LUZP1, a novel regulator of primary cilia and the actin cytoskeleton, is a contributing factor in Townes-Brocks Syndrome
Figures
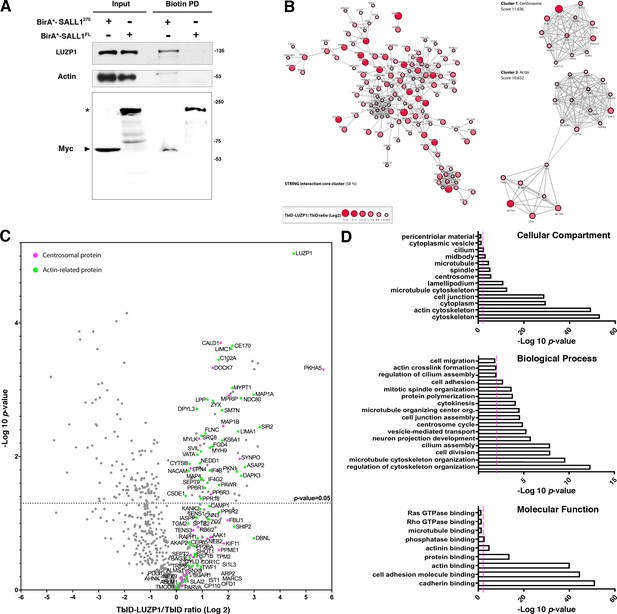
Proximity proteomics reveal LUZP1 interaction with truncated SALL1 and with centrosome- and actin cytoskeleton-associated proteins.
(A) Western blot analysis of BioID, streptavidin pulldown (PD) of HEK 293FT cells transfected with Myc-tagged BirA*-SALL1275 or BirA*-SALL1FL. Specific antibodies (LUZP1, actin, Myc) were used as indicated. Anti-Myc antibody detected the self-biotinylated form of BirA*-SALL1FL (asterisk) or BirA*-SALL1275 (black arrowhead). (B) STRING core cluster network analysis of LUZP1 interactors (confidence 0.7 or higher), visualized using Cytoscape software. Color and size of the nodes indicate Log2 of the TurboID-LUZP1 (TbID-LUZP1) versus TbID alone ratio. The most highly interconnected clusters, centrosome and actin, are indicated separately. (C) Volcano plot representing the distribution of the candidates identified by proximity proteomics in three independent experiments. Proteins with more than 1-fold change in TbID-LUZP1 intensity with respect to the TbID (Log2 ≥ 0) were considered as LUZP1-associated candidates (grey dots). Proteins associated with the actin cytoskeleton and the centrosome were colored in green and pink colors, respectively. (D) Graphical representation of the -Log10 of the p-value for each of the represented GO terms of the TbID experiment performed on RPE1 stably expressing near endogenous levels of TbID-LUZP1 vs TbID. Pink dotted line represents the cutoff of p-value<0.01.
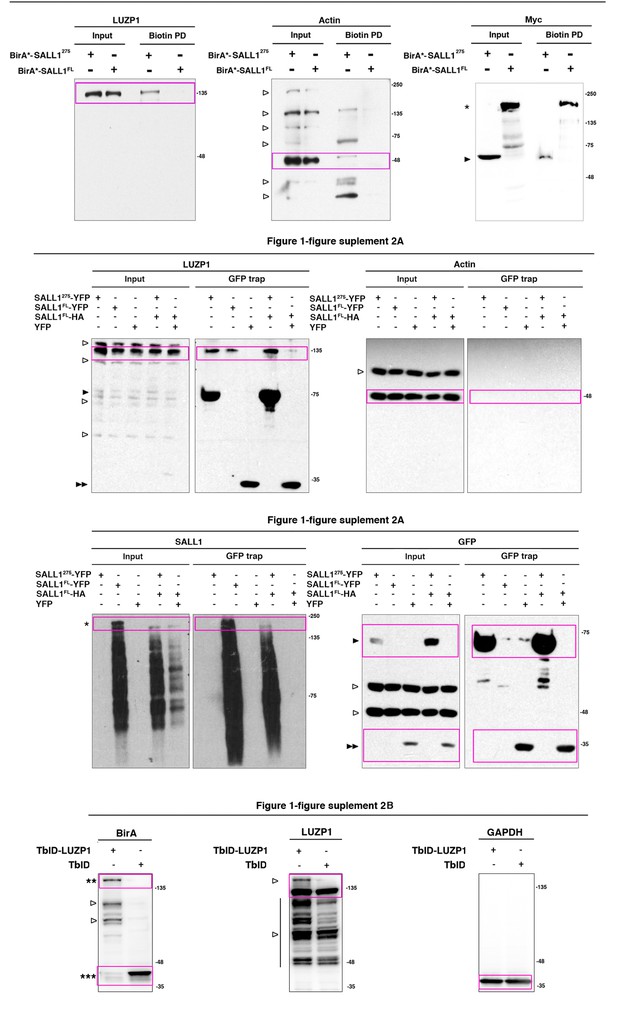
Full western blot images for Figure 1.
Title indicates the Figure to which the western blot corresponds; magenta boxes show the region of the gel that was used to build the indicated figures. SALL1FL protein is indicated by one asterisk, SALL1 truncated forms by one black arrowhead, YFP alone by two black arrowheads. One empty arrowhead indicates truncated forms, bands from previous probing or unspecific bands. Molecular weight markers are shown to the right.
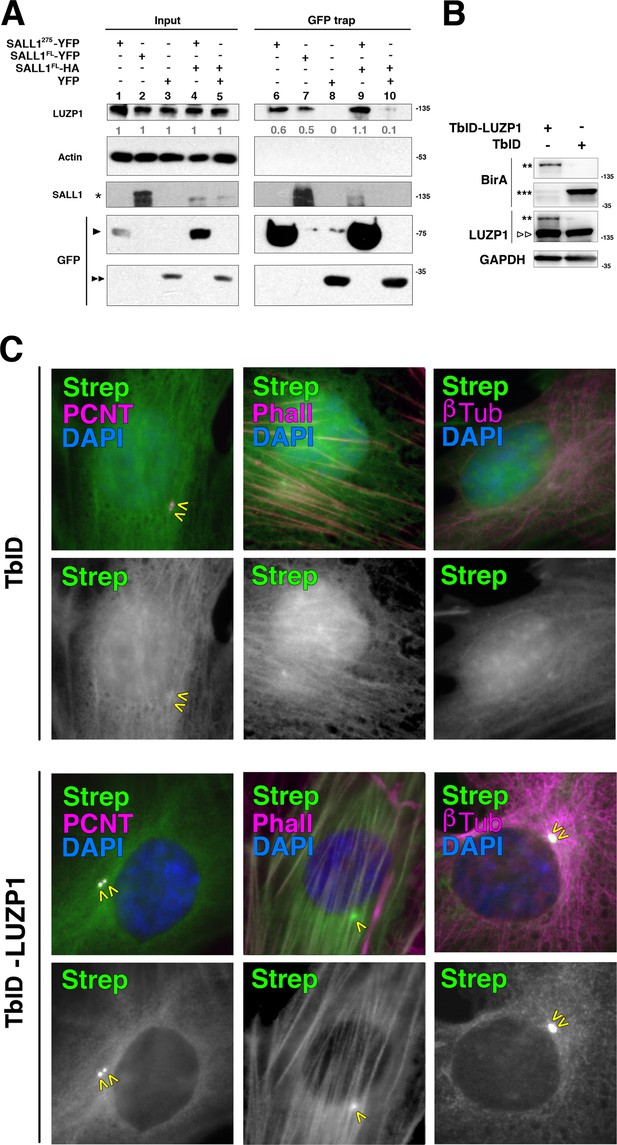
Analysis of TbID constructs expression and biotinylation localization.
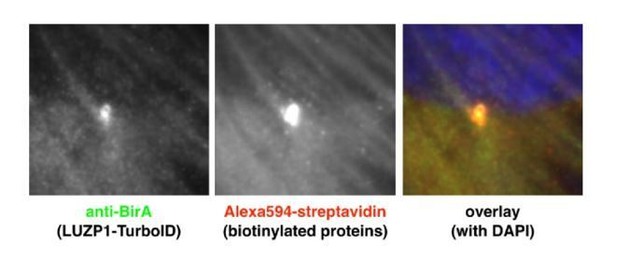
(A) Western blot of inputs or GFP-Trap pulldowns performed in HEK 293FT cells transfected with SALL1275-YFP (lanes 1 and 6), SALL1FL-YFP (lanes 2 and 7), YFP alone (lanes 3 and 8), SALL1275-YFP together with SALL1FL-2xHA (SALL1FL-HA; lanes 4 and 9) or SALL1FL-HA together with YFP alone (lanes 5 and 10). Specific antibodies (LUZP1, actin, SALL1) were used as indicated. Numbers under LUZP1 panel result from dividing band intensities of each pulldown by their respective input levels. One asterisk indicates BirA*-SALL1FL or SALL1FL-YFP, one black arrowhead SALL1275-YFP and two black arrowheads YFP alone. Molecular weight markers (kDa) are shown to the right. Actin was used as loading control. Blots shown are representative of three independent experiments. (B) Western blot analysis of RPE1 cells transfected with TbID-LUZP1 or TbID alone. Specific antibodies (BirA, LUZP1 and GAPDH) were used as indicated. Anti-BirA antibody detected the fusion form of TbID-LUZP1 (two asterisks) or TbID (three asterisks). LUZP1 antibodies detected endogenous LUZP1 (two empty arrowheads) and TbID-LUZP1 (two arrowheads). Molecular weight markers are shown to the right. (C) In RPE1 cells transfected with TbID-LUZP1, streptavidin (green) localizes to the centrosome labelled with Pericentrin (PCNT, magenta) or actin fibers labelled with phalloidin (Phall, magenta). No biotinylation in microtubules, labelled by βTubulin (βTub, magenta), was observed. By contrast, cells transfected with TbID show streptavidin labeling in nucleus and cytoplasm. Green channels are shown in black and white. Arrowheads indicate the centrioles. Note that basal actin stress fibers are less evident when apical centrioles are in focus.
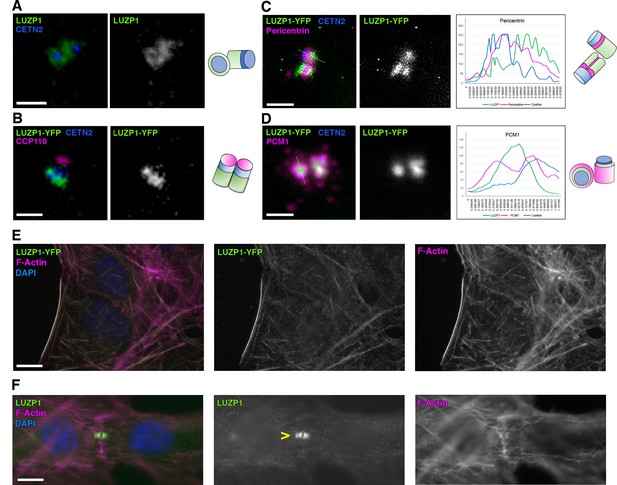
LUZP1 localizes to the centrosome and the actin cytoskeleton.
(A) Immunofluorescence micrographs of LUZP1 (green) and Centrin-2 (CETN2, blue) in RPE1 cells. (B–D) Immunofluorescence micrographs of U2OS cells expressing LUZP1-YFP (green) stained with antibodies against CETN2 (blue) and CCP110 (B), Pericentrin (C) and PCM1 (D) in magenta. Plot profile of the different fluorophore intensities along the yellow lines in (C, D). Schematic representation of LUZP1 localization at the centrosome according to their respective micrographs in (A–D). Scale bar, 1 µm. Imaging in (A–D) was performed using confocal microscopy (Leica SP8, 63x objective). Lightning software (Leica) was applied. (E, F) Immunofluorescence micrographs of U2OS cells stained with an antibody against endogenous LUZP1 (green), phalloidin to detect F-actin (magenta), and counterstained with DAPI (blue). Single green and magenta channels are shown in black and white. (F) LUZP1 at the midbody in dividing cells (yellow arrowhead). Scale bar, 10 µm (E, F). Imaging in (E, F) was performed using widefield fluorescence microscopy (Zeiss Axioimager D1, 63x objective).
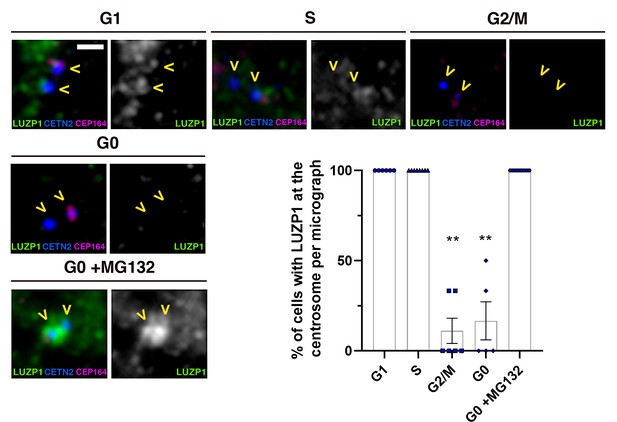
Centrosomal localization of LUZP1 at different cell cycle stages.
Immunofluorescence micrographs showing LUZP1 in the centrosome during cell cycle in RPE1 cells. Cells were treated with mimosine (G1 phase), thymidine (S phase), RO-3306 (G2/M phase) or starved (G0) with and without the proteasome inhibitor MG132. Cells were stained in green with antibodies against endogenous LUZP1, and in blue with DAPI. Scale bar 0.5 µm. Imaging was performed using widefield fluorescence microscopy (Zeiss Axioimager D1, 63x objective). On the lower right panel, graphical representation of the percentage of cells showing the presence of LUZP1 at the centrosome per micrograph, corresponding to the experiment in (A); n > 6 micrographs. Note a general decrease of LUZP1 during G2/M and upon starvation (G0), which is recovered by MG132 addition. Graphs represent Mean and SEM of three independent experiments pooled together. P-values were calculated using Kruskall-Wallis and Dunn's multiple comparisons test. ** over G2/M and G0 columns indicate that they are significantly different from the rest of the columns and not significant amongst them.
LUZP1 localization in the centrosome.
3D reconstruction of Z-stack micrographs of human control fibroblasts (ESCTRL#2) stained with antibodies against endogenous LUZP1 (green) and acetylated alpha and gamma tubulin to label the cilia and centrosomes, respectively (magenta). Image was taken using Confocal Super-resolution microscopy (LSM 980, Zeiss).
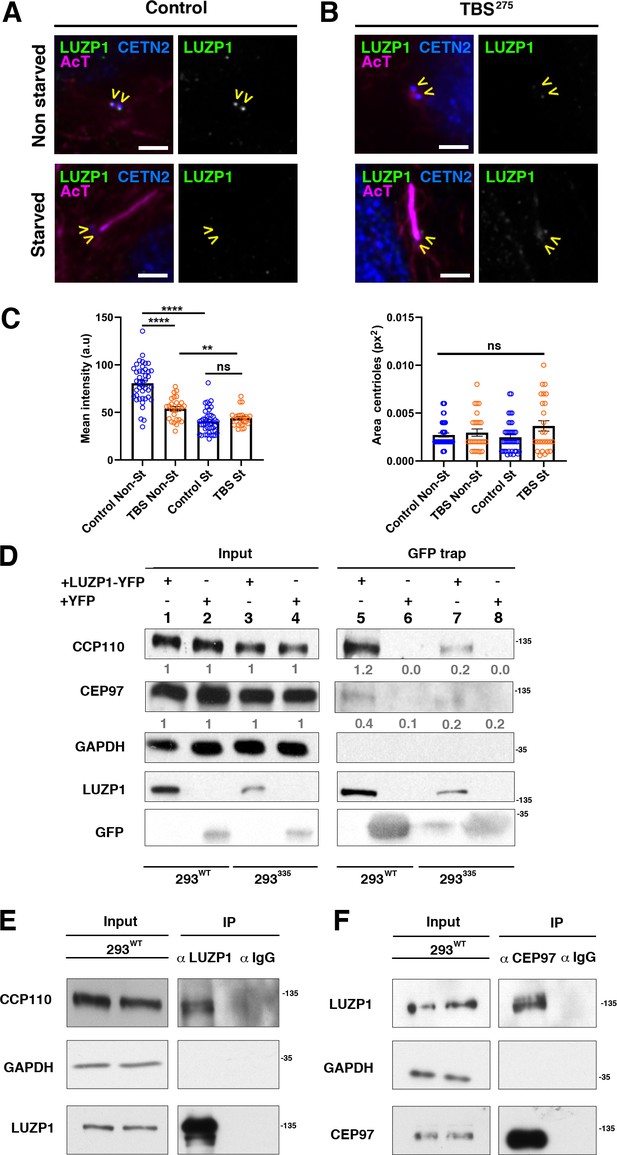
TBS cells show reduced LUZP1 levels at the centrosome.
(A, B) Immunofluorescence micrographs of non-starved and starved human-derived Control (A) and TBS275 fibroblasts (B) stained with antibodies against endogenous LUZP1 (green, yellow arrowheads), Centrin-2 (CETN2, blue) and acetylated alpha-tubulin (magenta). Black and white images show the isolated green channel. Note the reduction of LUZP1 in starved cells and in non-starved TBS275 compared to non-starved control fibroblasts. Imaging was performed using widefield fluorescence microscopy (Zeiss Axioimager D1, 63x objective). Scale bar, 4 µm. (C) Graphical representation of the LUZP1 mean intensities (left panel) or the centrosome area (right panel), corresponding to the experiments shown in (A–B); n ≥ 6 micrographs. Three independent experiments were pooled together. P-values were calculated using the unpaired two-tailed Student´s test or U- Mann-Whitney test. (D) Western blot of inputs (lanes 1 to 4) and GFP-Trap pulldowns (lines 5 to 8) performed in WT HEK 293FT cells or in 293335SALL1 mutant cells transfected with LUZP1-YFP (lanes 1, 3, 5 and 7) or YFP alone (lanes 2, 4, 6 and 8). Numbers under CCP110 and CEP97 panels result from dividing band intensities of each pulldown by their respective input levels. GAPDH was used as loading control. (E, F) Co-immunoprecipitation experiments show LUZP1-CCP110 (E) and CEP97-LUZP1 (F) interactions. Rabbit IgG was used for immunoprecipitation controls. GAPDH was used as loading and specificity control. In all panels, specific antibodies (LUZP1, GAPDH, CCP110, CEP97, GFP) were used as indicated. Blots shown here are representative of three independent experiments. Molecular weight markers are shown to the right.
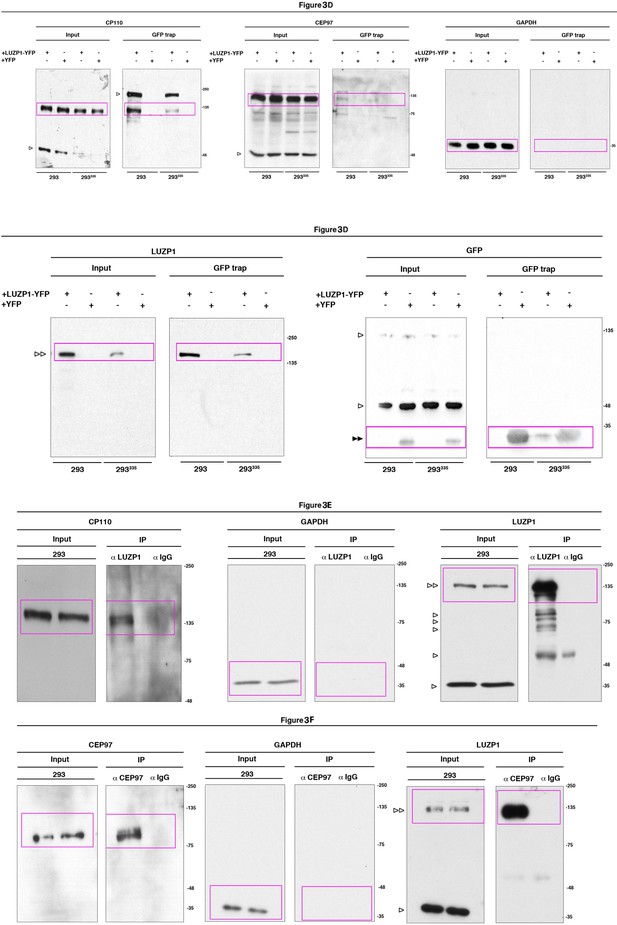
Full western blot images for Figure 3.
Title indicates the Figure to which the western blot corresponds; magenta boxes show the region of the gel that was used to build the indicated figures. LUZP1-YFP is indicated by two empty arrowheads and GFP or YFP alone by two black arrowheads. One empty arrowhead indicates truncated forms, bands from previous probing or unspecific bands. Molecular weight markers are shown to the right.
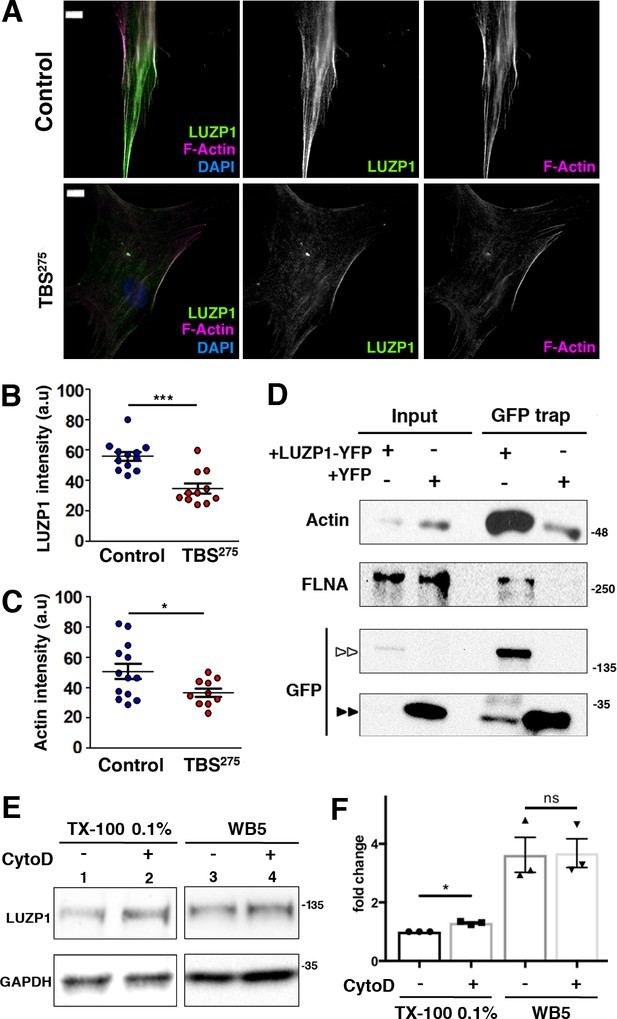
Reduction in LUZP1 coincides with decreased F-actin.
(A) Immunofluorescence micrographs of Control and TBS275 human fibroblasts stained with an antibody against endogenous LUZP1 (green), phalloidin to label F-actin (magenta), and counterstained with DAPI to label the nuclei (blue). Black and white images show the single green and magenta channels. Note the overall reduction in LUZP1 and F-actin levels in TBS275 compared to control fibroblasts. Scale bar, 10 µm. Imaging was performed using widefield fluorescence microscopy (Zeiss Axioimager D1, 63x objective). (B, C) Graphical representation of the LUZP1 (B) and F-actin (C) mean intensities, corresponding to the experiments shown in (A); n ≥ 6 micrographs. Three independent experiments were pooled together. P-values were calculated using the unpaired two-tailed Student´s test or U- Mann-Whitney test. (D) Western blot of inputs or GFP-Trap pulldowns performed in HEK 293FT cells transfected with LUZP1-YFP or YFP alone. Anti-GFP antibody detected YFP alone (two black arrowheads) and LUZP1-YFP (two white arrowheads). Blots shown here are representative of three independent experiments. Molecular weight markers are shown to the right. Specific antibodies (LUZP1, GAPDH, CCP110, CEP97, GFP) were used as indicated. (E) Western blot of total cell lysates of HEK 293FT treated or not with cytochalasin D (CytoD) in a mild lysis buffer (TX-100 0.1%, lanes 1, 2) or a strong lysis buffer (WB5, lanes 3, 4). Note the increase in LUZP1 levels upon actin polymerization blockage with CytoD, exclusively when cells were lysed on 01% TX-100-based lysis buffer. GAPDH was used as loading control. In (D) and (E) panels, specific antibodies (LUZP1, GAPDH, actin, FLNA, GFP) were used as indicated. (F) Graphical representation of LUZP1 vs GAPDH band intensities in (E) normalized to lane 1. Graphs represent Mean and SEM of three independent experiments. P-value was calculated using two tailed unpaired Student´s t-test. Molecular weight markers in (D) and (E) are shown to the right.
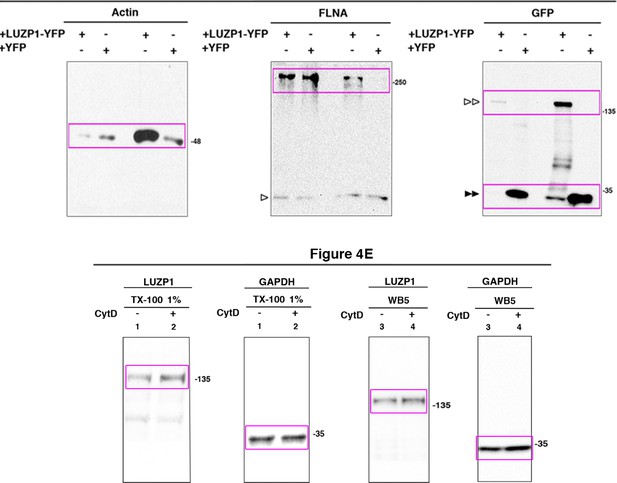
Full western blot images for Figure 4.
Titles indicate the Figure to which the western blot corresponds; magenta boxes show the region of the gel that was used to build the indicated figures. LUZP1-YFP is indicated by two empty arrowheads and YFP alone by two black arrowheads. Bands from previous probing or unspecific bands are indicated by one empty arrowhead. Molecular weight markers are shown to the right.
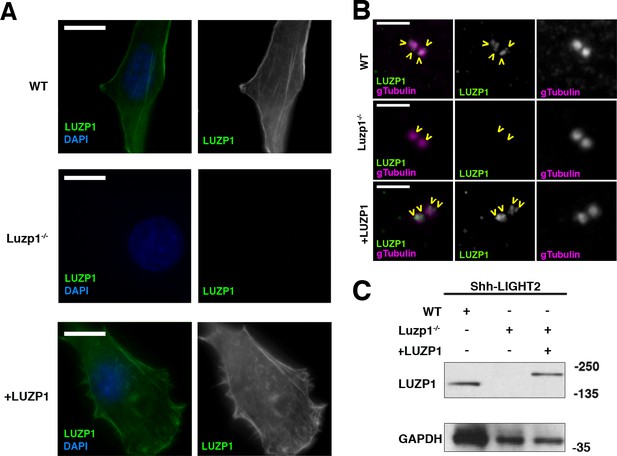
Generation of LUZP1 mutant cells.
(A, B) Immunofluorescence micrographs of Shh-LIGHT2 control cells (WT), Luzp1 depleted Shh-LIGHT2 cells (Luzp1-/-) and Luzp1-/- cells rescued with human LUZP1 (+LUZP1 cells) stained with a specific antibody against endogenous LUZP1 (green) and DAPI (blue) (A) or gamma-tubulin (magenta) (B). Single green and magenta channels are shown in black and white. Scale bars, 10 µm (A) or 2.5 µm (B). Images were taken using widefield fluorescence microscopy (Zeiss Axioimager D1, 63x objective). Note the lack of LUZP1 in Luzp1-/- cells. (C) Western blot analysis of total lysates of WT, Luzp1-/- and +LUZP1 cells using anti-LUZP1 antibodies. Molecular weight markers are shown to the right. Note the lack of LUZP1 signal in Luzp1-/- cells by immunofluorescence and western blot.
Full western blot images for Figure 5.
Title indicates the Figure to which the western blot corresponds; magenta boxes show the region of the gel that was used to build the indicated figures. Molecular weight markers are shown to the right.
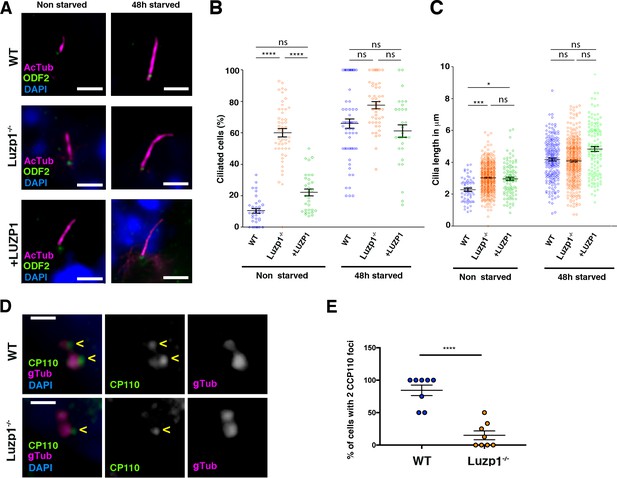
Loss of Luzp1 causes aberrant cilia frequency and length and Shh signaling.
(A) Micrographs of Shh-LIGHT2 cells (WT), Shh-LIGHT2 cells lacking Luzp1 (Luzp1-/-) and Luzp1-/- cells rescued with human LUZP1-YFP (+LUZP1) analyzed in cycling conditions (non-starved), or during cilia assembly (48 hr starved). Cilia were visualized by acetylated alpha-tubulin (magenta), basal body by ODF2 (green) and nuclei by DAPI (blue). Scale bar 2.5 µm. (B, C) Graphical representation of percentage of ciliated cells per micrograph (B) and cilia length (C) measured in WT (blue circles, n > 34 micrographs), Luzp1-/- (orange circles, n > 44 micrographs) or +LUZP1 cells (green circles, n > 30 micrographs) from three independent experiments. No starvation: WT 2.3 µm; Luzp1-/- cells 3.0 µm; +LUZP1 cells 2.9 µm; 48 hr starvation: WT 4.2 µm; Luzp1-/- cells 4.1 µm; +LUZP1 cells 4.8 µm; all average measures. (D) Immunofluorescence micrographs of WT and LUZP1-/- cells stained with antibodies against endogenous CCP110 (green), gamma-tubulin (gTub) to label the centrioles (magenta) and DAPI to label the nuclei (blue). Black and white images show the single green and magenta channels. Note the different distribution of CCP110 to the centrosome in LUZP1-/- compared to WT cells. Scale bar, 1 µm. (E) Graphical representation of the percentage of cells showing the presence of CCP110 to both centrioles per micrograph corresponding to the experiments in (D); n = 10 micrographs. Three independent experiments were pooled together.
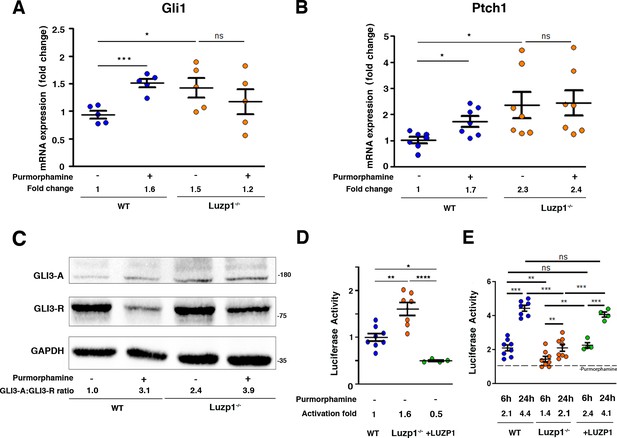
Luzp1-/-cells show aberrant Shh signaling.
(A, B) Graphical representation of the fold-change in the expression of Gli1 (n = 5) (A) and Ptch1 (n = 7) (B) obtained by qRT-PCR from wild-type Shh-LIGHT2 cells (WT; blue dots) or Shh-LIGHT2 cells lacking Luzp1 (Luzp1-/-; orange dots), treated (+) or not (-) with purmorphamine for 24 hr. (C) Western blot analysis of lysates from WT and Luzp1-/- cells. Samples were probed against GLI3 using an antibody that detects both GLI3-activator form (GLI3-A) and GLI3-repressor form (GLI3-R); GAPDH was used as loading control. Numbers under the lanes are the Mean of 3 independent experiments, resulting of dividing the activator by the repressor intensities, taking WT non-induced value as 1. Molecular weight markers are shown to the right. (D, E) Graphical representation of fold-change in luciferase activation when WT (n > 7; blue dots), Luzp1-/- (n > 7; orange dots) or +LUZP1 (n = 4; green dots) cells are treated for 6 and 24 hr or not (-) with purmorphamine. (E) Each treated condition was normalized against its respective non treated condition, taking the non-induced value as 1 (dashed line). All graphs represent the Mean and SEM. P-values were calculated using two-tailed unpaired Student´s t-test or One-way ANOVA and Bonferroni post-hoc test.
Full western blot images for Figure 7.
Title indicates the Figure to which the western blot corresponds; magenta boxes show the region of the gel that was used to build the indicated figures. Molecular weight markers are shown to the right.
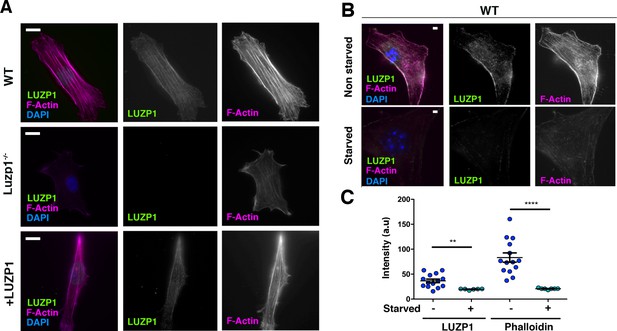
Cells missing Luzp1 show reduced F-actin levels.
(A) Immunofluorescence micrographs of WT, Luzp1-/- and +LUZP1 cells stained with an antibody against endogenous LUZP1 (green), phalloidin to detect F-actin (magenta), and counterstained with DAPI (blue). Single green and magenta channels are shown in black and white. Note the lack of LUZP1 signal in Luzp1-/- cells. Scale bar, 10 µm. (B) Immunofluorescence micrographs of non-starved and starved WT cells stained with antibodies against endogenous LUZP1 (green), phalloidin (magenta) and DAPI (blue). Single green and magenta channels are shown in black and white. Scale bar, 5 µm. Imaging was performed using widefield fluorescence microscopy (Zeiss Axioimager D1, 63x objective). (C) Graphical representation of the LUZP1 or F-actin mean intensity (arbitrary units) as shown in (B). Graphs represent Mean and SEM of three independent experiments pooled together. P-values were calculated using One-way ANOVA and Bonferroni post-hoc test.
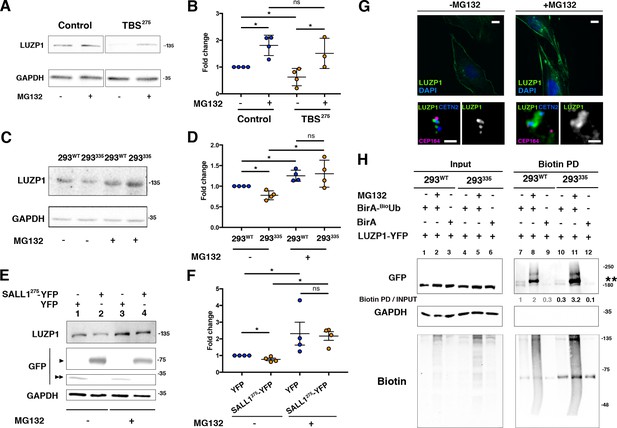
Truncated SALL1 leads to LUZP1 degradation through the UPS.
(A) Representative western blot of Control and TBS275 total cell lysates treated or not with MG132. A specific antibody detected endogenous LUZP1, and GAPDH was used as loading control. (B) Graphical representation of the fold changes of LUZP1/GAPDH ratios obtained in (A) for of Control (blue dots) and TBS275 (orange dots) treated (+) or not (-) with the proteasome inhibitor MG132. Note the increase of LUZP1 until reaching control levels in TBS275 cells upon MG132 treatment. (C) Representative western blot of 293WT and 293335 total cell lysates treated or not with MG132. A specific antibody against LUZP1 detected endogenous LUZP1, and GAPDH was used as loading control. (D) Graphical representation of the fold changes of LUZP1/GAPDH ratios obtained in panel C for 293WT (blue dots) and 293335 (orange dots) treated (+) or not (-) with MG132. Note that LUZP1 in 293335 reaches control levels with MG132 treatment. (E) Representative western blot of total lysates of HEK 293FT cells transfected with SALL1275-YFP (lanes 1 and 3) or YFP alone (lanes 2 and 4) treated (+) or not (-) with MG132. Specific antibodies against LUZP1, GFP and GAPDH were used. One black arrowhead indicates SALL1275-YFP, two back arrowheads YFP alone. (F) Graphical representation of the fold changes of LUZP1/GAPDH ratios obtained in (E) for HEK 293FT cells transfected with SALL1275-YFP (orange dots) or YFP alone (blue dots) treated (+) or not (-) with MG132. Note that LUZP1 increases in the presence of MG132 when SALL1275-YFP was transfected. Data from at least three independent experiments pooled together are shown. P-values were calculated using two-tailed unpaired Student´s t-test. (G) Immunofluorescence micrographs of RPE1 cells treated (+MG132) or not (-MG132) with proteasome inhibitor showing LUZP1 associated with the cytoskeleton (upper panels) or in the centrosome (lower panels). Antibodies against endogenous LUZP1 (green), Centrin-2 (CETN2, blue) and CEP164 (magenta) were used. DAPI labelled the nuclei (blue). Single green channels are shown in black and white. Note the overall increase of LUZP1 upon MG132 treatment. Scale bar 10 µm (cytoskeleton panels) or 0.5 µm (centrosome panels). Imaging was performed using widefield fluorescence microscopy (Zeiss Axioimager D1, 63x objective). (H) Western blot analysis of input and biotin pulldown (PD) of HEK 293FT cells transfected with CMV-LUZP1-YFP and BioUb or BirA alone treated (+) or not (-) with MG132. Specific antibodies (GFP, GAPDH, Biotin) were used as indicated. Numbers under GFP panel are the result of dividing each biotin PD band intensity by the respective input band intensity and normalize them to lane 1. Molecular weight markers in kDa are shown to the right. Two asterisks indicate monoubiquitinated LUZP1.
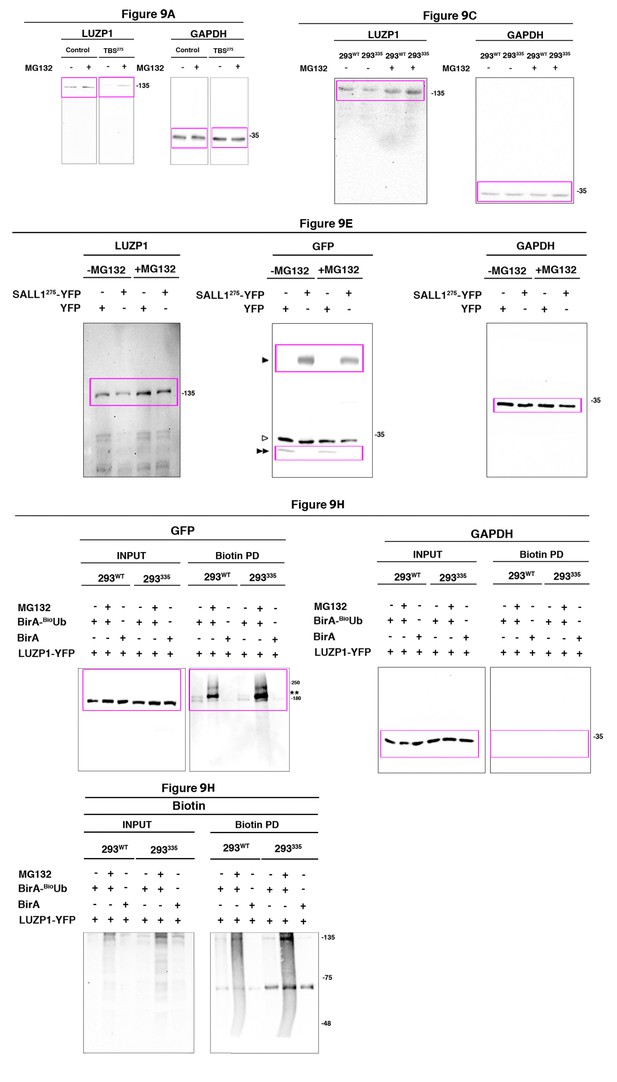
Full western blot images for Figure 9.
Titles indicate the Figure to which the western blot corresponds; magenta boxes show the region of the gel that was used to build the indicated figures. Ubiquitinated LUZP1 is indicated by two asterisks, LUZP1-YFP by two empty arrowheads and YFP alone by two black arrowheads. One empty arrowhead indicates truncated forms, bands from previous probing or unspecific bands. Molecular weight markers are shown to the right.
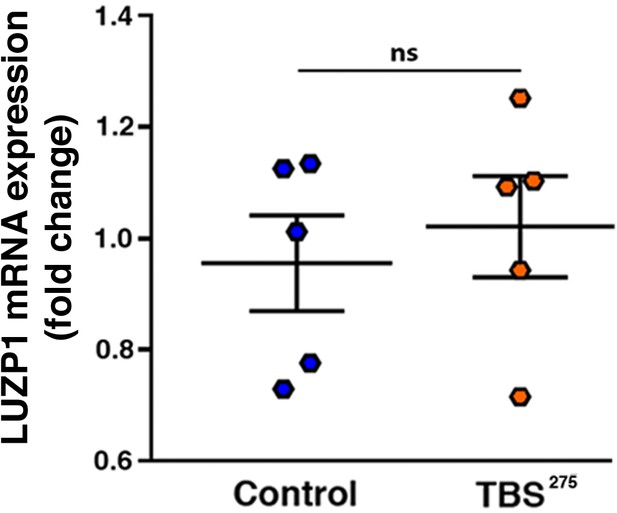
LUZP1 mRNA expression levels.
Quantification of LUZP1 expression in control (ESCTRL2) vs TBS275 cells by qRT-PCR. Graphs represent Mean and SEM from five independent experiments. P-values were calculated using the Mann Whitney test.
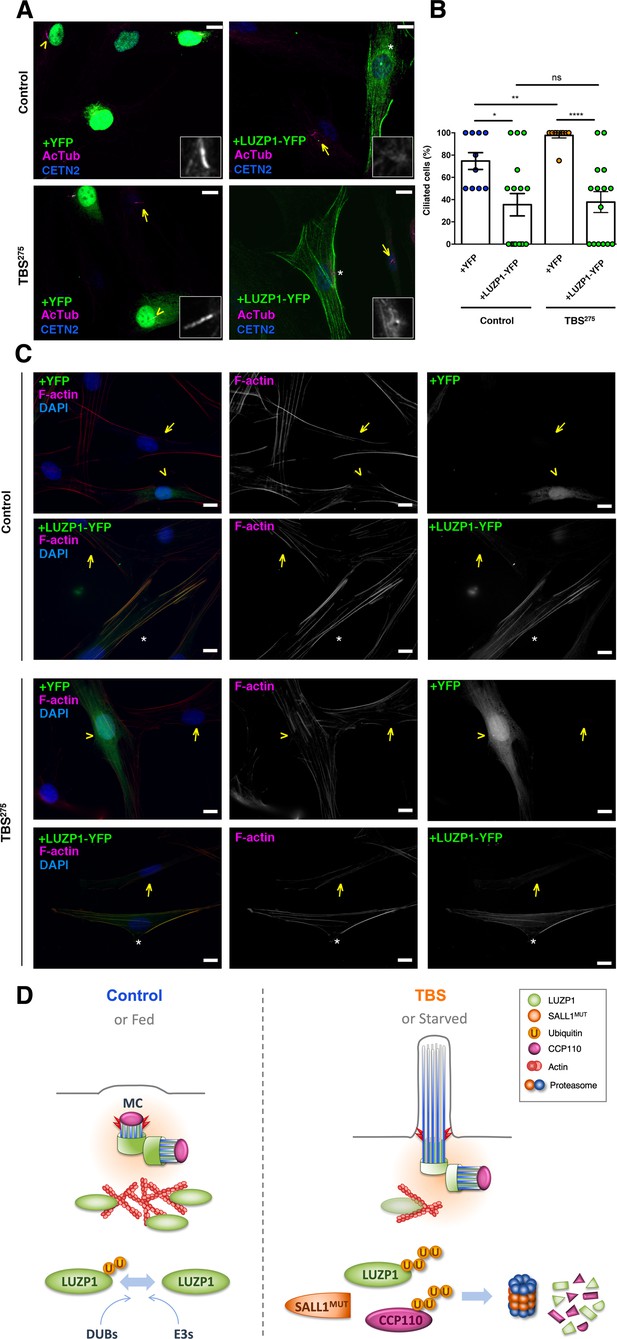
LUZP1 overexpression suppresses ciliogenesis and increases F-actin levels.
(A) Representative micrographs of ciliated Control and TBS275 cells transduced with YFP or LUZP1-YFP expressing lentivirus. Yellow arrowhead and white asterisk point at a magnified region shown in the lower right panel in black and white. Note the lack of cilia in cells transduced with LUZP1-YFP (white asterisk) compared to non-transduced cells (yellow arrow). AcTub: acetylated alpha-tubulin (magenta); CETN2: Centrin-2 (blue). (B) Graphical representation of the number of ciliated cells per micrograph in Control and TBS275 cells overexpressing YFP or LUZP1-YFP (n > 10 micrographs). Graphs represent Mean and SEM of ciliation frequencies per micrograph in three independent experiments pulled together. P-values were calculated using One-way ANOVA and Bonferroni post-hoc test or two-tailed unpaired Student´s t-test. (C) Representative micrographs of Control and TBS275 cells transduced with YFP (yellow arrowhead) or LUZP1-YFP (white asterisk) co-stained with phalloidin to label F-actin (magenta) and DAPI (blue). Note the increase in F-actin levels in cells transduced with LUZP1-YFP (white asterisk) compared to non-transfected cells (yellow arrow). Scale bar, 10 µm. Imaging was performed using widefield fluorescence microscopy (Zeiss Axioimager D1, 63x objective). (D) Schematic model representing how the presence of truncated SALL1 might cause cilia and actin malformations in TBS through LUZP1 interaction and UPS-mediated degradation. In control (or fed) cells (left), LUZP1 (in green) localizes to F-actin and to the proximal ends of the two centrioles, inhibiting cilia formation. LUZP1 proteostasis is maintained by the Ubiquitin Proteasome System (UPS). By contrast, in TBS (or starved) cells (right) the truncated form of SALL1 interacts with LUZP1 and promotes its UPS-mediated degradation. Likewise, others have shown ubiquitination and proteasome-mediated degradation of CCP110 at the mother centriole (MC) is permissive for ciliogenesis (Li et al., 2013). LUZP1 reduction at the centrosome and at the cytoskeleton favors the formation of the primary cilia.
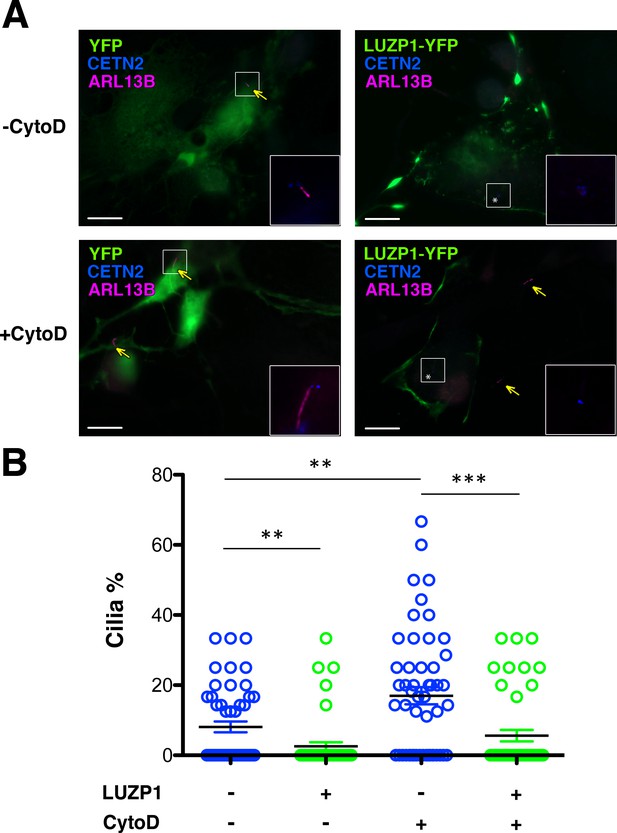
Exogenous LUZP1 expression suppresses the positive effects of CytoD on ciliogenesis.
(A) Representative micrographs of ciliated RPE1 cells transduced with YFP or LUZP1-YFP treated with DMSO (-) or 50 nM CytoD for 16 hr (+). Yellow arrowhead point to cilia. Note the lack of cilia in cells transduced with LUZP1-YFP (white asterisk) compared to non-transduced cells. ARL13B labelled the cilia (magenta) and Centrin-2 the centrosomes (CETN2, blue). Scale bars indicate 10 μm. (B) Graphical representation of the number of ciliated cells per micrograph in RPE1 cells transduced with YFP or LUZP1-YFP (n > 10 micrographs). Graphs represent Mean and SEM of ciliation frequencies per micrograph in three independent experiments pulled together. P-values were calculated using two-tailed unpaired Student´s t-test.
Additional files
-
Source data 1
Identification of LUZP1 interactors by proximity proteomics.
- https://cdn.elifesciences.org/articles/55957/elife-55957-data1-v2.xlsx
-
Source data 2
Values used for graphical representations and statistical analysis.
- https://cdn.elifesciences.org/articles/55957/elife-55957-data2-v2.xlsx
-
Supplementary file 1
Key Resources Table.
- https://cdn.elifesciences.org/articles/55957/elife-55957-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/55957/elife-55957-transrepform-v2.pdf