Intrinsic control of neuronal diversity and synaptic specificity in a proprioceptive circuit
Figures
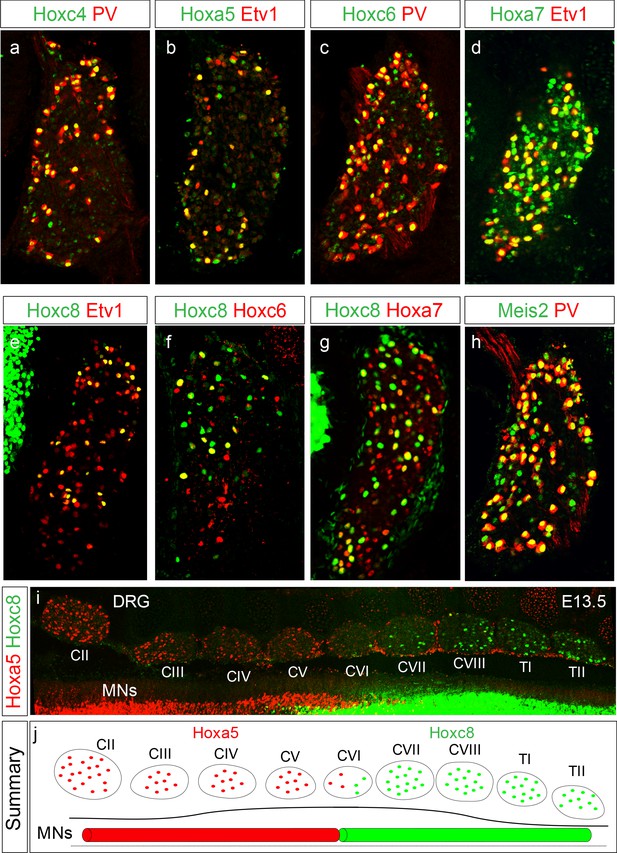
Restricted Hox expression defines pSN subtype identity.
(a–e) Expression of indicated Hox proteins in cervical DRG. Images show cross sections of DRG located between segmental levels C2-C8 of E14.5 mouse embryos stained with Hox and the pSN markers PV or Etv1. Images are representative of at least three embryos analyzed at this age. (f) Coexpression of Hoxc6 and Hoxc8 in subsets of pSNs in caudal cervical DRG. (g) Coexpression of Hoxc8 and Hoxa7 in sensory neurons. (h) Meis2 coexpression with PV in sensory neurons. (i) Top-down view of E13.5 mouse spinal cord showing mutually exclusive expression of Hoxa5 and Hoxc8 in DRG. Spinal MNs also abide by restricted rostrocaudal domains of Hoxa5 and Hoxc8 expression. (j) Summary of Hoxa5 and Hoxc8 expression pattern in MNs and pSNs. See also Figure 1—figure supplement 1.
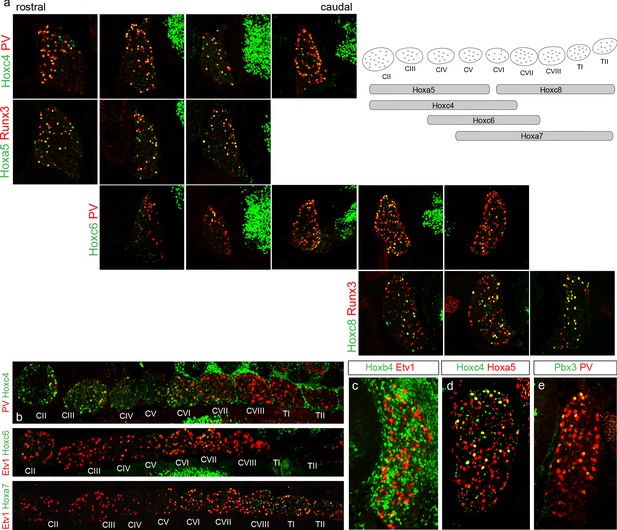
Hox expression pattern of sensory neurons in cervical DRG.
(a) Spatial profile of Hoxc4, Hoxa5, Hoxc6, and Hoxc8 expression in DRG along the rostrocaudal axis. Images show coincident expression of Hox proteins with pSN markers. Hoxa5 (with Runx3) and Hoxc4 (with PV) are present in rostral cervical DRG. Hoxc6 (with PV) spans from DRG CIV to CVII while Hoxc8 (with Runx3) is confined to caudal cervical DRG levels. Images shown were obtained from a single embryo, and are representative of N = 3 animals analyzed at this age. Schematic on left indicates Hox expression pattern in cervical level DRG. (b) Longitudinal sections show expression of indicated Hox protein with a pSN markers (PV or Etv1). DRG segmental levels are indicated (CII-TII). (c) Hoxb4+ cells are excluded from Etv1+ sensory neurons at E13.5. (d) A subpopulation of Hoxa5+ SNs coexpresses Hoxc4. (e) Pbx3+ colocalizes with PV+ SNs.
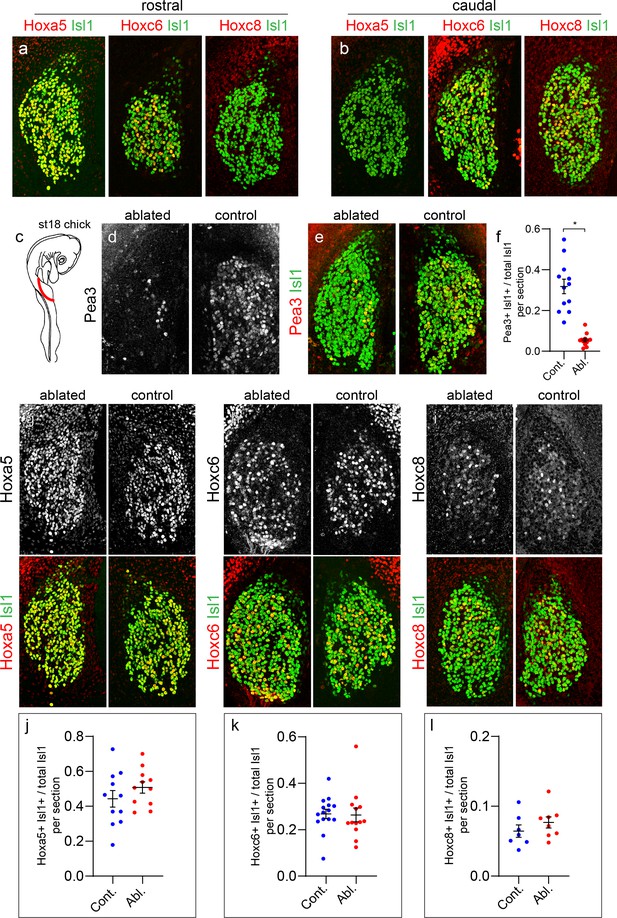
Limb-independent expression of Hox genes in sensory neurons.
(a and b) Expression of Hoxa5, Hoxc6, and Hoxc8 in chick forelimb level DRG shown with pan-sensory neuron marker Islet1 (Isl1). Images show cross sections of chick DRG at ~St 26. Hoxa5 and Hoxc6 are present at rostral cervical segments while Hoxc8 is absent from this region (a). Hoxa5 is not found in caudal cervical DRG whereas Hoxc6 and Hoxc8 are present within this region (b). Images are representative of at least three embryos analyzed at this age. (c) Unilateral forelimb bud ablation of chick at St 16–18. Embryos harvested at St 26–28. (d–e) Loss of Pea3 expression on the limb-ablated side in relation to the non-ablated side. Expression of Isl1 is unaffected at this stage. Images show cervical DRG of an individual embryo at the same segmental levels between ablated and non-ablated sides for each panel. (f) Quantification of loss of Pea3, as a fraction of total Isl1+ SNs. Controls, 31.8 ± 3.6%, N = 12 sections from three animals, ablated 5.5 ± 0.1%, N = 12 sections from three animals, p<0.0001, Student’s t-test. (g–i) Top panels show expression of individual Hox expression. Bottom panels show Hox expression with Isl1. There is no difference in Hox expression between the ablated and non-ablated side for Hoxa5+ SNs in rostral cervical segments (g), Hoxc6+ SNs in rostral and caudal cervical segments (h), or Hoxc8+ SNs in caudal cervical segments (i). (j–k) Quantification of fraction of Hox+Isl1+ over total Isl1+ SNs in control and limb-ablated chick embryos. For each Hox protein, sections were obtained from three limb-ablated embryos, with non-ablated side of embryo serving as the control. Hoxa5 (44.3 ± 4.7% in N = 11 control sections, 50.7 ± 3.2% in N = 11 limb-ablated sections, p=0.27, Student’s t-test), Hoxc6 (26.8 ± 2.0% in N = 15 control sections, 26.3 ± 3.0% in N = 13 limb-ablated sections, p=0.90), and Hoxc8 (6.4 ± 0.9% in N = 7 control sections, 7.6 ± 0.8% in N = 8 sections ablated, p=0.32). See also Figure 2—figure supplement 1.
-
Figure 2—source data 1
Quantification of markers proteins after limb bud ablation.
- https://cdn.elifesciences.org/articles/56374/elife-56374-fig2-data1-v2.xlsx
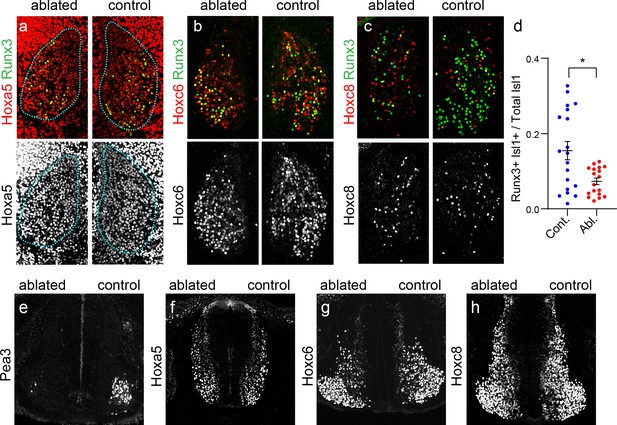
Hox expression in MNs and SNs is limb-independent.
(a–g) Cross sections of St. 28 chick embryos after unilateral limb ablation. (a–c) Expression of Hoxa5 (a), Hoxc6 (a), and Hoxc8 (a) expressing SNs. Runx3 marks pSNs. There is a noticeable reduction in Runx3 expression on ablated side, especially in the Hoxc8+ domain. (d) Loss of Runx3 expression in SNs after limb ablation. The fraction of Isl1+ SNs expressing Runx3 was reduced from 15.5 ± 2.4% in controls (N = 3 animals) to 7.3 ± 0.9% after limb ablation (N = 3 animals, p=0.0038, Student’s t-test). (e) Limb ablation results in a loss of Pea3+ MNs compared to the contralateral side. (f–h) Similar patterns of Hoxa5 (e), Hoxc6 (f), and Hoxc8 (g) expression in MNs after limb ablation.
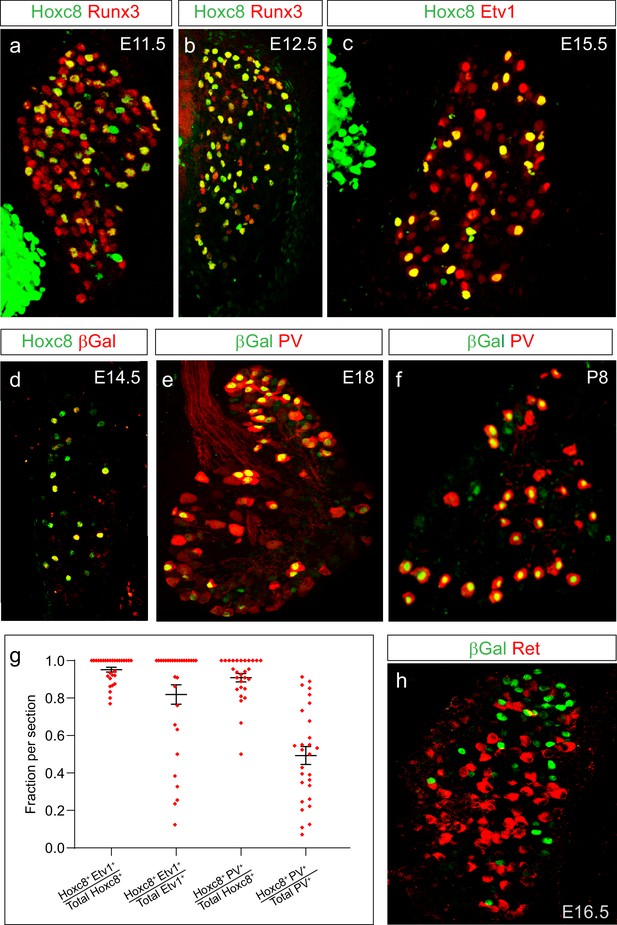
Profile of Hoxc8 in pSNs during sensory-motor circuit development.
(a, b) Expression of Hoxc8 with the pSN marker Runx3 at E11.5 (a) and E12.5 (b). (c) Hoxc8 and Etv1 expression in pSNs at E15.5. (d) Colocalization of Hoxc8 and βGal in PLAT::Cre; Hoxc8LacZ-flox/+ mice at E14.5. (e, f) Colocalization of βGal with the pSN marker PV at E18 (e) and P8 (f). (g) Quantification of Hoxc8 with either Etv1 or PV at E14.5. Each data point shows individual sections taken from at least three mice. Hoxc8+Etv1+/total Hoxc8+ cells = 0.95 ± 0.01 (mean ± SEM, N = 30 sections); Hoxc8+Etv1+/total Etv1+ cells = 0.82 ± 0.01 (N = 30 sections); Hoxc8+PV+/total Hoxc8+ cells = 0.91 ± 0.02 (N = 28 sections); Hoxc8+PV+/total PV+ cells = 0.50 ± 0.04 (N = 28 sections). (h) Non-overlapping expression of βGal and Ret, a marker for subpopulations of cutaneous sensory neurons at E16.5. See also Figure 3—figure supplement 1.
-
Figure 3—source data 1
Quanification of marker proteins in sensory neurons.
- https://cdn.elifesciences.org/articles/56374/elife-56374-fig3-data1-v2.xlsx
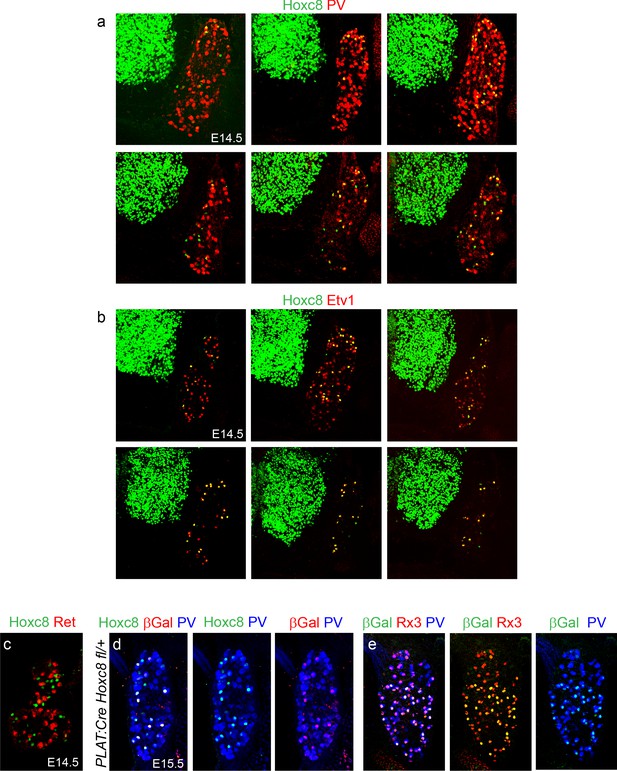
Detailed expression profile of Hoxc8.
(a, b) Hoxc8 colocalizes with PV and Etv1 in caudal cervical DRG. Hoxc8+ pSNs parallel the domain of Hoxc8 expression in MNs. Cross sections of E14.5 wildtype mice showing expression from rostral (top left) to caudal (bottom right) levels. (c) Hoxc8+ DRG neurons do not express Ret at E14.5. (d, e) Coexpression of βGal, Hoxc8, PV and βGal, Runx3 (Rx3), PV in sensory neurons of PLAT::Cre; Hoxc8LacZ-flox/+ mice (cross sections from E15.5 mice).
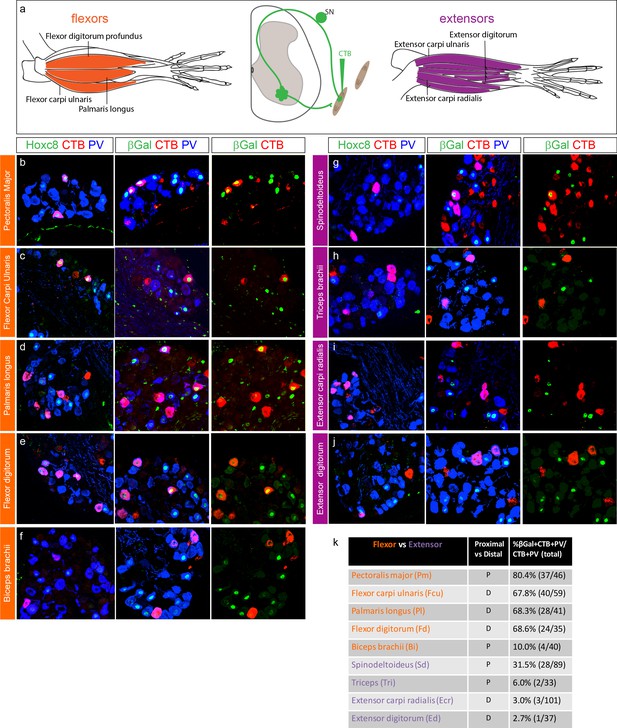
Hoxc8 pSNs preferentially target distal flexor limb muscles.
(a) Schematic of distal forelimb muscles of mouse. Limb flexor muscles shown in orange, extensors in purple. Muscles were injected with CTB at P4 and collected at P7. (b–j) Cross sections of caudal cervical DRG of PLAT::Cre; Hoxc8LacZ-flox/+ mice injected with CTB in an individual forelimb muscle. First column shows colocalization of Hoxc8, CTB, and PV; second column colocalization of βGal, CTB, and PV; and third column βGal and CTB. (k) Table of injected forelimb muscles with quantification of βGal+ pSN innervation shown as percentage (βGal+CTB + PV+ SNs over total CTB+PV+SNs in all sections) with raw numbers next to percentages. Values in table show cumulative data from CTB-labeled DRG sections collected from at least three mice per muscle. See also Figure 4—figure supplement 1.
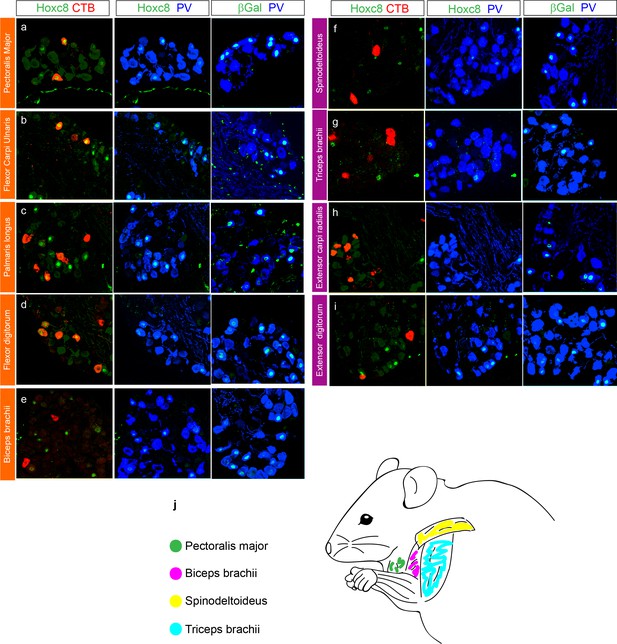
Characterization of Hoxc8+ pSN innervation of forelimb muscles.
(a–i) Cross sections of cervical level DRG in PLAT::Cre; Hoxc8LacZ-flox/+ mice injected with CTB in individual limb muscles (N ≥ 3). First column shows colocalization of Hoxc8 and CTB, 2nd column shows colocalization of the same sections in the 1st column with Hoxc8 and PV labeling and 3rd column βGal and PV (see also Figure 4) for CTB injected flexors: PM (a), FCU (b), PL (c), FD (d), Bic (e), and extensors: SD (f), Tri (g), ECR (h), ED (iI). (j) Schematic of injected proximal forelimb muscles: PM, Bic, SD, Tri.
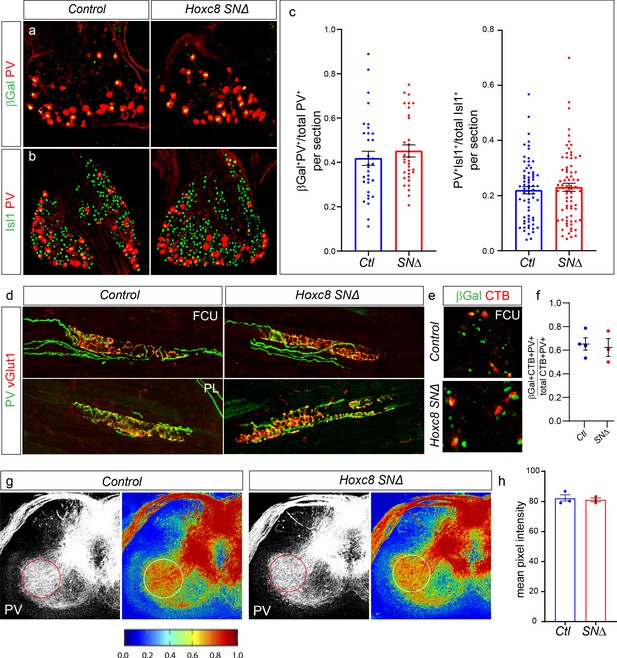
Hoxc8 is dispensable for pSN survival and differentiation.
(a, b) Example images showing expression of βGal and PV (a) or PV and Isl1 (b) in controls and Hoxc8SN∆ mice at P7. (c) Quantification of fraction of βGal+ PV+ SNs over total PV+ population: 42 ± 3% (mean ± SEM) in N = 33 sections from three mice, versus 45 ± 3% in N = 32 sections from 3 Hoxc8SN∆ mice (p=0.43, Student’s t test). Quantification of PV+ Isl1+ SNs over total Isl1+ population: 22 ± 1% in N = 68 sections from three control mice, versus 23 ± 1% for N = 74 sections from 4 Hoxc8SN∆ mice (p=0.59, Student’s t test). Data points in graphs show results from individual sections. Lines in graph show mean ± SEM. (d) Distal forelimb flexors FCU and PL still receive normal pSN innervation and muscle spindles develop normally in Hoxc8SN∆ mice compared to controls shown with PV and vGlut1 staining. (e) Retrograde labeling of βGal+ SNs after CTB injection of FCU in control and Hoxc8SN∆ mice. Muscle injection performed at ~P4 and DRG collected at ~P7 P8. (f) Quantification of CTB+βGal+PV+/total CTB+PV+ SNs after FCU retrograde tracing: 65.2 ± 5.1% for N = 4 control mice; 62.3 ± 7.6% for N = 3 Hoxc8SN∆ mice, p=0.75, Student’s t test. (g) No difference in PV fiber density in the ventral spinal cord between control and Hoxc8SN∆ mice. PV fiber stain with heat map below. (h) Quantification of the average PV pixel intensity at DRG C8 level. PV fiber density calculated only in ROI created in ventral spinal cord region. Lines indicate mean ± SEM. Average intensity for control: 82.1 ± 2.4, N = 3 mice. Average intensity for Hoxc8SN∆: 81.1 ± 1.3, N = 3 mice (p=0.74, Student’s t test). See also Figure 5—figure supplement 1.
-
Figure 5—source data 1
Quantification of sensory markers in control and Hoxc8 mutants.
- https://cdn.elifesciences.org/articles/56374/elife-56374-fig5-data1-v2.xlsx
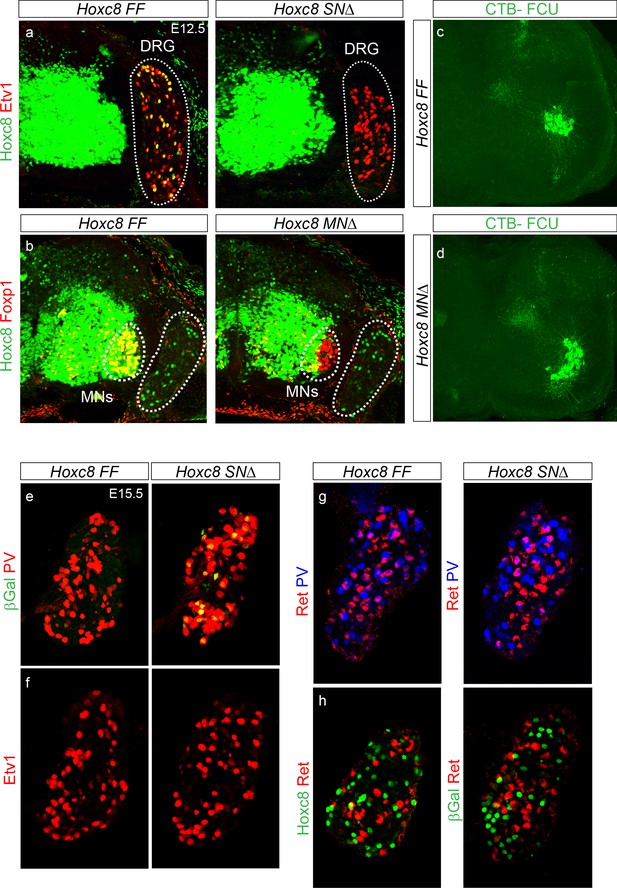
Preservation of sensory neuron identities in Hoxc8SN∆ mice.
(a and b) Genetic approaches to study sensory-motor circuit development in the absence of Hoxc8. Cross sections of spinal cord with adjacent DRG in the caudal cervical region of E12.5 mice. In Hoxc8SN∆ mice, Hoxc8 is selectively removed from SNs in the DRG but expression is retained in MNs (a). Deletion of Hoxc8 in MNs of Olig2::Cre; Hoxc8LacZ-flox/ LacZ-flox mice (Hoxc8MN∆) with Hoxc8 expression in SNs comparable to that of control mice (b). (c–d) Altered location of FCU MNs in Hoxc8MN∆ mice. Images are maximum intensity projections of confocal Z-stacks encompassing the entire domain of the FCU motor pool. In Hoxc8MN∆ mice, FCU MNs shift ventrally. (e–h) Cross sections of caudal cervical DRG C8 in Hoxc8SN∆ and control mice at E15.5. (e) Comparable number of PV+ cells between Hoxc8SN∆ and control mice. βGal+ SNs survive and maintain PV expression after removal of Hoxc8. (f) No depletion of Etv1+ cells in SNs of Hoxc8SN∆ mice compared to that of control mice. (g) Similar profile of Ret+PV-, Ret+PV+, and Ret-PV+ neurons in Hoxc8SN∆ and control mice. (h) βGal+ SNs do not express Ret in the absence of Hoxc8.
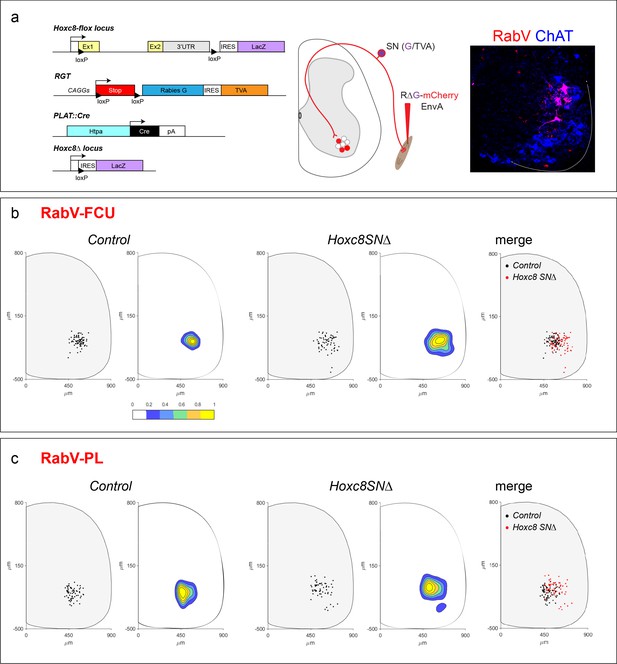
Altered central targeting of distal flexor muscle pSNs in Hoxc8SN∆ mice.
(a) Cre-dependent excision of Hoxc8 coding sequence and expression of LacZ reporter driven by PLAT promoter. Removal of the stop cassette permits expression of 2 rabies helper proteins, TVA and glycoprotein G, after Cre-recombination thus directing rabies infection in a cell type specific manner (left). Modified rabies virus labeling experimental design: Env/TVA system permits primary infection of SNs; SNs also express glycoprotein G which enables monosynaptic transfer of injected modified rabies virus (RV∆G-mCherry-EnvA) anterogradely to spinal cord MNs by secondary infection (middle). Representative spinal cord cross-section of RV∆G-mCherry-EnvA monosynaptic labeling in MNs via pSN transfer after injection into FCU muscle of PLAT::Cre; Hoxc8LacZ-flox/+ mice (right). (b and c) Dot plot showing RV∆G-mCherry-EnvA labeled MNs position of a representative spinal cord hemi-section in the caudal cervical region. Distances from the central canal are shown on x and y coordinate axes (in micrometers). Contour plots to the right depicting labeled MN density in relation to position in spinal cord. Area of greatest labeling density in yellow. Overlay of labeled MN dot plots for both control and Hoxc8SN∆ mice. Total number of labeled MNs in which the FCU is injected in control mice; N = 3 mice, 79 cells. Total number of labeled MNs in Hoxc8SN∆ mice; N = 3 mice, 66 cells (b). Total number of labeled MNs in which the PL is injected in control mice; N = 3 mice, 71 cells. Total number of labeled MNs in Hoxc8SN∆ mice; N = 3 mice, 62 cells. See also Figure 6—figure supplement 1.
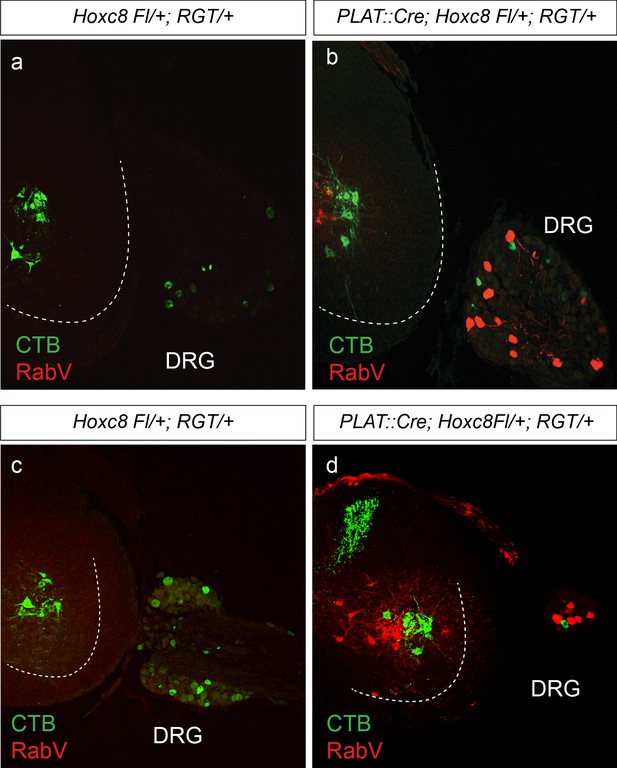
Cre-dependent rabies infection of sensory neurons.
(a–d) Cross sections of spinal cord with adjacent DRG in caudal cervical region of P12 mice. RV∆G-mCherry-EnvA injected into FCU muscle with concurrent injection of CTB in ECR/ED. No mCherry was detectable in SNs or MNs of Cre- mice (a, c). RV∆G-mCherry-EnvA injected into FCU of Cre+ Hoxc8LacZ-flox/+ mice exhibited mCherry expression in predominately large diameter SNs, INs, and MNs in the ipsilateral ventral spinal cord (b, d).
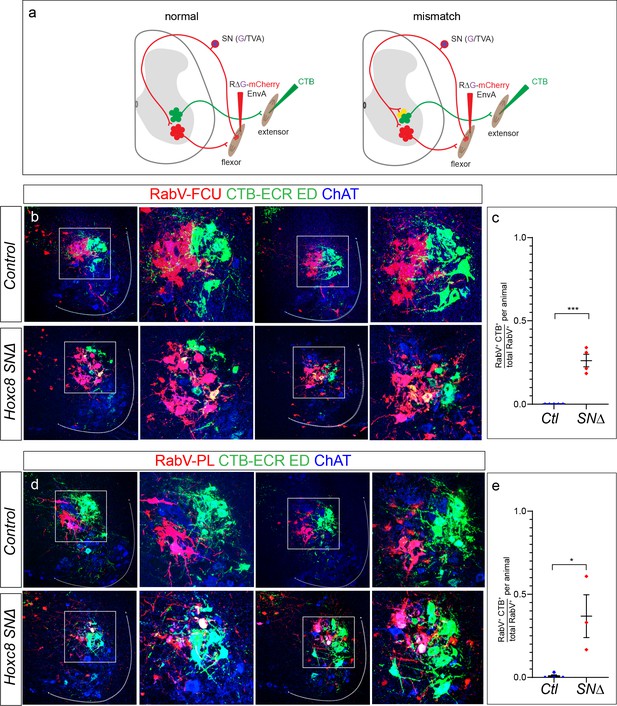
Flexor pSNs form ectopic synapses on extensor MNs in Hoxc8SN∆ mice.
(a) Schematic illustrating muscle injection of RV∆G-mCherry-EnvA (RabV). Selective infection of SNs and anterograde transsynaptic transport leads to secondary infection of MNs in the ventral spinal cord but not direct infection of MNs. CTB injected into a muscle labels directly connected MNs. In a normal condition, pSNs avoid synapsing onto antagonist MNs. In a mismatch condition, pSNs form ectopic contacts onto antagonist muscle MNs yielding colocalization of RabV (red) and CTB (green). Individual distal forelimb flexor muscles were injected with RabV and distal forelimb extensors were injected with CTB. Injections were performed at ~P6 P7 and spinal cords were collected at ~P12-13. (b) Colocalization of RabV, CTB, and ChAT signifying ectopic contacts in Hoxc8SN∆ mice where RabV was injected into the FCU and CTB was injected into the ECR and ED. (c) Quantification of the average percentage of RabV+CTB+ChAT+ cells over total RabV+ChAT+ cells where RabV was injected into the FCU. Lines indicate mean ± SEM. Average from control mice: N = 5 mice; 0 ± 0. Average from Hoxc8SN∆: N = 4 mice; 0.26 ± 0.04. (p<0.0001, Student’s t test). (d) Colocalization of RabV, CTB, and ChAT signifying ectopic contacts in Hoxc8SN∆ mice where RabV was injected into the PL and CTB was injected into the ECR and ED. (e) Quantification of the average percentage of RabV+CTB+ChAT+ cells over total RabV+ChAT+ cells where RabV was injected into the PL. Lines indicate mean ± SEM. Average from control mice: N = 4 mice; 0.008 ± 0.008. Average from Hoxc8SN∆: N = 3 mice; 0.37 ± 0.13. (p=0.02, Student’s t test). For both Hoxc8SN∆ mice and controls in which the FCU was injected, a total of ~150 RabV MNs were counted while 120 RabV MNs were counted for control mice injected in the PL and 101 RabV MNs for Hoxc8SN∆ mice. See also Figure 7—figure supplement 1.
-
Figure 7—source data 1
Quantification of rabies labeled neurons in control and Hoxc8 mutants.
- https://cdn.elifesciences.org/articles/56374/elife-56374-fig7-data1-v2.xlsx
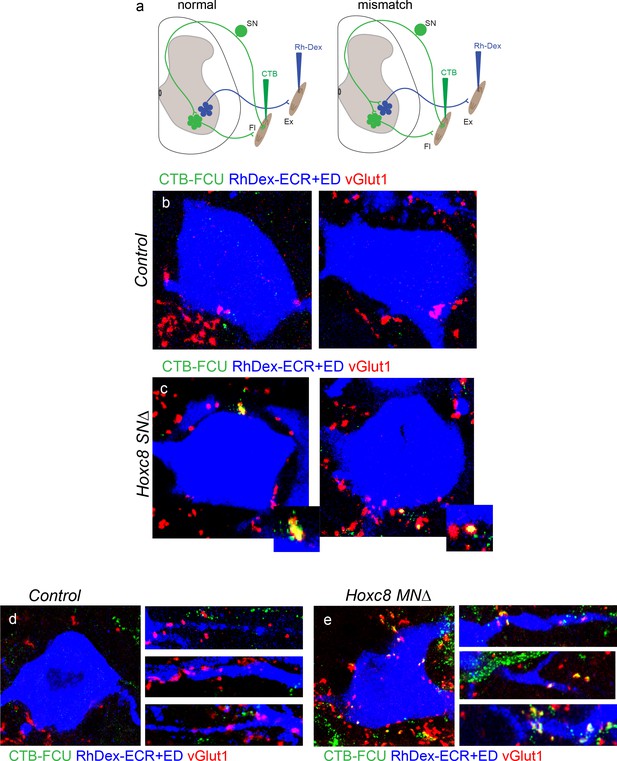
Fidelity of sensory-motor specificity compromised is in Hoxc8SN∆ and Hoxc8MN∆ mice.
(a) Schematic depicting synaptic assay design to detect ectopic pSN synapses onto MNs. CTB is injected into a distal forelimb flexor muscle and Rh-Dex is concurrently injected into distal forelimb extensor muscles. In injected control mice, vGluT1+ sensory boutons contact CTB-labeled MNs of the homonymous (flexor) muscle but do not form synapses onto antagonist (extensor) MNs. Erosion of sensory-motor connection specificity results in CTB labeled vGluT1+ sensory boutons ectopic contacts onto Rh-Dex labeled antagonist MNs. (b) No CTB labeled vGluT1+ FCU sensory boutons are found on ECR/ED MNs in control mice at P8. N = 2 mice analyzed. (c) Ectopic CTB labeled vGluT1+ FCU sensory boutons detected on ECR/ED MNs in Hoxc8SN∆ mice at P8. N = 2 mice analyzed. (d) CTB labeled vGluT1+ FCU sensory boutons are not observed on the soma or dendrites of ECR/ED MNs in a control animal at P8. (e) CTB labeled vGluT1+ FCU sensory boutons form on the soma and proximal dendrites of ECR/ED MNs in a Hoxc8MN∆ mutant.
Tables
Expression of Hox4-Hox8 paralog proteins in SNs.
Table lists each of the tetrapod Hox4-Hox8 gene paralogs and their expression pattern in SNs between segmental levels CII-TII. Not all antibody combinations were tested for Hox co-expression. ND, not detected; NT, not tested.
| Hox protein | DRG expression | pSN expression | RC level | Hox co-expression |
|---|---|---|---|---|
| Hoxc4 | yes | yes | CII-CVII | Hoxa5 |
| Hoxc5 | NT | - | - | - |
| Hoxc6 | yes | yes | CIV-CVIII | Hoxc8 |
| Hoxc8 | yes | yes | CVI-TII | Hoxc6, Hoxa7 |
| Hoxa4 | NT | - | - | - |
| Hoxa5 | yes | yes | CII-CVI | Hoxc4 |
| Hoxa6 | ND | - | - | - |
| Hoxa7 | yes | yes | CV-TII | Hoxc8 |
| Hoxb4 | yes | no | NT | - |
| Hoxb5 | yes | no | NT | - |
| Hoxb6 | NT | - | - | - |
| Hoxb7 | NT | - | - | - |
| Hoxb8 | NT | - | - | - |
| Hoxd4 | NT | - | - | - |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (M. musculus) | Hoxc8 flox | PMID:19621436 | MGI: 4365797 | |
| Genetic reagent (M. musculus) | PLAT::Cre | PMID:12812797 | MGI: 3052515 | |
| Genetic reagent (M. musculus) | Olig2::Cre | PMID:18046410 | MGI: 3774124 | |
| Genetic reagent (M. musculus) | Gt(ROSA)26Sor::CAG-loxp-STOP-loxp-rabies-G-IRES-TVA | PMID:23352170 | MGI: J:206510 | |
| Biological sample (rabies virus) | EnvA-RabV-mCherry (pseudotyped G-deleted rabies virus) | PMID:21867879 PMID:17329205 PMID:26844832 | ~108 IU/mL | |
| Biological sample (chicken eggs) | SPF Eggs | Charles River | 10100332 | |
| Antibody | anti-Hoxc4 (Rabbit polyclonal) | PMID:16269338 | (1:16000) | |
| Antibody | anti-Hoxa5 (Rabbit polyclonal) | PMID:16269338 | (1:16000) | |
| Antibody | anti-Hoxc6 (Guinea pig polyclonal) | PMID:11754833 | RRID:AB_2665443 | (1:16000) |
| Antibody | anti-Hoxc6 (Rabbit polyclonal) | Aviva Systems Biology | Cat# ARP38484; RRID:AB_10866814 | (1:32000) |
| Antibody | anti-Hoxa7 (Guinea pig polyclonal) | PMID:16269338 | (1:32000) | |
| Antibody | anti-Hoxc8 (Mouse monoclonal) | Covance | RRID:AB_2028778 | (1:4000) |
| Antibody | anti-Hoxb4 (Rat monoclonal) | Developmental Studies Hybridoma Bank | Cat# I12; RRID:AB_2119288 | (1:100) |
| Antibody | anti-Hoxb5 (Rabbit polyclonal) | PMID:23103965 | (1:32000) | |
| Antibody | anti-Foxp1 (Rabbit polyclonal) | PMID:18662545 | RRID:AB_2631297 | (1:32000) |
| Antibody | anti-Isl1/2 (Mouse monoclonal) | Developmental Studies Hybridoma Bank | Cat# 39.4D5, RRID:AB_2314683 | (1:50) |
| Antibody | anti-Isl1/2 (Rabbit polyclonal) | Jessell lab | (1:5000) | |
| Antibody | anti-Meis2 (Rabbit polyclonal) | PMID:16269338 | (1:16000) | |
| Antibody | anti-Pbx3 (Rabbit polyclonal) | PMID:16269338 | (1:16000) | |
| Antibody | anti-Pea3 (Rabbit polyclonal) | PMID:9814709 | RRID:AB_2631446 | (1:32000) |
| Antibody | anti-βGal (Goat polyclonal) | Abcam | Cat# 9361; RRID:AB_307210 | (1:1000) |
| Antibody | anti-βGal (Goat polyclonal) | Santa Cruz | Cat# sc-19119; RRID:AB_2111604 | (1:2000) |
| Antibody | anti-Ret (Goat polyclonal) | Santa Cruz | Cat# sc-1290; RRID:AB_631316 | (1:100) |
| Antibody | anti-CTB (Goat polyclonal) | List Biological Laboratories | Cat# 703; RRID:AB_10013220 | (1:4000) |
| Antibody | anti-vGlut1 (Guinea pig polyclonal) | Millipore | Cat# AB5905; RRID:AB_2238022 | (1:1000) |
| Antibody | anti-PV (Rabbit polyclonal) | Swant | Cat# PV27; RRID:AB_2631173 | (1:1000) |
| Antibody | anti-Runx3 (Rabbit polyclonal) | Abcam | Cat# ab68938; RRID:AB_1141661 | (1:16000) |
| Antibody | anti-TRITC (Rabbit polyclonal) | Thermofisher | Cat# A6397; RRID:AB_2536196 | (1:1000) |
| Antibody | anti-Ret (Rabbit polyclonal) | Cell Signaling | Cat# 3223; RRID:AB_2238465 | (1:100) |
| Antibody | anti-ChAT (Rabbit polyclonal) | Jessell Lab | (1:16000) | |
| Antibody | anti-Etv1 (Rabbit polyclonal) | Jessell Lab | (1:8000) | |
| Antibody | anti-βGal (Chick polyclonal) | Jessell Lab | (1:5000) | |
| Antibody | anti-cRunx3 (Guinea pig polyclonal) | Jessell Lab | (1:5000) | |
| Antibody | Alexa 488-, Cy3-, Alexa 647- secondaries (Donkey polyclonal) | Jackson ImmunoResearch | (1:1000) | |
| Peptide, recombinant protein | 1% Cholera Toxin B subunit | Sigma-Aldrich | Cat# C9903 | |
| Chemical compound, drug | Dextran, Tetramethyl- rhodamine | ThermoFisher | Cat# D3308 | |
| Software, algorithm | Fiji | PMID:22743772 | RRID:SCR_002285 | http://imagej.net/Fiji |
| Software, algorithm | Imaris | Bitplane/Oxford Instruments | v8.1.2 RRID:SCR_007370 | |
| Software, algorithm | Matlab | Mathworks | R2019b RRID:SCR_001622 | |
| Software, algorithm | GraphPad Prism | GraphPad Software | 8.0.2 (263) |





