Caenorhabditis elegans processes sensory information to choose between freeloading and self-defense strategies
Figures
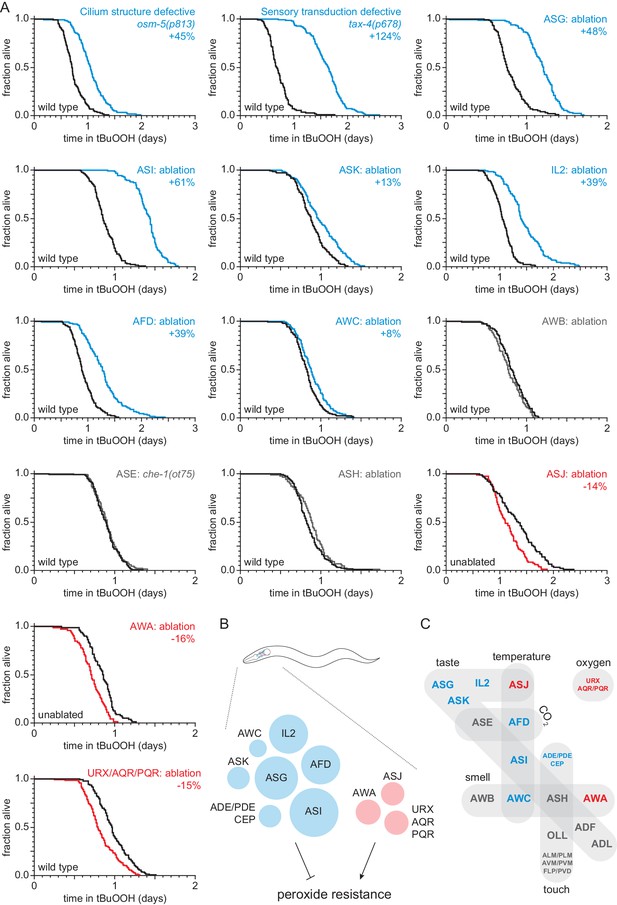
Sensory neurons regulate peroxide resistance in C. elegans.
(A) Peroxide resistance of nematodes with defects in sensory cilia and sensory transduction, or with genetic ablation of specific sensory neurons. The fraction of nematodes remaining alive in the presence of 6 mM tert-butyl hydroperoxide (tBuOOH) is plotted against time. Interventions that increased, decreased, or did not affect survival are denoted in blue, red, and gray, respectively, and their effects on mean peroxide resistance are noted. (B) Specific sensory neurons normally reduce (blue) or increase (red) peroxide resistance. Circle area denotes the effect of ablation of the respective neurons on mean peroxide resistance. (C) Sensory neurons are grouped by the stimuli they sense. Neurons that normally reduce (seven classes) or increase (three classes) peroxide resistance are shown in blue and red, respectively. See also Figure 1—figure supplement 1. Statistical analyses are in Supplementary file 1.
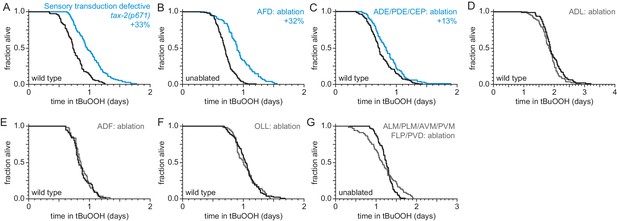
Sensory neurons regulate peroxide resistance in C. elegans.
(A–G) Peroxide resistance of nematodes with defects in sensory transduction, or with genetic ablation of specific sensory neurons. The fraction of nematodes remaining alive in the presence of 6 mM tert-butyl hydroperoxide (tBuOOH) is plotted against time. Interventions that increased or did not affect survival are denoted in blue and gray, respectively, and their effect on mean peroxide resistance is noted. Statistical analyses are in Supplementary file 1.
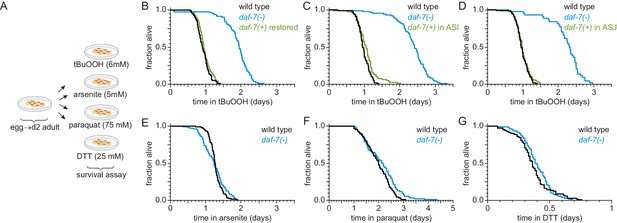
ASI sensory neurons secrete DAF-7/TGFβ to specifically lower the nematode’s peroxide resistance.
(A) Diagram summarizing experimental strategy. (B–D) Peroxide resistance of wild type, daf-7(ok3125), and daf-7(ok3125) with daf-7(+) reintroduced with (B) its endogenous promoter, (C) the ASI-specific str-3 promoter, or (D) the ASJ-specific trx-1 promoter. (E–G) Resistance to 5 mM arsenite, 75 mM paraquat, and 25 mM dithiothreitol (DTT) of wild type and daf-7(ok3125). See also Figure 2—figure supplement 1. Statistical analyses are in Supplementary file 2.
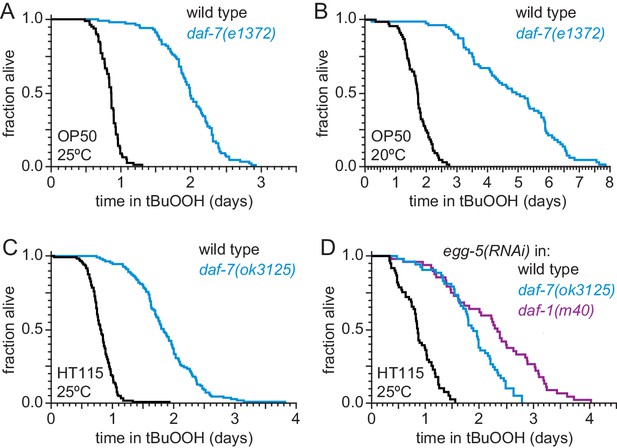
DAF-7 regulates peroxide resistance robustly.
(A–B) The daf-7(e1372) loss-of-function mutation increased peroxide resistance at (A) 25°C, and (B) 20°C. (C) daf-7(ok3125) increased peroxide resistance when C. elegans was fed E. coli HT115(DE3) during development, adulthood, and the survival assay. (D) daf-7(ok3125) and daf-1(m40) increase peroxide resistance when we inhibited formation of the eggshell of fertilized C. elegans embryos with RNAi of egg-5 and did not expose nematodes to FUDR at any point. Statistical analyses are in Supplementary file 2.
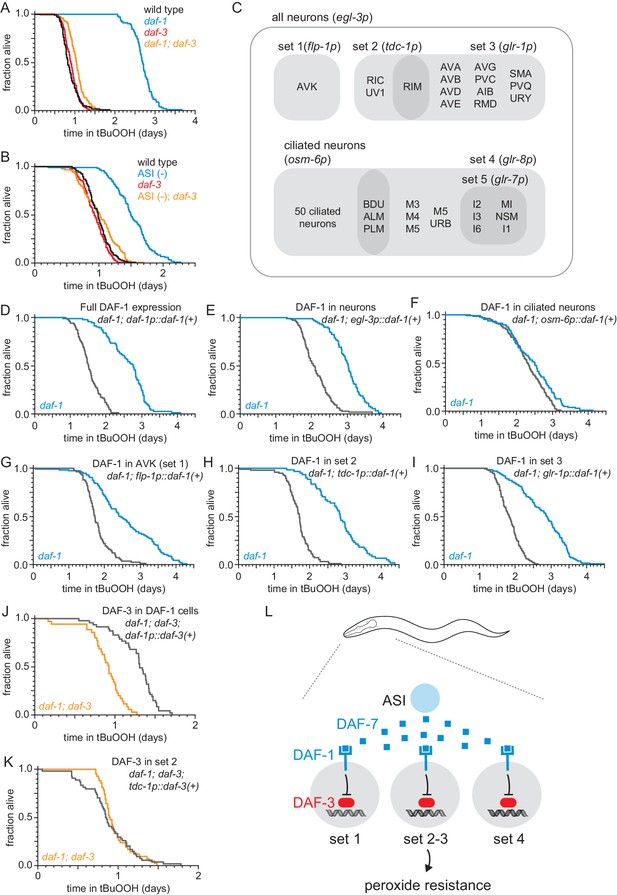
The DAF-1/TGFβ receptor functions redundantly in interneurons to regulate peroxide resistance in response to DAF-7/TGFβ from ASI.
(A–B) daf-3(mgDf90) almost completely abrogated the increased peroxide resistance of (A) daf-1(m40) and of (B) genetic ablation of ASI. (C) Diagram of the subsets of neurons where daf-1(+) or daf-3(+) were expressed in transgenic rescue experiments shown in panels (D–K) and in Figure 3—figure supplement 1. The promoter elements used to drive gene expression in those subsets of neurons are shown in parentheses. The tdc-1 promoter also drives gene expression in the sheath cells of the somatic gonad. (D–I) Peroxide resistance of transgenic nematodes expressing daf-1(+) in specific subsets of cells and daf-1(m40) controls. (J–K) Peroxide resistance of transgenic nematodes expressing daf-3(+) in specific subsets of cells and daf-1(m40); daf-3(mgDf90) controls. (L) ASI signals to three sets of interneurons to lower the nematode’s peroxide resistance. To increase peroxide resistance, all of these sets of neurons must independently activate the DAF-3/DAF-5 transcriptional complex. See also Figure 3—figure supplement 1. Statistical analyses are in Supplementary file 3.
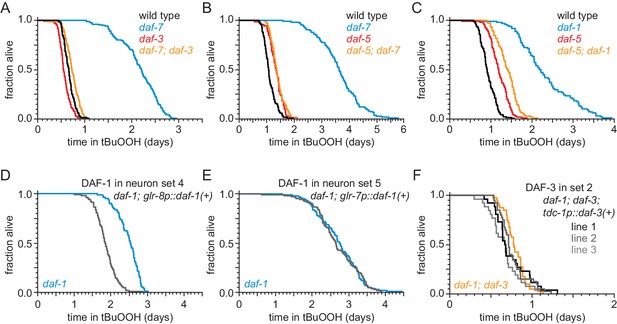
The DAF-1/TGFβ receptor functions redundantly in interneurons to regulate peroxide resistance in response to DAF-7/TGFβ from ASI.
(A) daf-3(e1376) almost completely abrogated the increased peroxide resistance of daf-7(ok3125). (B) daf-5(e1386) abrogated the increased peroxide resistance of daf-7(e1372). (C) daf-5(e1386) abrogated part of the increased peroxide resistance of daf-1(m40). (D–E) Peroxide resistance of transgenic nematodes expressing daf-1(+) in specific subsets of cells and daf-1(m40) controls. (F) Peroxide resistance of independent transgenic lines expressing daf-3(+) in tdc-1-expressing cells and daf-1(m40); daf-3(mgDf90) controls. Statistical analyses are in Supplementary file 3.
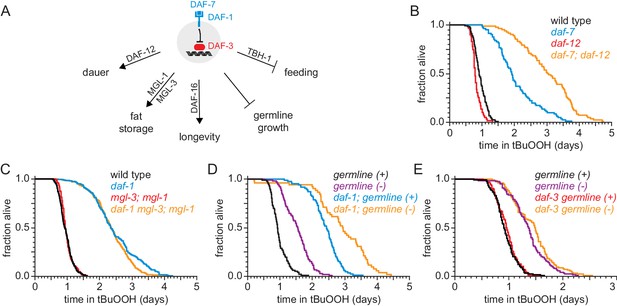
DAF-1/TGFβ-receptor signaling regulates peroxide resistance separately from its role in dauer formation, fat storage, and germline growth.
(A) Different mechanisms operate downstream of DAF-3 to mediate the effects of DAF-7/TGFβ signaling on dauer-larva formation, fat storage, germline size, lifespan, and feeding. (B) daf-12(rh61rh411) did not suppress the increased peroxide resistance of daf-7(e1372). (C) mgl-1(tm1811) and mgl-3(tm1766) did not jointly suppress the increased peroxide resistance of daf-1(m40). (D) Genetic ablation of the germline and daf-1(m40) independently increased peroxide resistance. (E) daf-3(mgDf90) did not suppress the increased peroxide resistance of genetic ablation of the germline. See also Figure 4—figure supplement 1. Statistical analyses are in Supplementary file 4.
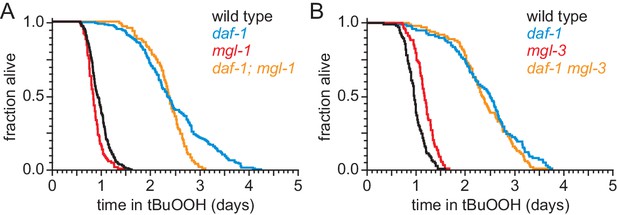
DAF-1/TGFβ-receptor signaling regulates peroxide resistance separately from its role in fat storage.
(A–B) The increased peroxide resistance of daf-1(m40) was not suppressed by (A) mgl-1(tm1811), or (B) mgl-3(tm1766). Statistical analyses are in Supplementary file 4.
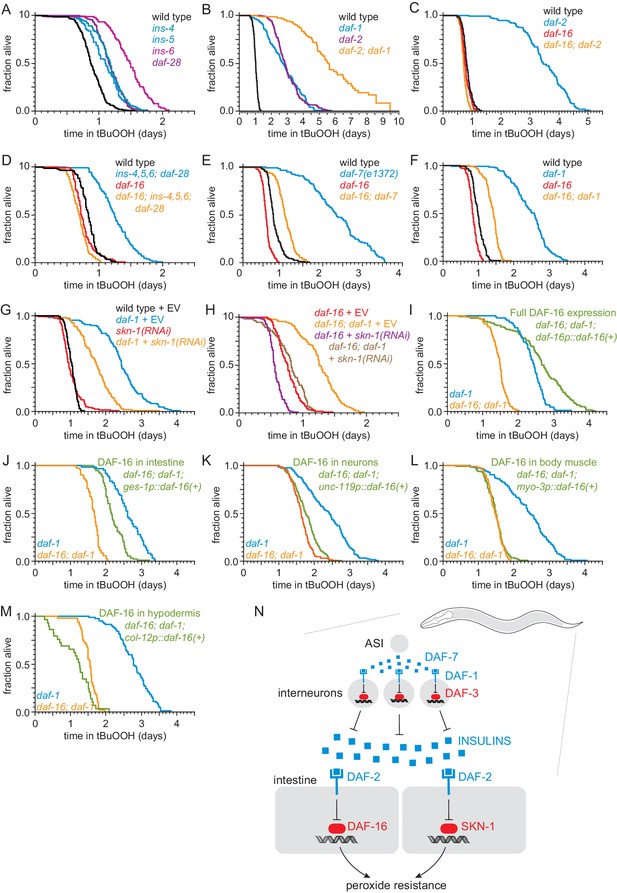
ASI regulates the nematode’s peroxide resistance via a TGFβ-Insulin/IGF1 hormone relay.
(A) Deletions in ins-4, ins-5, ins-6, and daf-28 insulin-coding genes increased peroxide resistance. (B) daf-2(e1370) and daf-1(m40) independently increased peroxide resistance. (C–D) daf-16(mu86) abrogated the increased peroxide resistance of (C) daf-2(e1370) and (D) an ins-4 ins-5 ins-6; daf-28 quadruple mutant. (E–F) daf-16(mu86) suppressed part of the increased peroxide resistance of (E) daf-7(e1372) and (F) daf-1(m40). (G) skn-1(RNAi) suppressed part of the increased peroxide resistance of daf-1(m40). Control RNAi consisted of feeding the nematodes the same bacteria but with the empty vector (EV) plasmid pL4440 instead of a plasmid targeting skn-1. (H) skn-1(RNAi) lowered the peroxide resistance of daf-16(mu86); daf-1(m40). (I–M) Peroxide resistance of transgenic nematodes expressing daf-16(+) in specific subsets of cells, daf-16(mu86); daf-1(m40) controls, and daf-1(m40) reference. (N) ASI sensory neurons make nematodes more sensitive to hydrogen peroxide via a multistep hormonal relay. DAF-7/TGFβ from ASI is received by interneurons. These interneurons act redundantly to relay this signal to target tissues by promoting transcription of insulin genes. These insulins activate the DAF-2 insulin/IGF1 receptor, leading to inhibition of DAF-16-dependent peroxide protection services by the intestine and neurons. SKN-1 acts independently of DAF-16 to promote peroxide resistance in response to reduced DAF-1 signaling. SKN-1 likely acts in the intestine, because skn-1(+) promotes peroxide resistance in daf-2 mutants and induces oxidative-stress defenses in this tissue (An et al., 2005; Tullet et al., 2008). See also Figure 5—figure supplement 1. Statistical analyses are in Supplementary file 5.
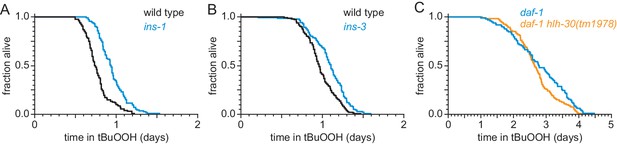
ASI regulates the nematode’s peroxide resistance via a TGFβ-Insulin/IGF1 hormone relay.
(A–B) Peroxide resistance was increased by deletions in (A) ins-1, and (B) ins-3 insulin-coding genes. (C) hlh-30(tm1978) did not affect the increased peroxide resistance of daf-1(m40). Statistical analyses are in Supplementary file 5.
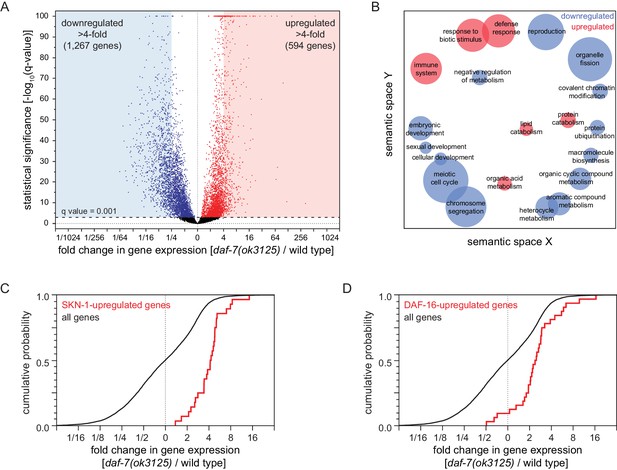
Reduced DAF-7/TGFβ signaling upregulates expression of DAF-16/FOXO and SKN-1/NRF target genes.
(A) Volcano plot showing the level and statistical significance of changes in gene expression induced by the daf-7(ok3125) null mutation. Genes up- and down-regulated significantly (q value < 0.001) are shown in red and blue, respectively. (B) Gene Ontology (GO) term enrichment analysis of biological processes associated with the set of 594 upregulated genes (blue bubbles) and the set of 1267 downregulated genes (red bubbles) with a statistically significant and greater than four-fold change in expression in daf-7(ok3125) mutants relative to wild-type animals. Bubble size is proportional to the statistical significance [-log10(P value)] of enrichment. (C) The daf-7(ok3125) mutation increased the expression of genes upregulated by skn-1(+) in wild type animals (Oliveira et al., 2009). (D) The daf-7(ok3125) mutation increased the expression of genes directly upregulated by DAF-16 (Kumar et al., 2015). See also Figure 6—figure supplement 1. Additional statistical analyses are in Supplementary file 6.
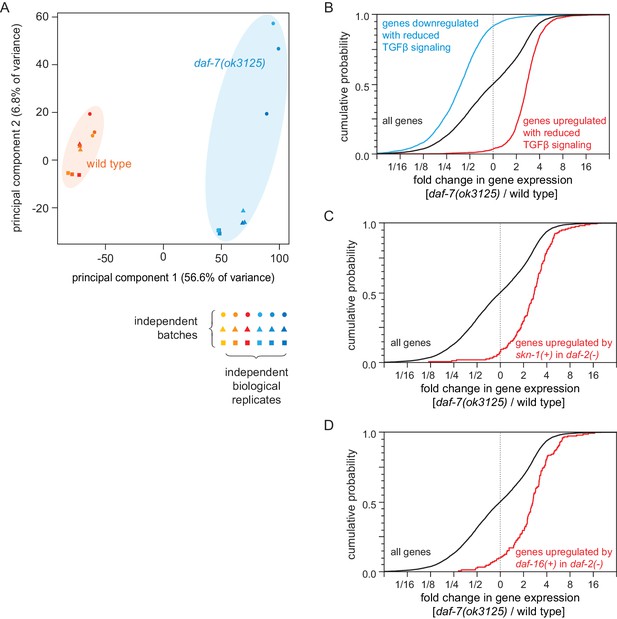
Reduced DAF-7/TGFβ signaling upregulates expression of DAF-16/FOXO and SKN-1/NRF target genes.
(A) Principal component analysis (PCA) of the sequenced samples of daf-7(ok3125) mutants and wild-type animals. (B) daf-7(ok3125) increased the expression of genes upregulated (red) in partial loss-of-function TGFβ signaling pathway mutants (Shaw et al., 2007) and decreased the expression of genes downregulated by those TGFβ signaling pathway mutants (blue) (Shaw et al., 2007). (C) daf-7(ok3125) increased the expression of genes upregulated (red) by skn-1(+) in daf-2(-) mutants (Ewald et al., 2015). (D) daf-7(ok3125) increased the expression of genes upregulated (red) by daf-16(+) in daf-2(-) mutants (Murphy et al., 2003). Statistical analyses are in Supplementary file 6.
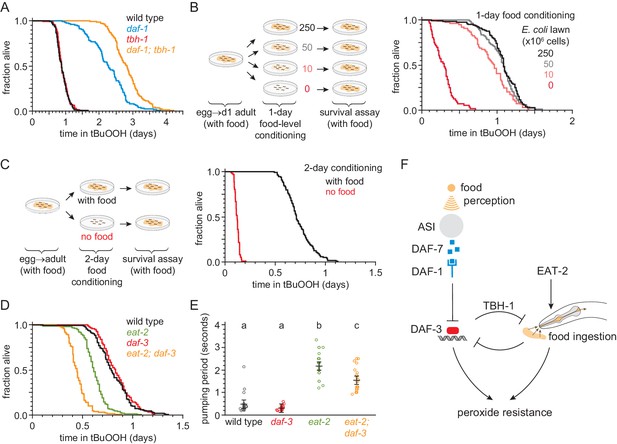
Food ingestion regulates the nematode’s peroxide resistance via DAF-3/coSMAD.
(A) tbh-1(ok1196) increased the peroxide resistance of daf-1(m40). (B–C) The E. coli level before the assay affected C. elegans peroxide resistance in a dose-dependent manner. (D) eat-2(ad1116) caused a more severe reduction in peroxide resistance in daf-3(mgDf90) than in wild type. (E) eat-2(ad1116) caused a less severe reduction in feeding in daf-3(mgDf90) than in wild type. Lines mark the mean pumping period and its 95% confidence interval. Genotypes labeled with different letters exhibited significant differences in pumping period (p < 0.0001, Turkey HSD test) otherwise (p > 0.05). (F) DAF-3 and feeding increase peroxide resistance but attenuate each other’s effects. Feeding inhibits DAF-3; this attenuates the reduction in peroxide resistance caused by reduced feeding. DAF-3 inhibits feeding via TBH-1; this attenuates the increase in peroxide resistance of daf-1 mutants. Sensory perception of E. coli induces DAF-7 expression (Chang et al., 2006; Gallagher et al., 2013) in a concentration-dependent manner (Entchev et al., 2015; Ren et al., 1996) leading to DAF-3 repression by the DAF-7 receptor DAF-1. Therefore, both ingestion and perception of E. coli inhibit DAF-3. See also Figure 7—figure supplement 1. Additional statistical analyses are in Supplementary file 7.
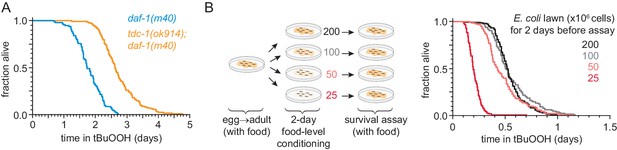
DAF-3/coSMAD increases the nematode’s peroxide resistance in response to reduced feeding.
(A) tdc-1(ok914) increased the peroxide resistance of daf-1(m40). (B) The E. coli level for two days before the assay affected C. elegans peroxide resistance in a dose-dependent manner. Statistical analyses are in Supplementary file 7.
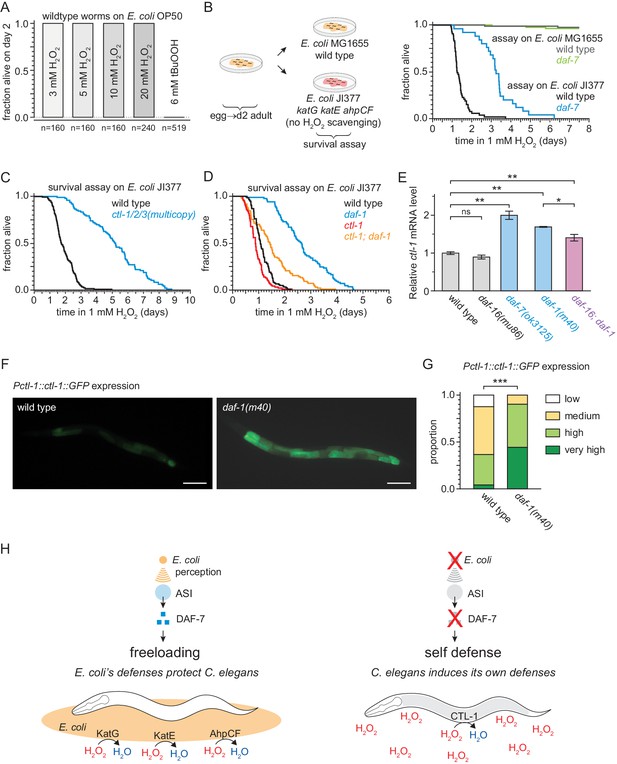
DAF-7/TGFβ signals that hydrogen-peroxide protection will be provided by catalases from E. coli and not by catalases from C. elegans.
(A) C. elegans was sensitive to killing by tert-butyl hydroperoxide (tBuOOH), but not by hydrogen peroxide, in the presence of E. coli OP50. (B) Hydrogen peroxide resistance of wild type and daf-7(ok3125) C. elegans in assays with wild type and Kat – Ahp – E. coli. (C) Overexpression of the three endogenous catalases protects nematodes from hydrogen peroxide in assays with Kat – Ahp – E. coli. (D) The cytosolic catalase ctl-1(ok1242) mutation suppressed part of the increased hydrogen peroxide resistance of daf-1(m40) in assays with Kat – Ahp – E. coli. (E) The DAF-7/TGFβ-pathway regulates ctl-1 mRNA expression via DAF-16/FOXO, determined by quantitative RT-PCR. Data are represented as mean ± s.e.m of three independent biological replicates, each with three technical replicates. For comparisons of ctl-1 mRNA expression between pairs of genotypes, ** indicates p < 0.001, * indicates p < 0.05, and ‘ns’ indicates p > 0.05 (Turkey HSD test). (F) Representative pictures of the expression of the chIs166[Pctl-1::ctl-1::gfp] reporter in wild type animals (left picture; category: medium) or daf-1(m40) mutants (right picture; category: very high). Scale bar = 100 µm. (G) The expression of the promoter of ctl-1 fused with GFP (chIs166[Pctl-1::ctl-1::gfp]) is higher in daf-1(m40) mutants (237 animals) than in wild type animals (145 animals), *** indicates p < 0.0001 (ordinal logistic regression). Scoring is described in Materials and methods. See Figure 8—figure supplement 1D for representative pictures of each expression category. (H) DAF-7/TGFβ signaling enables C. elegans to decide whether to induce its own hydrogen-peroxide defenses or, instead, freeload on protection provided by molecularly orthologous hydrogen-peroxide defenses from E. coli. See also Figure 8—figure supplement 1. Additional statistical analyses are in Supplementary file 8.
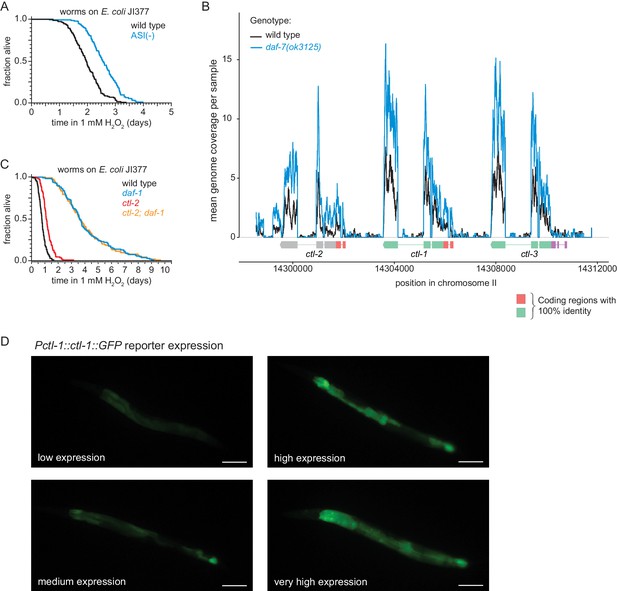
DAF-7/TGFβ signals that hydrogen-peroxide protection will be provided by catalases from E. coli and not by catalases from C. elegans.
(A) Genetic ablation of ASI sensory neurons increased peroxide resistance. (B) Gene expression levels within the genomic region encoding the three C. elegans catalase genes. Because these genes contain multiple regions with perfect identity, we determined the relative genome coverage of reads (including multimaps) between daf-7(ok3125) mutants and wild-type animals using BEDTools (Quinlan and Hall, 2010) and plotted the mean read count across six samples normalized using scran (Lun et al., 2016). Gene models show the positions and splicing pattern of each catalase gene, regions with 100% nucleotide identity (orange and green), and unique regions (grey and purple). (C) The peroxisomal catalase ctl-2(ok1137) did not suppresses the increased peroxide resistance of daf-1(m40). (D) Representative pictures of the low, medium, high and very high expression categories of the chIs166[Pctl-1::ctl-1::gfp] reporter. Scale bar = 100 µm. Statistical analyses are in Supplementary file 8.
Additional files
-
Supplementary file 1
Statistical analysis for Figure 1 and Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp1-v2.docx
-
Supplementary file 2
Statistical analysis for Figure 2 and Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp2-v2.docx
-
Supplementary file 3
Statistical analysis for Figure 3 and Figure 3—figure supplement 1.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp3-v2.docx
-
Supplementary file 4
Statistical analysis for Figure 4 and Figure 4—figure supplement 1.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp4-v2.docx
-
Supplementary file 5
Statistical analysis for Figure 5 and Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp5-v2.docx
-
Supplementary file 6
Statistical analysis for Figure 6 and Figure 6—figure supplement 1.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp6-v2.docx
-
Supplementary file 7
Statistical analysis for Figure 7 and Figure 7—figure supplement 1.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp7-v2.docx
-
Supplementary file 8
Statistical analysis for Figure 8 and Figure 8—figure supplement 1.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp8-v2.docx
-
Supplementary file 9
Bacterial strains.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp9-v2.docx
-
Supplementary file 10
C. elegans strains.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp10-v2.docx
-
Supplementary file 11
PCR genotyping primers and enzymes, and phenotypes used for strain construction.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp11-v2.docx
-
Supplementary file 12
Key resources table.
- https://cdn.elifesciences.org/articles/56186/elife-56186-supp12-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56186/elife-56186-transrepform-v2.docx





