Sequential activation of transcriptional repressors promotes progenitor commitment by silencing stem cell identity genes
Figures
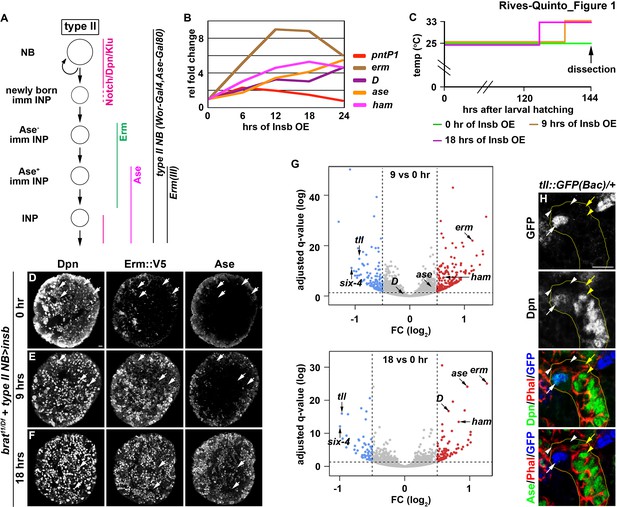
Identification of candidate regulators of type II neuroblast functional identity.
(A) A diagram of the type II neuroblast lineage showing the expression patterns of genes and Gal4 drivers used throughout this study. (B) Gene transcription profiles of brat-null brains transiently overexpressing Insb driven by a type II neuroblast Gal4. Supernumerary type II neuroblasts in brat-null brains transiently overexpressing Insb displayed patterns of gene transcription that are indicative of immature INPs undergoing INP commitment in wild-type brains. (C) A strategy to induce synchronized INP commitment in supernumerary type II neuroblasts in brat-null brains. Larvae carrying a UAS-insb transgene and a type II neuroblast Gal4 were collected and aged at 25°C. One third of larvae were shifted to 33°C at 126 or 135 hr after hatching to induce high levels of transient Insb expression, and the last one-third remained at 25°C serving as the source enriched for type II neuroblast-specific transcripts (time 0). (D–F) Images of brat-null brains transiently overexpressing Insb driven by a type II neuroblast Gal4 for 0, 9, or 18 hr. Transient overexpression of Insb first induced Erm and then Ase expression in supernumerary type II neuroblasts in brat-null brains. (G) Volcano plots showing fold-change of gene expression in brat-null brains transiently overexpressing Insb for 9 or 18 hr. (H) Tll expression pattern in the type II neuroblast lineage. The tll::GFP(Bac) transgene revealed the expression of endogenous Tll in type II neuroblasts but not in immature INPs and INPs. The following labeling applies to all images in this figure: yellow dashed line encircles a type II neuroblast lineage; white arrow: type II neuroblast; white arrowhead: Ase- immature INP; yellow arrow: Ase+ immature INP; yellow arrowhead: INP. Scale bar, 10 μm.
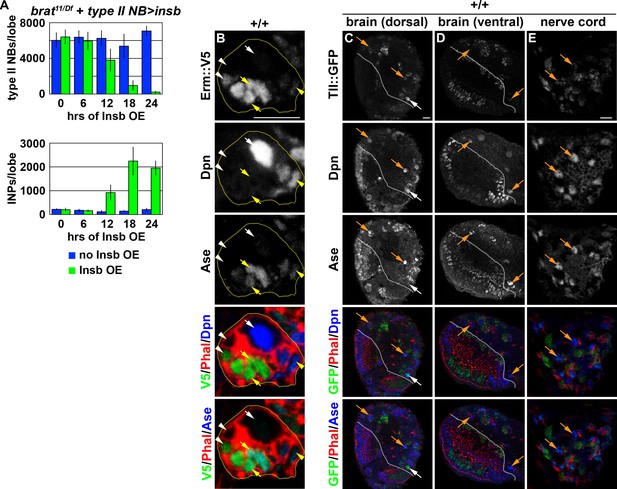
Time-course analysis of transient Insb overexpression in brat-null brains.
(A) Quantification of total type II neuroblasts (top) or INPs (bottom) per brat-null brain lobe that transiently overexpressed Insb driven by a type II neuroblast Gal4. Insb overexpression led to progressive decrease in supernumerary type II neuroblasts but increase in INPs in brat-null brains. (B) Images of Erm::V5 expression in a type II neuroblast lineage. Erm::V5 is strongly expressed in immature INPs and weakly in INPs. (C–E) Images of Tll::GFP expression in the larval central nervous system. Tll::GFP is expressed at high levels in type II neuroblasts, and much lower levels in type I neuroblasts in the ventral brain region and in the ventral nerve cord. The following labeling is applicable to all panels of images in this figure: white dashed lines separates the optic lobe from the brain; yellow dashed line encircles a type II neuroblast lineage; white arrow: type II neuroblast; white arrowhead: Ase- immature INP; yellow arrow: Ase+ immature INP; yellow arrowhead: INP; orange arrow: type I neuroblast. Bar graphs are represented as mean ± standard deviation.
-
Figure 1—figure supplement 1—source data 1
Quantification of total type II neuroblasts or INPs per brat-null brain lobe that transiently overexpressed Insb.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig1-figsupp1-data1-v2.xlsx
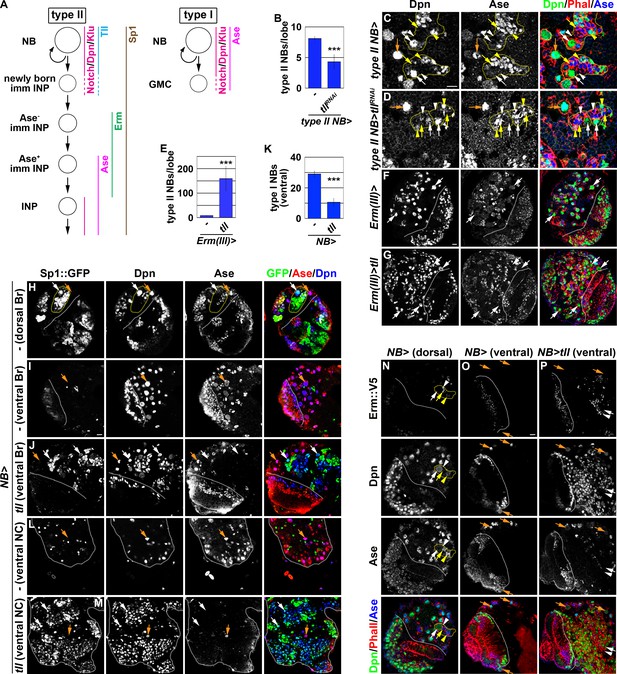
tll is necessary and sufficient for type II neuroblast functional identity.
(A) A diagram showing the expression patterns of genes in the type I and II neuroblast lineages. (B) Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-tllRNAi transgene driven by a type II neuroblast Gal4. Knocking-down tll function reduced the number of type II neuroblasts from 8 to 4 per brain lobe. (C–D) Images of brains that overexpressed a UAS-tllRNAi transgene driven by a type II neuroblast Gal4. Knocking-down tll function led to premature differentiation in type II neuroblast as indicated by reduced cell diameter and ectopic Ase expression but did not affect the maintenance of type I neuroblasts. (E) Quantification of total type II neuroblasts per brain that overexpressed a UAS-tll transgene driven by an INP Gal4. tll overexpression in INPs led to supernumerary type II neuroblast formation. (F–G) Images of brains that overexpressed a UAS-tll transgene driven by an INP Gal4. (H–I) Images of Sp1::GFP(Bac) brains. Sp1::GFP is detected in most cells in all type II neuroblast lineages that are exclusively located in the dorsal brain region, and is detected in few neurons in type I neuroblast lineages in the ventral brain region. (J) Images of the ventral region of Sp1::GFP(Bac) brains that overexpressed a UAS-tll transgene driven by a pan-neuroblast Gal4 (Wor-Gal4). Tll overexpression transforms type I neuroblasts (Ase+Sp1::GFP-) in the ventral brain region into type II neuroblasts (Ase-Sp1::GFP+). (K) Quantification of total ventral type I neuroblasts per brain lobe that overexpressed a UAS-tll transgene driven by a pan-neuroblast Gal4. Tll overexpression transforms 66% of type I neuroblasts in the ventral brain region into type II neuroblasts. (L–M) Images of the thoracic segments on the ventral nerve cord of Sp1::GFP(Bac) larvae that overexpressed a UAS-tll transgene driven by a pan-neuroblast Gal4 (Wor-Gal4). Tll overexpression transforms type I neuroblasts in the thoracic segments into type II neuroblasts. (N–O) Images of dorsal and ventral regions of erm::V5 brains. Erm::V5 is detected in immature INPs in type II neuroblast lineages but is undetectable in type I neuroblast lineages in the ventral region of larval brain. (P) Images of erm::V5 brains that overexpressed a UAS-tll transgene driven by a pan-neuroblast Gal4. Tll overexpression induced type I neuroblasts in the ventral-medial region of the brain to generate supernumerary type II neuroblasts interspersed with Erm::V5+ immature INPs. The following labeling applies to all images in this figure: white dashed line separates the optic lobe from the brain; yellow dashed line encircles a type II neuroblast lineage; white arrow: type II neuroblast; white arrowhead: Ase- immature INP; yellow arrow: Ase+ immature INP; yellow arrowhead: INP; orange arrow: type I neuroblast. Br: brain. NC: nerve cord. Scale bar, 10 μm. Bar graphs are represented as mean ± standard deviation. p-values: ***<0.005.
-
Figure 2—source data 1
Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-tllRNAi transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Quantification of total type II neuroblasts per brain that overexpressed a UAS-tll transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig2-data2-v2.xlsx
-
Figure 2—source data 3
Quantification of total ventral type I neuroblasts per brain lobe that overexpressed a UAS-tll transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig2-data3-v2.xlsx
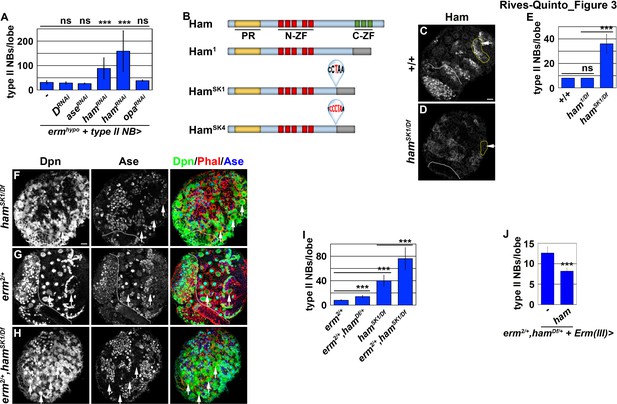
Ham is a novel regulator of INP commitment.
(A) Quantification of total type II neuroblasts per ermhypo brain lobe that overexpressed a UAS-RNAi transgene driven by a type II neuroblast Gal4. Knocking-down ham function consistently enhanced the supernumerary type II neuroblast phenotype in ermhypo brains. (B) A diagram summarizing the lesions in ham alleles. The molecular lesions in hamSk1 and hamSK4 alleles were not independently verified. (C–D) Ham expression in wild-type or hamSK1 homozygous brains. Ham was detected in immature INPs and INPs in wild-type brains, but was undetectable in hamSK1 homozygous brains. (E) Quantification of total type II neuroblasts per ham-mutant brain lobe. hamSK1 homozygous but not ham1 homozygous brains displayed a supernumerary type II neuroblast phenotype. (F–H) Images of ham single mutant or ham,erm double mutant brains. The heterozygosity of erm alone had no effect on type II neuroblasts, but enhanced the supernumerary type II neuroblast phenotype in hamSK1 homozygous brains. (I) Quantification of total type II neuroblasts per brain lobe of the indicated genotypes. erm and ham function synergistically to prevent supernumerary type II neuroblast formation. (J) Quantification of total type II neuroblasts per erm,ham double heterozygous brain lobe that overexpressed a UAS-ham transgene driven by an INP Gal4. Overexpressing Ham in INPs rescued the supernumerary type II neuroblast phenotype in erm,ham double heterozygous brains. The following labeling applies to all images in this figure: white dashed line separates the optic lobe from the brain; yellow dashed line encircles a type II neuroblast lineage; white arrow: type II neuroblast. Scale bar, 10 μm. Bar graphs are represented as mean ± standard deviation. p-values: ***<0.005. ns: not significant.
-
Figure 3—source data 1
Quantification of total type II neuroblasts per ermhypo brain lobe that overexpressed a UAS-RNAi transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Quantification of total type II neuroblasts per ham-mutant brain lobe.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Quantification of total type II neuroblasts per brain lobe of the indicated genotypes.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig3-data3-v2.xlsx
-
Figure 3—source data 4
Quantification of total type II neuroblasts per erm,ham double heterozygous brain lobe that overexpressed a UAS-ham transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig3-data4-v2.xlsx
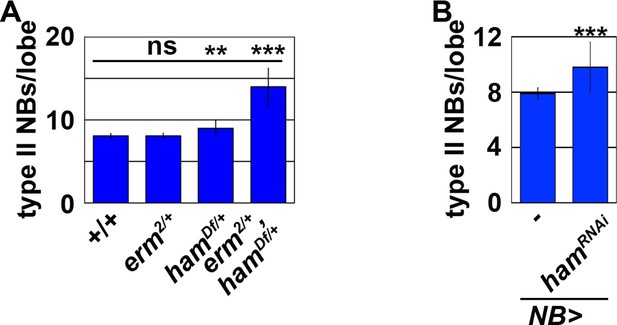
Ham functions synergistically with Erm to suppress supernumerary type II neuroblast formation.
(A) Quantification of total type II neuroblasts per brain lobe of the indicated genotypes. erm,ham double heterozygous brains displayed the supernumerary type II neuroblast phenotype. (B) Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-hamRNAi transgene driven by a pan-neuroblast Gal4. Knock-down of ham function in type II neuroblasts led to a mild supernumerary neuroblast phenotype. Bar graphs are represented as mean ± standard deviation. p-values: **<0.05, ***<0.005. ns: not significant.
-
Figure 3—figure supplement 1—source data 1
Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-hamRNAi transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig3-figsupp1-data1-v2.xlsx
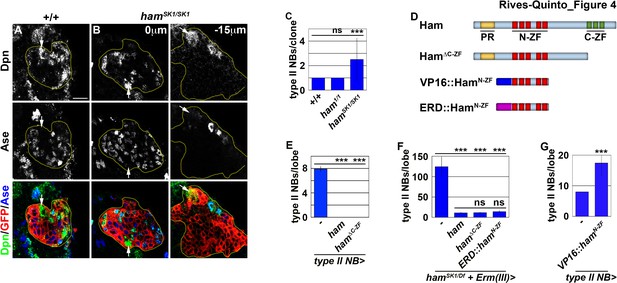
Ham suppresses INP reversion by repressing gene transcription.
(A–B) Images of wild-type or hamSK1 homozygous type II neuroblast mosaic clones. Supernumerary neuroblasts (−15 μm) in hamSK1 homozygous clones were always located far from the parental neuroblast (0 μm), and were surrounded by Ase+ cells. (C) Quantification of total neuroblasts per ham1 or hamSK1 homozygous type II neuroblast clone. hamSK1 homozygous clones contained supernumerary neuroblasts but ham1 homozygous clones did not. (D) A diagram summarizing UAS-ham transgenes used in this study. (E) Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-ham transgene driven by a type II neuroblast Gal4. Overexpressing full-length Ham or HamΔC-ZF led to premature differentiation in type II neuroblasts. (F) Quantification of total type II neuroblasts per hamSK1 homozygous brain lobe that overexpressed various UAS-ham transgenes driven by an INP Gal4. Overexpressed full-length Ham, HamΔC-ZF or ERD::HamN-ZF rescued the supernumerary neuroblast phenotype in hamSK1 homozygous brains. (G) Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-VP16::hamN-ZF transgene driven by a type II neuroblast Gal4. Overexpressing VP16::HamN-ZF led to supernumerary type II neuroblast formation. The following labeling applies to all images in this figure: yellow dashed line encircles a type II neuroblast lineage; white arrow: type II neuroblast. Scale bar, 10 μm. Bar graphs are represented as mean ± standard deviation. Ppvalues: ***<0.005. ns: not significant.
-
Figure 4—source data 1
Quantification of total neuroblasts per ham1 or hamSK1 homozygous type II neuroblast clone.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-ham transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig4-data2-v2.xlsx
-
Figure 4—source data 3
Quantification of total type II neuroblasts per hamSK1 homozygous brain lobe that overexpressed various UAS-ham transgenes.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig4-data3-v2.xlsx
-
Figure 4—source data 4
Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-VP16::hamN-ZF transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig4-data4-v2.xlsx
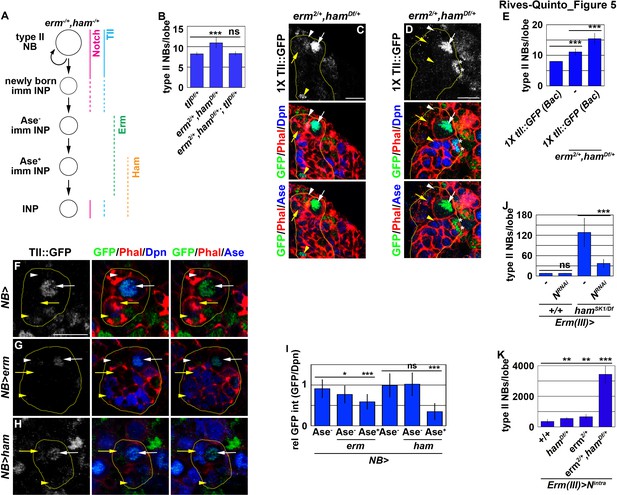
Erm- and Ham-mediated repression renders tll refractory for activation by Notch.
(A) A diagram depicting our hypothesis that ectopic activation of tll in INPs leads to supernumerary type II neuroblasts in erm,ham double heterozygous brains. (B) Quantification of total type II neuroblasts per brain lobe that was erm,ham double heterozygous or erm,ham,tll triple heterozygous. Heterozygosity of tll suppressed the supernumerary neuroblast phenotype in erm,ham double heterozygous brains. (C–D) Images of erm,ham double heterozygous brains that carries a tll::GFP(BAC) transgene. Tll::GFP becomes ectopically expressed in INPs and supernumerary type II neuroblasts (*) in erm,ham double heterozygous brains. (E) Quantification of total type II neuroblasts erm,ham double heterozygous brain lobe that carried one copy of the tll::GFP(BAC) transgene. One copy of the tll::GFP(BAC) transgene was sufficient to enhance the supernumerary type II neuroblast phenotype in erm,ham double heterozygous brains. (F–H) Images of type II neuroblasts that mis-expressed a UAS-erm or UAS-ham transgene driven by a pan-neuroblast Gal4. Erm or Ham mis-expression drastically reduced Tll::GFP expression in type II neuroblasts. (I) Quantification of Tll::GFP expression relative to Dpn expression in type II neuroblasts that mis-expressed a UAS-erm or UAS-ham transgene driven by a pan-neuroblast Gal4. Erm mis-expression reduced Tll::GFP expression in type II neuroblasts before the onset of Ase expression, whereas Ham mis-expression reduced Tll::GFP expression in type II neuroblasts after the onset of Ase expression. (J) Quantification of total type II neuroblasts per wild-type or hamSK1 homozygous brain lobe that overexpressed a UAS-NRNAi transgene driven by an INP Gal4. Knocking-down Notch function in INPs suppressed the supernumerary type II neuroblast phenotype in hamSK1 homozygous brains. (K) Quantification of total type II neuroblasts per erm or ham heterozygous brain lobe that overexpressed a UAS-Nintra transgene driven by an INP Gal4. Overexpressing Nintra in INPs more efficiently induced supernumerary neuroblasts in erm,ham double heterozygous brains than in erm or ham heterozygous brains. The following labeling is applicable to all panels of images in this figure: yellow dashed line encircles a type II neuroblast lineage; white arrow: type II neuroblast; white arrowhead: Ase- immature INP; yellow arrow: Ase+ immature INP; yellow arrowhead: INP; *: supernumerary type II neuroblasts. Scale bar, 10 μm. Bar graphs are represented as mean ± standard deviation. p-values: **<0.05, ***<0.005. ns: not significant.
-
Figure 5—source data 1
Quantification of total type II neuroblasts per brain lobe that was erm,ham double heterozygous or erm,ham,tll triple heterozygous.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Quantification of total type II neuroblasts erm,ham double heterozygous brain lobe that carried one copy of the tll::GFP(BAC) transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Quantification of Tll::GFP expression relative to Dpn expression in type II neuroblasts that mis-expressed a UAS-erm or UAS-ham transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig5-data3-v2.xlsx
-
Figure 5—source data 4
Quantification of total type II neuroblasts per wild-type or hamSK1 homozygous brain lobe that overexpressed a UAS-NRNAi transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig5-data4-v2.xlsx
-
Figure 5—source data 5
Quantification of total type II neuroblasts per erm or ham heterozygous brain lobe that overexpressed a UAS-Nintra transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig5-data5-v2.xlsx
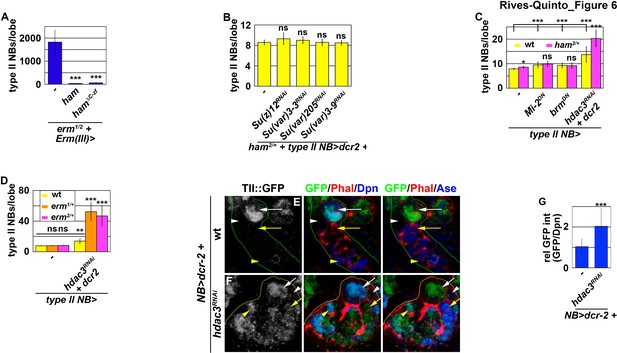
Erm and Ham function through Hdac3 to prevent INPs from reverting to type II neuroblasts.
(A) Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-ham transgene driven by an INP Gal4. Overexpressing full-length Ham or HamΔC-ZF in INPs suppressed the supernumerary type II neuroblast phenotype in erm-null brains. (B) Quantification of total type II neuroblasts per ham heterozygous brain lobe that overexpressed a UAS-RNAi transgene driven by a type II neuroblast Gal4. Reducing activity of the indicated chromatin complex did not increase INP reversion into supernumerary type II neuroblasts in ham heterozygous brains. (C) Quantification of total type II neuroblasts per ham heterozygous brain lobe that overexpressed a UAS transgene driven by a type II neuroblast Gal4. Reducing Hdac3 activity increased INP reversion into supernumerary type II neuroblasts in ham heterozygous brains. (D) Quantification of total type II neuroblasts per erm heterozygous brain lobe that overexpressed a UAS-hdac3RNAi transgene driven by a type II neuroblast Gal4. Reducing Hdac3 activity increased INP reversion into supernumerary type II neuroblasts in erm heterozygous brains. (E–F) Images of tll::GFP(Bac) brains that overexpressed a UAS-hdac3RNAi transgene driven by a pan-neuroblast Gal4. Reducing Hdac3 activity in type II neuroblasts led to ectopic Tll::GFP expression in immature INPs and INPs. (G) Quantification of Tll::GFP expression relative to Dpn expression in INPs derived from type II neuroblasts that overexpressed a UAS-hdac3RNAi transgene. Reducing Hdac3 activity in type II neuroblasts led to ectopic Tll::GFP expression in INPs. The following labeling is applicable to all panels of images in this figure: yellow dashed line encircles a type II neuroblast lineage; white arrow: type II neuroblast; white arrowhead: Ase- immature INP; yellow arrow: Ase+ immature INP; yellow arrowhead: INP. Bar graphs are represented as mean ± standard deviation. p-values: **<0.05, ***<0.005. ns: not significant.
-
Figure 6—source data 1
Quantification of total type II neuroblasts per brain lobe that overexpressed a UAS-ham transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Quantification of total type II neuroblasts per ham heterozygous brain lobe that overexpressed various UAS transgenes.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Quantification of total type II neuroblasts per erm heterozygous brain lobe that overexpressed a UAS-hdac3RNAi transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig6-data3-v2.xlsx
-
Figure 6—source data 4
Quantification of Tll::GFP expression relative to Dpn expression in INPs derived from type II neuroblasts that overexpressed a UAS-hdac3RNAi transgene.
- https://cdn.elifesciences.org/articles/56187/elife-56187-fig6-data4-v2.xlsx
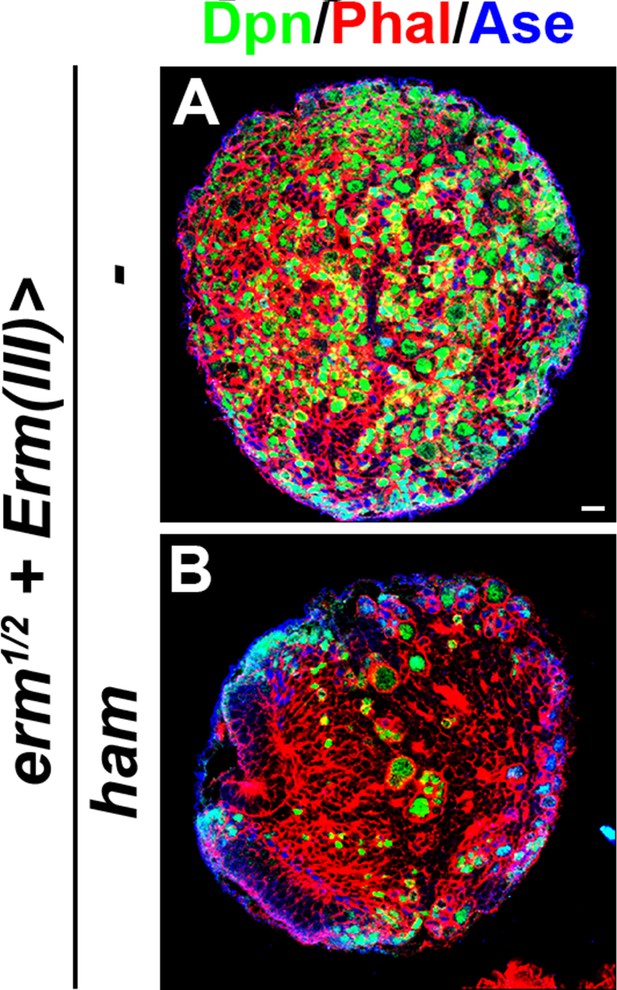
Ham overexpression in Ase+ immature INPs suppressed INP reversion in erm-null brains.
(A–B) Images of erm-null brains that overexpressed a UAS-ham transgene driven by an INP Gal4. Ham overexpression in INPs suppressed the supernumerary type II neuroblast phenotype in erm-null brains. Scale bar, 10 μm.
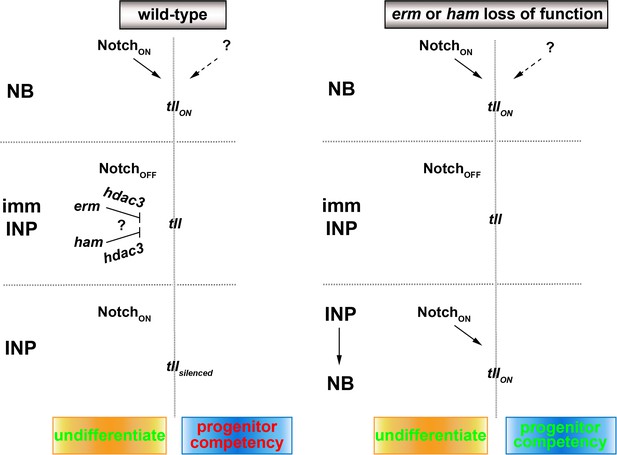
A proposed model for the regulation of type II neuroblast functionality.
We propose that Erm and Ham recruits Hdac3 to silence tll during INP commitment, preventing re-activation of Notch signaling in INPs from triggering Tll expression in wild-type brains. The tll locus remains in an activatable state in erm- or ham-null brains, and re-activation of Notch signaling in INPs triggers aberrant Tll expression driving INP reversion to supernumerary type II neuroblasts.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | anti-GFP (Chicken polyclonal) | Aves Labs, INC. | Cat#GFP-1020, RRID:AB_2307313 | IF(1:2000) |
| Antibody | anti-V5 (Mouse monoclonal) | ThermoFisher Scientific | Cat#R960-25, RRID:AB_2556564 | IF(1:500) |
| Antibody | anti-Ase (Rabbit polyclonal) | Weng et al., 2010 doi: 10.1016/j.devcel.2009.12.007. | IF(1:400) | |
| Antibody | anti-Hamlet (Rabbit polyclonal) | Eroglu et al., 2014 doi: 10.1016/j.cell.2014.01.053. | IF(1:50) | |
| Antibody | anti-Dpn (Rat monoclonal) | Lee et al., 2006a doi: 10.1038/nature04299. | clone 11D1BC7.14 | IF(1:2) |
| Antibody | Alexa Fluor 488 AffiniPure Anti-Chicken IgY (IgG) (H+L) (Donkey polyclonal) | Jackson Immuno Research Laboratories, INC. | Cat#703-545-155, RRID:AB_2340375 | IF(1:500) |
| Antibody | Alexa Fluor 647 AffiniPure anti-Rat IgG (H+L) (Goat polyclonal) | Jackson Immuno Research Laboratories, INC. | Cat#112-605-167 RRID:AB_2338404 | IF(1:500) |
| Antibody | anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Goat polyclonal) | ThermoFisher Scientific | Cat#A-11029, RRID:AB_2534088 | IF(1:500) |
| Antibody | anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 (Goat polyclonal) | ThermoFisher Scientific | Cat#A-11034, RRID:AB_2576217 | IF(1:500) |
| Antibody | anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 546 (Goat polyclonal) | ThermoFisher Scientific | Cat#A-11035, RRID:AB_2534093 | IF(1:500) |
| Other | Rhodamine Phalloidin | ThermoFisher Scientific | Cat#R415 | IF(1:100) |
| Genetic reagent (D. melanogaster) | brat11/CyO, Actin-GFP | Lee et al., 2006b doi: 10.1016/j.devcel.2006.01.017. | ||
| Genetic reagent (D. melanogaster) | w1118; Df(2L)Exel8040/CyO | Bloomington Drosophila Stock Center | BDSC: 7847 FlyBase: FBst0007847; RRID:BDSC_7847 | FlyBase symbol: Df(2L)Exel8040/CyO |
| Genetic reagent (D. melanogaster) | y1, M{vas-int.Dm}ZH-2A w*; M{UAS-insbFL-myc}ZH-86Fb | Komori et al., 2018 doi: 10.1101/gad.320333.118. | ||
| Genetic reagent (D. melanogaster) | Wor-Gal4(II) | Lee et al., 2006a doi: 10.1038/nature04299. | ||
| Genetic reagent (D. melanogaster) | Wor-Gal4(III) | Weng et al., 2010 doi: 10.1016/j.devcel.2009.12.007. | ||
| Genetic reagent (D. melanogaster) | y1, w*; P{tubPGAL80} LL10, P{neoFRT}40A/CyO | Bloomington Drosophila Stock Center | BDSC: 5192 FlyBase: FBst0005192; RRID:BDSC_5192 | FlyBase symbol: y1, w*; P{tubPGAL80} LL10, P{neoFRT}40A/CyO |
| Genetic reagent (D. melanogaster) | P{hsFLP}1, P{tubP-GAL80}LL1, w*, P{neoFRT}19A; P{UAS-mCD8::GFP.L}LL5 | Bloomington Drosophila Stock Center | BDSC: 5134 FlyBase: FBst0005134; RRID:BDSC_5134_ | FlyBase symbol: P{hsFLP}1, P{tubP-GAL80}LL1, w*, P{neoFRT}19A; P{UAS-mCD8::GFP.L}LL5 |
| Genetic reagent (D. melanogaster) | y1 w*; PBac{y[+mDint2] w[+mC]=tll EGFP.S}VK00037 | Bloomington Drosophila Stock Center | BDSC: 30874 FlyBase: FBst0030874; RRID:BDSC_30874 | FlyBase symbol: y1 w*; PBac{y[+mDint2] w[+mC]=tll EGFP.S}VK00037 |
| Genetic reagent (D. melanogaster) | Erm-Gal4 (II) | Pfeiffer et al., 2008 doi: 10.1073/pnas.0803697105. | ||
| Genetic reagent (D. melanogaster) | Erm-Gal4 (III) | Pfeiffer et al., 2008 doi: 10.1073/pnas.0803697105. | ||
| Genetic reagent (D. melanogaster) | tllRNAi: y1 sc* v1 sev21; P{TRiP.HMS01316}attP2 | Bloomington Drosophila Stock Center | BDSC: 34329 FlyBase: FBst0034329; RRID:BDSC_34329 | FlyBase symbol: y1 sc* v1 sev21; P{TRiP.HMS01316}attP2 |
| Genetic reagent (D. melanogaster) | M{UAS-tll.ORF-VN}ZH-86Fb | FlyORF | F004752 FBst0502964; RRID:FlyORF_ F004752 | FlyBase symbol: M{UAS-tll.ORF-VN}ZH-86Fb |
| Genetic reagent (D. melanogaster) | Ase-Gal80 (II) | Neumüller et al., 2011 doi: 10.1016/j.stem.2011.02.022. | ||
| Genetic reagent (D. melanogaster) | erm1/CyO, Act-GFP | Weng et al., 2010 doi: 10.1016/j.devcel.2009.12.007. | ||
| Genetic reagent (D. melanogaster) | erm2/CyO, Act-GFP | Weng et al., 2010 doi: 10.1016/j.devcel.2009.12.007. | ||
| Genetic reagent (D. melanogaster) | UAS-erm | Weng et al., 2010 doi: 10.1016/j.devcel.2009.12.007. | ||
| Genetic reagent (D. melanogaster) | PBac{erm-flag4C(g)}VK33 | Janssens and Lee, 2014 doi: 10.1242/dev.106534. | ||
| Genetic reagent (D. melanogaster) | DRNAi: y1 v1; P{TRiP.JF02115}attP2 | Bloomington Drosophila Stock Center | BDSC: 26217 FlyBase: FBst0026217; RRID:BDSC_26217 | FlyBase symbol: y1 v1; P{TRiP.JF02115}attP2 |
| Genetic reagent (D. melanogaster) | AseRNAi: y1 sc* v1 sev21; P{TRiP.HMS02847}attP2 | Bloomington Drosophila Stock Center | BDSC: 44552 FlyBase: FBst0044552; RRID:BDSC_44552 | FlyBase symbol: y1 sc* v1 sev21; P{TRiP.HMS02847}attP2 |
| Genetic reagent (D. melanogaster) | hamRNAi: y1 v1; P{TRiP.JF02270}attP2 | Bloomington Drosophila Stock Center | BDSC: 26728 FlyBase: FBst0026728; RRID:BDSC_26728 | FlyBase symbol: y1 v1; P{TRiP.JF02270}attP2 |
| Genetic reagent (D. melanogaster) | hamRNAi: y1 sc* v1 sev21; P{y[+t7.7] v[+t1.8]=TRiP.HMS00470}attP2 | Bloomington Drosophila Stock Center | BDSC: 32470 FlyBase: FBst0032470; RRID:BDSC_32470 | FlyBase symbol: y1 sc* v1 sev21; P{y[+t7.7] v[+t1.8]=TRiP.HMS00470}attP2 |
| Genetic reagent (D. melanogaster) | OpaRNAi: y1 sc* v1 sev21; P{TRiP.HMS01185}attP2/TM3, Sb1 | Bloomington Drosophila Stock Center | BDSC: 34706 FlyBase: FBst0034706; RRID:BDSC_34706 | FlyBase symbol: y1 sc* v1 sev21; P{TRiP.HMS01185}attP2/TM3, Sb1 |
| Genetic reagent (D. melanogaster) | w1118; Df(2L)Exel7071/CyO | Bloomington Drosophila Stock Center | BDSC: 7843 FlyBase: FBst0007843; RRID:BDSC_7843 | FlyBase symbol: w1118; Df(2L)Exel7071/CyO |
| Genetic reagent (D. melanogaster) | hamSKI, FRT40A/CyO | This paper | A new hamlet mutant fly line | |
| Genetic reagent (D. melanogaster) | P{w[+mW.hs]=GawB}elav[C155], P{w[+mC]=UAS-mCD8::GFP.L}Ptp4E[LL4], P{ry[+t7.2]=hsFLP}1, w[*] | Bloomington Drosophila Stock Center | BDSC: 5146 FlyBase: FBst0005146; RRID:BDSC_5146 | FlyBase symbol: P{w[+mW.hs]=GawB}elav[C155], P{w[+mC]=UAS-mCD8::GFP.L}Ptp4E[LL4], P{ry[+t7.2]=hsFLP}1, w[*] |
| Genetic reagent (D. melanogaster) | w1118; P{w[+mC]=UAS-Dcr-2.D}2 | Bloomington Drosophila Stock Center | BDSC: 24650 FlyBase: FBst00024650; RRID:BDSC_24650 | FlyBase symbol: w1118; P{w[+mC]=UAS-Dcr-2.D}2 |
| Genetic reagent (D. melanogaster) | w*; P{w[+mC]=tubP- GAL8ts}2/TM2 | Bloomington Drosophila Stock Center | BDSC: 7017 FlyBase: FBst00024650; RRID:BDSC_7017 | FlyBase symbol: w*; P{w[+mC]=tubP- GAL8ts}2/TM2 |
| Genetic reagent (D. melanogaster) | ham1,FRT40A/Cyo | Moore et al., 2002 doi: 10.1126/science.1072387. | ||
| Genetic reagent (D. melanogaster) | UAS-ham | Moore et al., 2002 doi: 10.1126/science.1072387. | ||
| Genetic reagent (D. melanogaster) | RNAi of Notch: y1, v1; P{y+t7.7v+t1.8=TRiP.HMS00001}attP2 | Bloomington Drosophila Stock Center | BDSC: 33611 FlyBase: FBst0033611; RRID:BDSC_33611 | FlyBase symbol: y1, v1; P{y+t7.7v+t1.8=TRiP.HMS00001}attP2 |
| Genetic reagent (D. melanogaster) | Oregon-R-C | Bloomington Drosophila Stock Center | BDSC: 5 FlyBase: FBst0000005; RRID:BDSC_5 | FlyBase symbol: Oregon-R-C |
| Genetic reagent (D. melanogaster) | P{hsFLP}1, y1 w*; P{UAS- N.intra.GS}2/CyO; MKRS/TM2 | Bloomington Drosophila Stock Center | BDSC: 52008 FlyBase: FBst0052008; RRID:BDSC_52008 | FlyBase symbol: P{hsFLP}1, y1 w*; P{UAS- N.intra.GS}2/CyO; MKRS/TM2 |
| Genetic reagent (D. melanogaster) | y1; M{vas-int.Dm}ZH-2A w*; M{UAS-ham∆C-ZF-myc}ZH-86Fb | This paper | Transgene expressing Hamlet mutant form of the C-terminal zinc finger deletion version | |
| Genetic reagent (D. melanogaster) | y1; M{vas-int.Dm}ZH-2A w*; M{UAS-ERD::hamN-ZF-myc}ZH-86Fb | This paper | Transgene expressing Hamlet the N-terminal zinc finger fused with ERD transcriptional repression domain | |
| Genetic reagent (D. melanogaster) | y1; M{vas-int.Dm}ZH-2A w*; M{UAS-VP-16::hamN-ZF-myc}ZH-86Fb | This paper | Transgene expressing Hamlet the N-terminal zinc finger fused with VP16 transcriptional activatoin domain | |
| Genetic reagent (D. melanogaster) | cu1, tll49/TM3, P{ftz/lacC}SC1, Sb1, Ser1 | Bloomington Drosophila Stock Center | BDSC: 7093 FlyBase: FBst007093; RRID:BDSC_7093 | FlyBase symbol: cu1, tll49/TM3, P{ftz/lacC}SC1, Sb1, Ser1 |
| Genetic reagent (D. melanogaster) | st1 e1 tll1/TM3, Sb1 | Bloomington Drosophila Stock Center | BDSC: 2729 FlyBase: FBst002729; RRID:BDSC_2729 | FlyBase symbol: st1 e1 tll1/TM3, Sb1 |
| Genetic reagent (D. melanogaster) | hamSK4, FRT40A/CyO | Bloomington Drosophila Stock Center | BDSC: 34329FlyBase: FBst0034329;RRID:BDSC_34329 | FlyBase symbol: y1 sc* v1 sev21; P{TRiP.HMS01316}attP2 |
| Genetic reagent (D. melanogaster) | hdac3RNAi: y1 sc* v1 sev21; P{TRiP.HMS00087}attP2 | Bloomington Drosophila Stock Center | BDSC: 34778 FlyBase: FBst0034778; RRID:BDSC_34778 | FlyBase symbol: hdac3RNAi: y1 sc* v1sev21; P{TRiP.HMS00087}attP2 |
| Genetic reagent (D. melanogaster) | Su(z)12RNAi: y1 sc* v1 sev21; P{TRiP.HMS00280}attP2/TM3, Sb1 | Bloomington Drosophila Stock Center | BDSC: 33402 FlyBase: FBst0033402; RRID:BDSC_33402 | FlyBase symbol: y1 sc* v1 sev21; P{TRiP.HMS00280}attP2/TM3, Sb1 |
| Genetic reagent (D. melanogaster) | Su(var)3-3RNAi: y1 sc* v1 sev21; P{TRiP.HMS00638}attP2 | Bloomington Drosophila Stock Center | BDSC: 32853 FlyBase: FBst0032853; RRID:BDSC_32853 | FlyBase symbol: y1 sc* v1 sev21; P{TRiP.HMS00638}attP2 |
| Genetic reagent (D. melanogaster) | Su(var)205RNAi: y1 sc* v1 sev21; P{TRiP.GL00531}attP40 | Bloomington Drosophila Stock Center | BDSC: 36792 FlyBase: FBti0146447; RRID:BDSC_36792 | FlyBase symbol: y1 sc* v1 sev21; P{TRiP.GL00531}attP40 |
| Genetic reagent (D. melanogaster) | UAS-Mi-2DN | Kovač et al., 2018 doi: 10.1038/s41467-018-04503-2. | ||
| Genetic reagent (D. melanogaster) | UAS-Mi-brmDN | Herr et al., 2010 doi: 10.1016/j.ydbio.2010.04.006. | ||
| Genetic reagent (D. melanogaster) | In(1)wm4; Su(var)3–91/TM3, Sb1 Ser1 | Bloomington Drosophila Stock Center | BDSC: 6209 FlyBase: FBst0006209; RRID:BDSC_6209 | FlyBase symbol: In(1)wm4; Su(var)3–91/TM3, Sb1 Ser1 |
| Genetic reagent (D. melanogaster) | w1118; PBac{Sp1- EGFP.S}VK00033 | Bloomington Drosophila Stock Center | BDSC: 38669 FlyBase: FBst0038669; RRID:BDSC_38669 | FlyBase symbol: w1118; PBac{Sp1- EGFP.S}VK00033 |
| Sequenced-based reagent | Ham_F | Eroglu et al., 2014 doi: 10.1016/j.cell.2014.01.053. | PCR primers | atagatcctttggccagcagac |
| Sequenced-based reagent | Ham_R | Eroglu et al., 2014 doi: 10.1016/j.cell.2014.01.053. | PCR primers | agtactcctccctttcggcaat |
| Sequenced-based reagent | Ase_F | Komori et al., 2014b doi: 10.7554/eLife.03502. | PCR primers | agcccgtgagcttctacgac |
| Sequenced-based reagent | Ase_R | Komori et al., 2014b doi: 10.7554/eLife.03502. | PCR primers | gcatcgatcatgctctcgtc |
| Sequenced-based reagent | D_F | This paper | PCR primers | gcggcggcggtcaacaat |
| Sequenced-based reagent | D_R | This paper | PCR primers | tgcggcgtacagcgaagggt |
| Sequenced-based reagent | Erm_F | Eroglu et al., 2014 doi: 10.1016/j.cell.2014.01.053. | PCR primers | gttacggccaggcatcgggtcaa |
| Sequenced-based reagent | Erm_R | Eroglu et al., 2014 doi: 10.1016/j.cell.2014.01.053. | PCR primers | gggccaggcgggattactcgtctc |
| Sequenced-based reagent | PntP1_F | Komori et al., 2014b doi: 10.7554/eLife.03502. | PCR primers | ggcagtacgggcagcaccac |
| Sequenced-based reagent | PntP1_R | Komori et al., 2014b doi: 10.7554/eLife.03502. | PCR primers | ctcaacgcccccaccagatt |
| Sequenced-based reagent | Dpn_F | Komori et al., 2014b doi: 10.7554/eLife.03502.Komori et al., 2014b | PCR primers | catcatgccgaacacaggtt |
| Sequenced-based reagent | Dpn_R | Komori et al., 2014b | PCR primers | gaagattggccggaactgag |
| Recombinant DNA reagent | pUAST-ham∆C-ZF-myc-attB (plasmid) | This paper | Plasmid DNA of a transgene expressing Hamlet mutant form of the C-terminal zinc finger deletion version | |
| Recombinant DNA reagent | pUAST-ERD::hamN-ZF-myc-attB (plasmid) | This paper | Plasmid DNA of a transgene expressing Hamlet the N-terminal zinc finger fused with ERD transcriptional repression domain | |
| Recombinant DNA reagent | pUAST-VP16::hamN-ZF-myc-attB (plasmid) | This paper | Plasmid DNA of a transgene expressing Hamlet the N-terminal zinc finger fused with VP16 transcriptional activatoin domain | |
| Software, algorithm | LAS AF | Leica Microsystems | RRID:SCR_013673 | |
| Software, algorithm | ImageJ 1.50 g | National Institute of Health | RRID:SCR_003070 |





