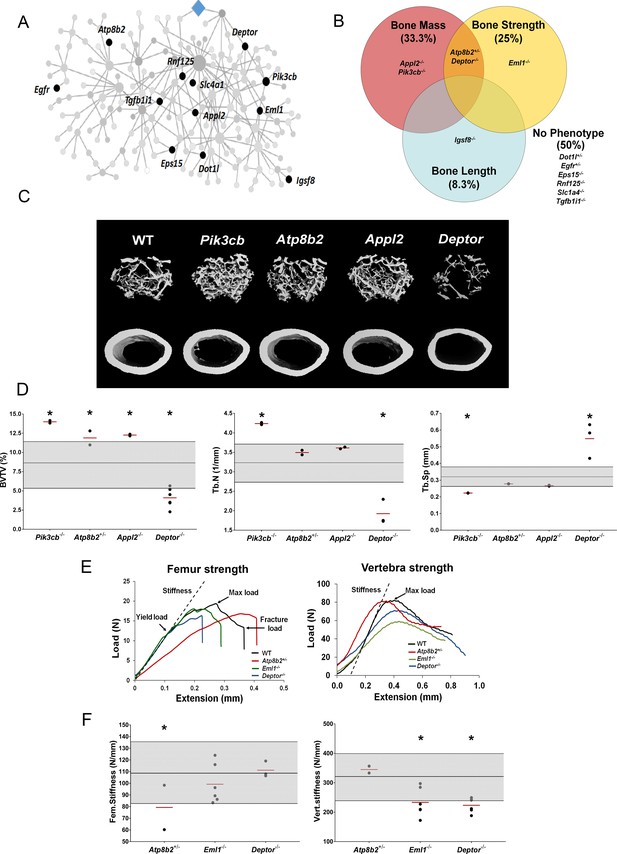
MMnet regulates bone mass.
(A) The location of the 12 MMnet genes for which knockout mice were available are highlighted in the network. (B) Venn diagram showing mutant strains with outlier phenotypes for bone mass, bone strength and bone length. (C) Representative micro-CT images of distal femur trabecular bone and mid diaphyseal cortical bone from WT and mutant mice (scale bar = 200 μm). (D) Graphs show trabecular bone volume (BV/TV), trabecular number (Tb.N) and trabecular separation (Tb.Sp). Grey box represents WT reference mean + / - 2 SD (n = 320, 16 week old, female C57BL/6 WT mice). (E) Representative load displacement curves for femur three-point bend testing and vertebral compression. (F) Femur stiffness (Fem.stiffness) and vertebral stiffness (vert.stiffness). Data from individual mice are shown as black dots and mean value by a red line (n = 2 or six per genotype). Parameters outside the WT reference range are indicated by an asterisk *.
-
Figure 3—source data 1
Results of high-throughput MMnet mouse knockout skeletal phenotyping.
- https://cdn.elifesciences.org/articles/55549/elife-55549-fig3-data1-v2.xlsx
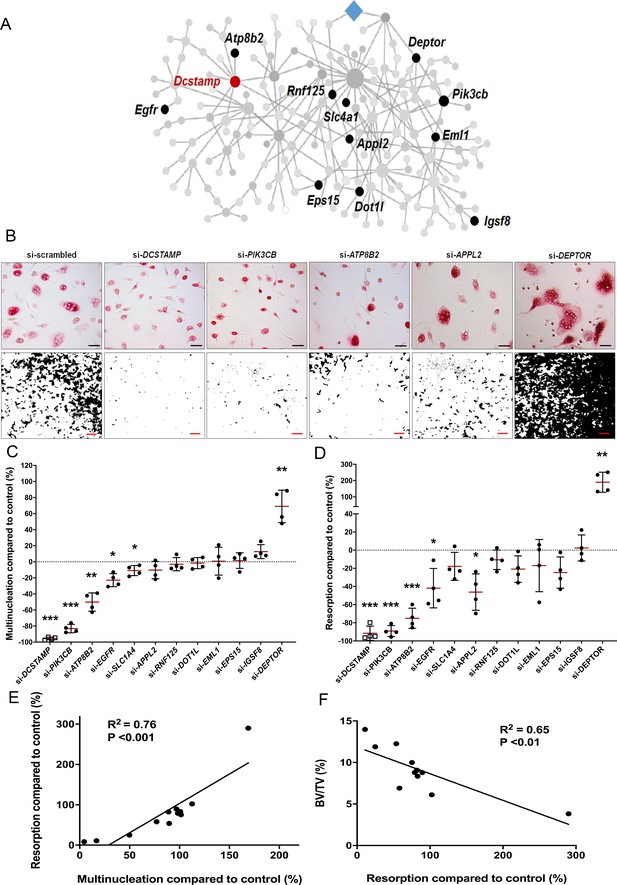
MMnet regulates human osteoclast multinucleation and resorption in vitro.
(A) The location of the 11 MMnet genes for which knockout mice were available, and for which there are human orthologues. The master regulator of macrophage fusion Dcstamp is highlighted in red. (B) Representative images of TRAP+ osteoclasts (Top panel) (scale bar = 40 µm) and hydroxyapatite resorption (Bottom panel) (scale bar = 1 mm) following siRNA knockdown for DCSTAMP, PIK3CB, ATP8B2, APPL2 and DEPTOR. (C) Graphs show the frequency of multinucleated cells (>4 nuclei) following siRNA knockdown as a percentage of the scramble siRNA control; n = 4 donors. (D) Hydroxyapatite resorption following siRNA knockdown as a percentage of the scramble siRNA control; n = 4 donors. (For (C) and (D): Mean ± SD from n = 4 donors, *p<0.05, **p<0.01, ***p<0.001 vs scrambled siRNA, one-sample-t test). (E) Pearson correlation between human osteoclast multinucleation and resorptive activity following siRNA knockdown of 12 MMnet genes compared to control (R2 = 0.76; p<0.001) (F) Pearson correlation between human osteoclast resorption activity following si-RNA knockdown of 11 MMnet genes compared to control and BV/TV in the corresponding 11 MMnet knockout mouse strains (R2 = 0.65; p<0.01).
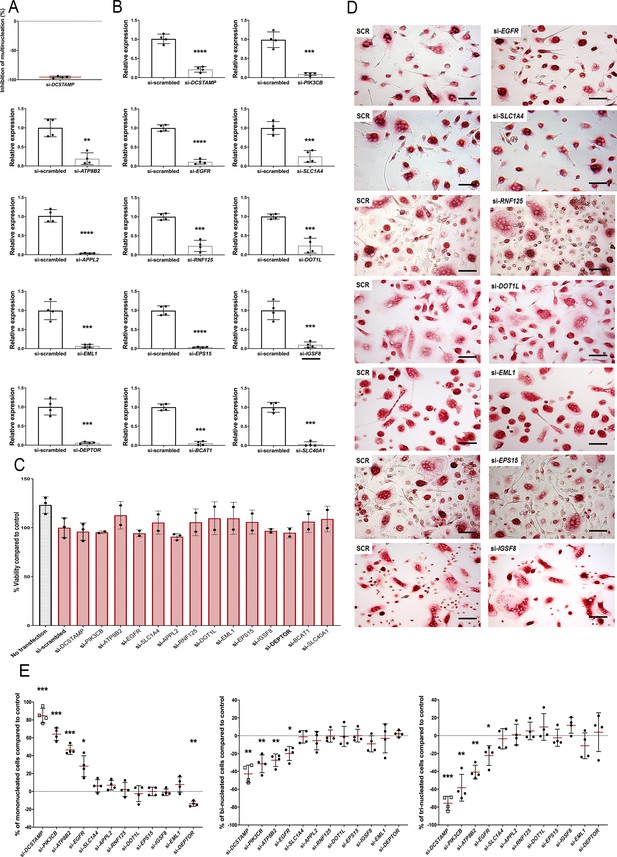
siRNA mediated knockdown of MMnet genes in human osteoclasts.
(A) Inhibition of osteoclast multinucleation following DCSTAMP siRNA knockdown compared with scrambled control (Mean ± SD; n = 4 donors, ****p<0.001 vs scrambled siRNA, one-sample-t test). (B) Relative mRNA expression of the targeted gene determined by qRT-PCR following siRNA knockdown (Mean ± SD; n = 4 donors per gene, **p<0.01, ***p<0.001, ****p<0.001 vs scrambled siRNA, Student’s t-test). (C) Cell viability following siRNA transfection determined using alamar blue staining (n = 2–3 donors per gene). (D) Representative images of TRAP+ multinucleated osteoclasts following siRNA knockdown. (E) Graphs show the percentage of mono, bi and tri-nucleated cells following siRNA knockdown compared to scramble siRNA control; (Mean ± SD from n = 4 donors, *p<0.05, **p<0.01, ***p<0.001 vs scrambled siRNA, one-sample-t test).
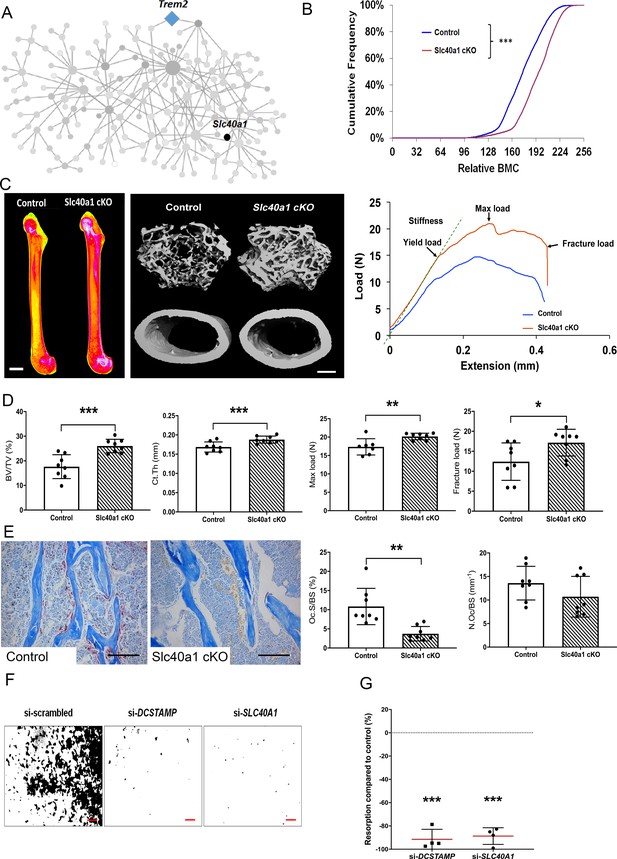
Myeloid-specific deletion of Ferroportin results in increased bone mass and strength due to impaired osteoclast multinucleation and function.
(A) Location of Slc40a1 in MMnet. (B) Cumulative frequency histogram of relative BMC; n = 8 per genotype. ***p<0.001 vs. WT by Kolmogorov–Smirnov test. (C) Representative X-ray microradiography images of femurs from male Control and Slc40a1 cKO mice. Pseudocoloured images indicating low BMC (blue and green) and high BMC (red and pink) are shown in the left panel (scale bar = 1 mm). Micro-CT images of distal femur trabecular bone and mid-diaphysis cortical bone (middle panel) (scale bar = 200 μm). Representative load displacement curves from femur three-point bend testing (right panel). (D) Trabecular bone volume (BV/TV) and cortical thickness (Ct.Th), femur maximum load and fracture load. (E) Representative TRAP-stained femur sections from Control and Slc40a1 cKO mice showing osteoclasts stained in red (left panel). Osteoclast surface (Oc.S/BS) and osteoclast number (N.Oc/BS) (right panel) (scale bar = 100 µm); (For (D) and (E): Mean ± SD; n = 8 per genotype, *p<0.05, **p<0.01, ***p<0.001 vs Control Student’s t-test). (F) Representative images of hydroxyapatite resorption (scale bar = 1 mm) following DCSTAMP and SLC40A1 si-RNA knockdown. (G) Inhibition of osteoclast hydroxyapatite resorption following DCSTAMP and SLC40A1 siRNA knockdown; (Mean ± SD from n = 4 donors, ***p<0.001 vs scrambled siRNA, one-sample-t test).
-
Figure 5—source data 1
Prioritisation pipeline to identify a novel and tractable MMnet gene.
- https://cdn.elifesciences.org/articles/55549/elife-55549-fig5-data1-v2.xlsx
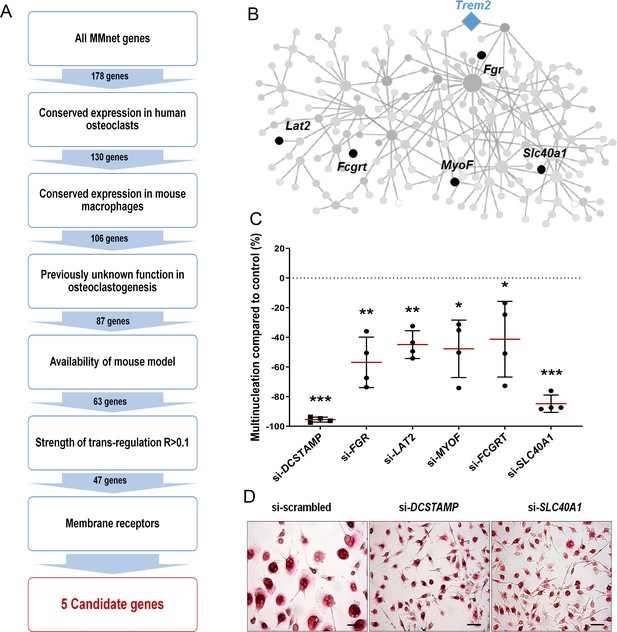
An In silico filtering strategy prioritises Slc40a1 (ferroportin) for myeloid-targeted gene deletion.
(A) Step-wise filtering workflow. (B) Five candidate genes within MMnet. (C) Percentage of osteoclasts with >4 nuclei after RNAi compared to non-targeting si-RNA; n = 4 donors. (D) Representative images of TRAP+ multinucleated osteoclasts (scale bar = 40 µm) following DCSTAMP and SLC40A1 si-RNA knockdown. (Mean ± SD from n = 4 donors, *p<0.05, **p<0.01, ***p<0.001 vs scrambled siRNA, one-sample-t test).
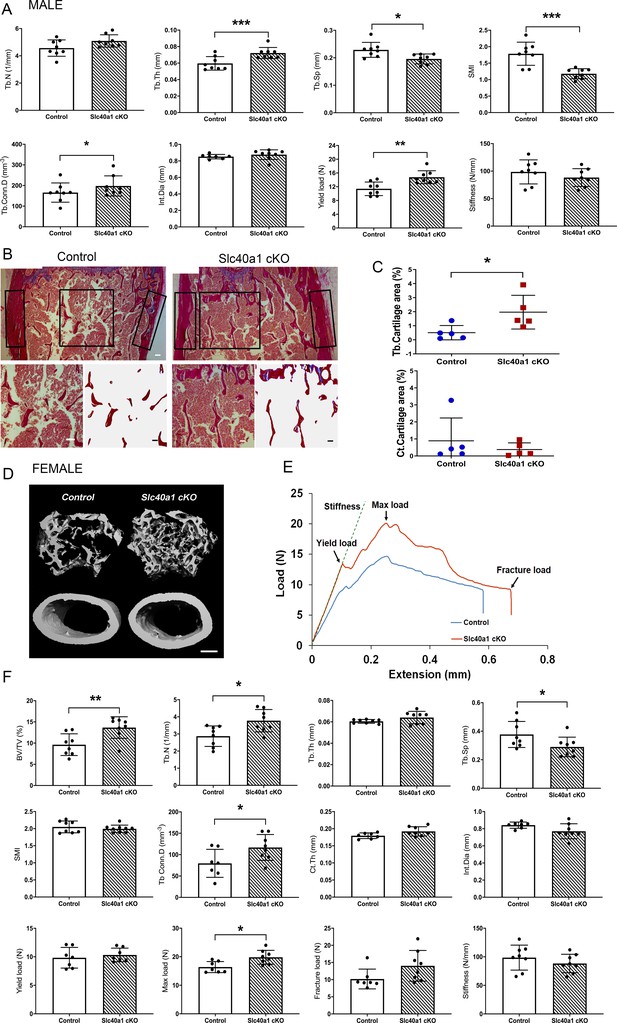
Myeloid-specific deletion of Ferroportin results in increased bone mass and strength due to impaired osteoclast multinucleation and function.
(A) Graphs show micro-CT analysis of distal femur trabecular bone, micro-CT analysis of mid-diaphysis cortical bone and femur three-point-bend testing in male mice. (trabecular bone volume/tissue volume (BV/TV), trabecular number (Tb.N), trabecular thickness (Tb.Th), trabecular separation (Tb.Sp), structure model index (SMI), trabecular connectivity density (Tb.Conn.D), cortical thickness, (Ct.Th), internal diameter (Int.Dia) and cortical bone mineral density (Ct.BMD)) (Mean ± SD; n = 8 per genotype as *p<0.05, **p<0.01, ***p<0.001 Slc40a1 cKO vs Control Student’s t-test). (B) Representative images of retained cartilage stained by Alcian Blue (blue-green) in trabecular bone stained with van Gieson (red) from the distal femurs of Control and Slc40a1 cKO mice. Top panel shows images of the distal femoral metaphyses. Black boxes indicate the region of interest (ROI) used to determine trabecular and cortical cartilage areas. Lower panel shows trabecular ROI before and after image analysis to highlight retained trabecular cartilage in blue (scale bar = 100 μm). (C) Quantification of the retained cartilage area as a proportion of total trabecular bone area (Tb.Cartilage area) and as a proportion of total cortical bone area (Ct.Cartilage area). (Mean ± SD; n = 5 per genotype *p<0.05 vs Control, Student’s t-test). (D) Representative micro-CT images of distal femur trabecular bone and mid-diaphysis cortical bone in Slc40a1 cKO and Control female mice (scale bar = 200 μm). (E) Representative load displacement curves from three-point bend testing of femurs from Slc40a1 cKO and Control female mice. (F) Graphs show micro-CT analysis of distal femur trabecular bone, micro-CT analysis of mid-diaphysis cortical bone and femur three-point-bend testing in female mice; (Mean ± SD; n = 8 per genotype, *p<0.05, **p<0.01 Slc40a1 cKO vs Control, Student’s t-test).
Tables
MMnet is significantly enriched for variants that associate with different bone traits.
Single nucleotide polymorphisms (SNPs) significantly associated with the reported GWAS traits were interrogated in two datasets: the background dataset which includes all the eQTLs (posterior probability >80% see Materials and methods) and MMnet gene set. The enrichment was then tested over the background using a hypergeometric test. The significantly enriched traits are shown in bold.
| GWAS trait | Number of eQTLs genes (background) with at least one GWAS hit | Number of MMnet genes with at least one GWAS hit | Frequency (%) eQTLs genes (background) with at least one GWAS hit | Frequency (%) MMnet genes with at least one GWAS hit | P-value MMnet enrichment over background (hypergeometric test) |
|---|---|---|---|---|---|
| All GWAS combined | 299 | 41 | 20.3% | 27.7% | 0.0085 |
| Heel bone mineral density | 117 | 20 | 8.0% | 13.5% | 0.0045 |
| Osteoarthritis | 9 | 3 | 0.6% | 2.0% | 0.0083 |
| Psoriatic arthritis | 2 | 1 | 0.1% | 0.7% | 0.0101 |
| Body height | 165 | 24 | 11.2% | 16.2% | 0.0186 |
| Bone quantitative ultrasound measurement | 4 | 1 | 0.3% | 0.7% | 0.0527 |
| Bone density | 16 | 3 | 1.1% | 2.0% | 0.0686 |
| Hip bone mineral density | 5 | 1 | 0.3% | 0.7% | 0.0821 |
| Spine bone mineral density | 7 | 1 | 0.5% | 0.7% | 0.1509 |
| Bone fracture | 3 | 0 | 0.2% | 0.0% | 0.2727 |
| Ankylosing spondylitis | 4 | 0 | 0.3% | 0.0% | 0.3460 |
| Periodontitis | 4 | 0 | 0.3% | 0.0% | 0.3460 |
| Rheumatoid arthritis | 15 | 1 | 1.0% | 0.7% | 0.4550 |
| Adolescent idiopathic scoliosis | 60 | 5 | 4.1% | 3.4% | 0.5723 |
| Bone mineral content measurement | 1 | 1 | 0.1% | 0.7% | - |
| Lumbar disc degeneration | 1 | 1 | 0.1% | 0.7% | - |
| Osteitis deformans | 1 | 0 | 0.1% | 0.0% | - |
Additional files
-
Supplementary file 1
List of primers used.
- https://cdn.elifesciences.org/articles/55549/elife-55549-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55549/elife-55549-transrepform-v2.docx





