Miga-mediated endoplasmic reticulum–mitochondria contact sites regulate neuronal homeostasis
Figures
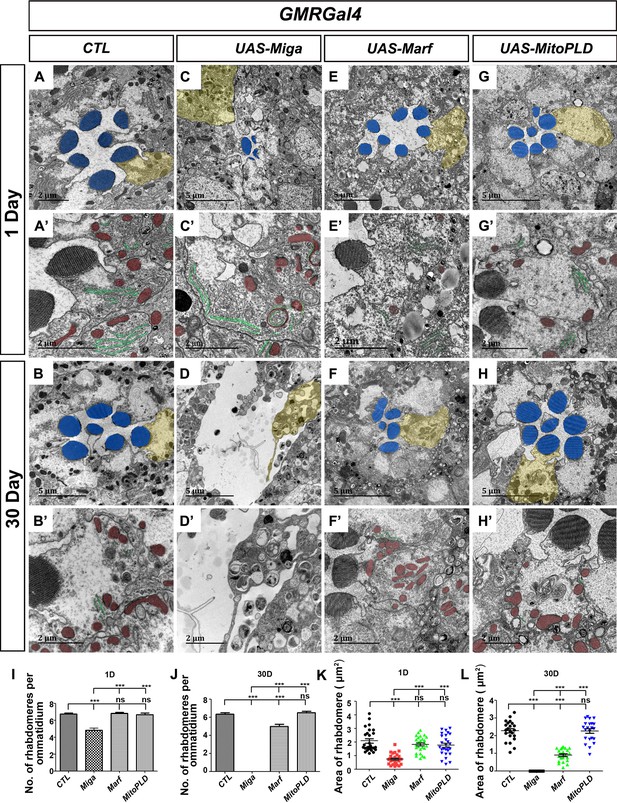
Miga overexpression led to severe retinal degeneration.
TEM analysis was performed for the retina thin sections of young (1 Day) and old (30 Day) flies with indicated genotypes. (A–B’) The ommatidia of the control (CTL) flies showed seven photoreceptor cells with intact rhabdomeres (highlighted with blue pseudo-color) at both the young (A, A’) and old (B, B’) stages. (C–D’) GMR-Gal4 driven Miga overexpression resulted in reduction of rhabdomere number and size in the 1-day-old flies (C, C’) and the loss of photoreceptor cells in 30-day-old flies (D, D’). Large amount of circular membrane structures accumulated in the photoreceptor cells. ER tubules (green) attached to the mitochondria (red) were increased in the photoreceptor cells (C’). (E–F) GMR-Gal4-driven Marf overexpression did not change the number and size of rhabdomeres in the 1-day-old flies (E, E’), but slightly reduced the number and size of rhabdomeres in the 30-day-old animals (F, F’). (G–H’) GMR-Gal4-driven MitoPLD overexpression did not affect the number and size of rhabdomeres in both 1-day and 30-day-old animals. (A’–H’) are enlarged views of the photoreceptor cells highlighted with yellow pseudo-color in (A–H). The ER tubules was marked with green pseudo-color and the mitochondria was marked by the red pseudo-color. (I) Quantification of the rhabdomere numbers per ommatidia in the 1-day-old flies with indicated genotypes. n = 12 for each genotype, data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (J) Quantification of the rhabdomere numbers per ommatidia in the 30-day-old flies with indicated genotypes. n = 12 for each genotype, data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (K) Quantification of the rhabdomere size in the 1-day-old flies with indicated genotypes. n = 27 for each genotype, data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (L) Quantification of the rhabdomere size in in the 30-day-old flies with indicated genotypes. n = 22 for each genotype, data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test.
-
Figure 1—source data 1
The numerical data that are represented as a graph in Figure 1I.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig1-data1-v2.xlsx
-
Figure 1—source data 2
The numerical data that are represented as a graph in Figure 1J.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig1-data2-v2.xlsx
-
Figure 1—source data 3
The numerical data that are represented as a graph in Figure 1K.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig1-data3-v2.xlsx
-
Figure 1—source data 4
The numerical data that are represented as a graph in Figure 1L.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig1-data4-v2.xlsx
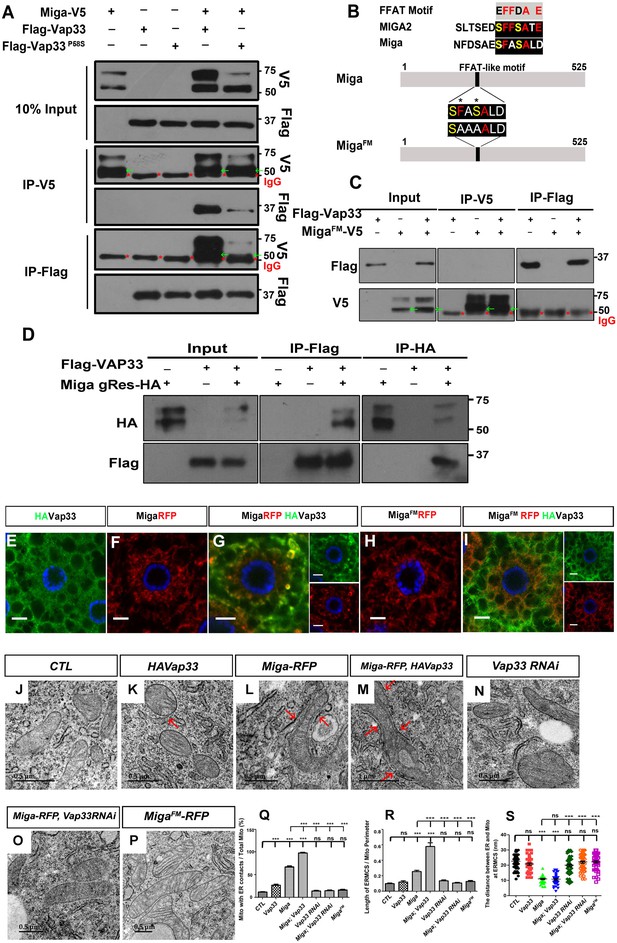
Miga forms complex with Vap33 and mediates ERMCSs.
(A) Miga-V5 and Vap33-Flag could pull down each other in both directions in the IP assay when both were overexpressed in S2 cells. The affinity between Miga-V5 and Vap33P58S-Flag was reduced compared with that between Miga-V5 and wildtype Vap33-Flag. Miga-V5 showed two bands in most of the blots. The lower bands have a molecular weight close to 50 KD (indicated with green arrows), which were often merged with the IgG heavy chain (indicated with red *) in the blots after IP experiments. (B) A scheme to show the typical FFAT motif, the FFAT-like motifs in human MIGA2 and Drosophila Miga protein, and the amino acid changed in MigaFM. (C) MigaFM-V5 and Vap33-Flag failed to pull down each other in the IP assay. MigaFM-V5 showed two bands in most of the blots. The lower bands have a molecular weight close to 50 KD (indicated with green arrows), which were often merged with the IgG heavy chain (indicated with red *) in the blots after IP experiments. (D) Genomic rescue fragment of Miga with HA tags (Miga gRes-HA, mimics the endogenous Miga expression level ) and Vap33-Flag were expressed in S2 cells and the IP assays were perfromed by IP with anti-HA or IP with anti-Flag antibodies. Miga and Vap33 could pull down each other in this condition. (E–I) HA-tagged Vap33 (green), RFP-tagged Miga or MigaFM (red) were overexpressed in fat body tissues with indicated combinations. When Vap33 and Miga were co-expressed, they colocalized with each other (G) and the patterns of both proteins were different from the patterns when they were expressed individually (E, F). (H, I) MigaFM over-expression failed to recruit Vap33. (J–P) TEM of the fat body tissues with indicated genotypes. Miga overexpression increased ERMCSs (L). Co-expression of Miga and Vap33 further increased ERMCSs (M). Vap33 RNAi did not affect ERMCSs (N). Miga overexpression could not induce ERMCS increase when Vap33 were knock down by RNAi (O). MigaFM overexpression did not affect ERMCSs (P). The red arrows indicate the ERMCSs. (Q) Quantification of the proportion of mitochondria with ERMCSs. n = 6 images for each genotype. Data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (R) Quantification of ratio between the length of ERMCSs and the mitochondrial perimeter. n = 16 for each genotype. Data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (S) Quantification of the distance between ER and mitochondria at ERMCSs. n = 50 for each genotype. Data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test.
-
Figure 2—source data 1
The numerical data that are represented as a graph in Figure 2Q.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig2-data1-v2.xlsx
-
Figure 2—source data 2
The numerical data that are represented as a graph in Figure 2R.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig2-data2-v2.xlsx
-
Figure 2—source data 3
The numerical data that are represented as a graph in Figure 2S.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig2-data3-v2.xlsx
-
Figure 2—source data 4
The mass spectrometry data to identify the binding partners of Miga.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig2-data4-v2.xlsx
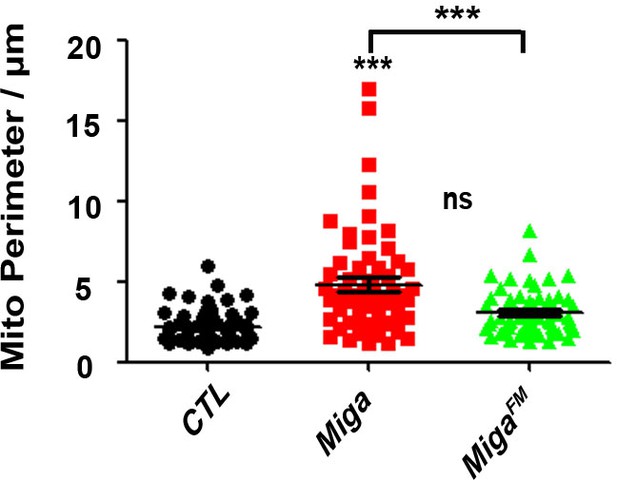
FFAT motif is required for the pro-fusion activity of Miga.
The quantification of mitochondrial perimeter in the early third instar larvae fat body tissues with indicated genotypes. n = 60 for each genotype. Data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. TEM images were collected for the early third instar larvae fat body tissues thin sections with indicated genotypes. The perimeter of the mitochondria was measured by Image J. Wildtype Miga overexpression but not MigaFM overexpression increased the mitochondrial perimeter.
-
Figure 2—figure supplement 1—source data 1
The numerical data that are represented as a graph in Figure 2—figure supplement 1.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig2-figsupp1-data1-v2.xlsx
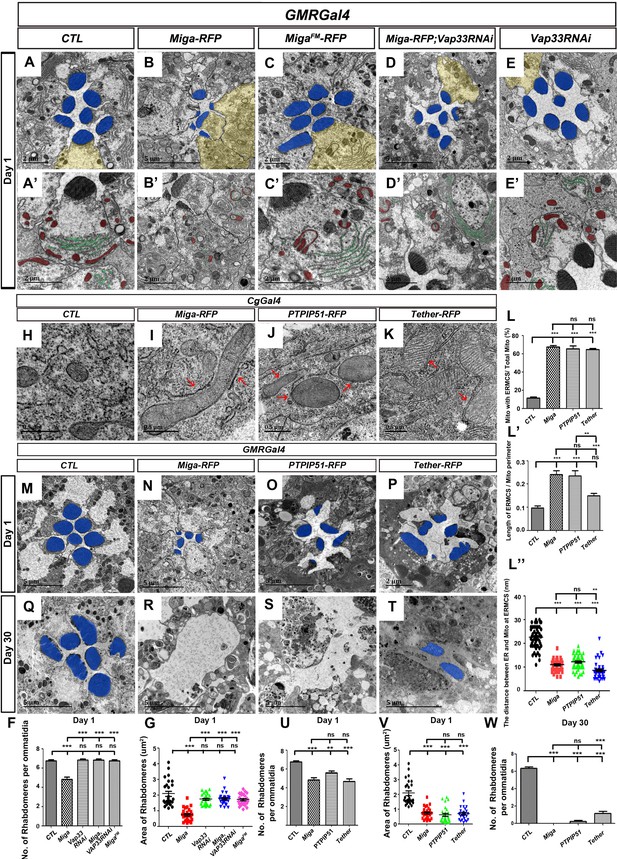
Increased ERMCSs led to neurodegeneration.
The interaction between Miga and Vap33 is critical for the eye degeneration caused by Miga overexpression. TEM analysis was performed for the retina thin sections of 1-day-old flies with indicated genotypes. (A, A’) The ommatidia of the control (CTL) flies showed seven photoreceptor cells with intact rhabdomeres (highlighted with blue pseudo-color). (B, B’) GMR-Gal4 driven Miga-RFP overexpression resulted in reduced number and size of rhabdomeres in the 1-day-old flies. (C, C’) GMR-Gal4-driven MigaFM-RFP overexpression did not affect the number and size of rhabdomeres in the 1-day-old flies. (D, D’) GMR-Gal4-driven Miga-RFP overexpression together with Vap33 RNAi did not affect the number and size of rhabdomeres in the 1-day-old flies. (E, E’) GMR-Gal4-driven Vap33 RNAi did not cause obvious defects in the 1-day-old fly eyes. (A'-E') were enlarged views of the photoreceptor cells highlighted with yellow pseudo-color in (A–E). (F) Quantification of the rhabdomere numbers per ommatidia of fly eyes with indicated genotypes. n = 12 for each genotype, data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (G) Quantification of the rhabdomere size of the fly eyes with indicated genotypes. n = 27 for each genotype, data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (H–K) TEM analysis was performed for the fat body thin sections of the early third instar larvae with indicated genotypes. Miga-RFP, human PTPIP51-RFP and the artificial ER- mitochondrial tether (Tether-RFP) overexpression led to increase of ERMCS. Red arrows indicate ERMCSs. (L–L’’) Quantification of the ERMCSs in the fat body tissues with indicated genotypes. Data are represented as mean + SD. ns, not significant; **, p<0.01, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (M–T) TEM analysis was performed for the retina thin sections of 1-day (M–P) and 30-day-old flies (Q–T) with indicated genotypes. Miga, PTPIP51 and Tether overexpression led to severe retinal degeneration in fly eyes with aging. (U) Quantification of the rhabdomere numbers per ommatidia of the fly eyes with indicated genotypes. n = 12 for each genotype, Data are represented as mean + SD. ns, not significant; **, p<0.01, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (V) Quantification of the rhabdomere size of the fly eyes with indicated genotypes. n = 27 for each genotype, data are represented as mean + SD. ns, not significant; **, p<0.01, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (W) Quantification of the rhabdomere numbers of the fly eyes with indicated genotypes. n = 12 for each genotype, data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test.
-
Figure 3—source data 1
The numerical data that are represented as a graph in Figure 3F.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-data1-v2.xlsx
-
Figure 3—source data 2
The numerical data that are represented as a graph in Figure 3G.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-data2-v2.xlsx
-
Figure 3—source data 3
The numerical data that are represented as a graph in Figure 3L.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-data3-v2.xlsx
-
Figure 3—source data 4
The numerical data that are represented as a graph in Figure 3L’.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-data4-v2.xlsx
-
Figure 3—source data 5
The numerical data that are represented as a graph in Figure 3L’’.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-data5-v2.xlsx
-
Figure 3—source data 6
The numerical data that are represented as a graph in Figure 3U.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-data6-v2.xlsx
-
Figure 3—source data 7
The numerical data that are represented as a graph in Figure 3V.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-data7-v2.xlsx
-
Figure 3—source data 8
The numerical data that are represented as a graph in Figure 3W.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-data8-v2.xlsx
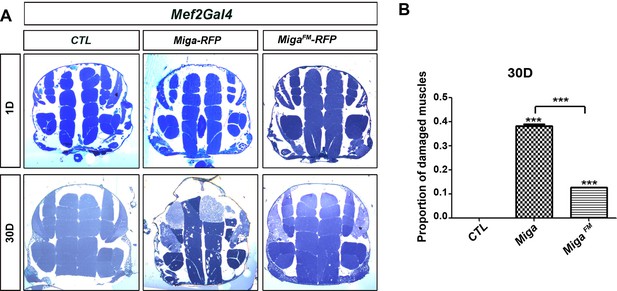
FFAT motif are required for the activity of Miga.
(A) The thick sections of adult fly flight muscles in the young (1 Day) and old (30 Day) flies with indicated genotypes. Mef2-Gal4 driven UAS-Miga-RFP overexpression induced severe muscle degeneration in the 30-day-old flies. However, MigaFM overexpression in muscles only led to mild muscle degeneration in the 30-day-old flies. (B) The quantification of the proportion of damaged muscle fragments in the thick sections of the 30-day-old flies with indicated genotype. n = 3 images for each genotype. Data are represented as mean + SD. ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test.
-
Figure 3—figure supplement 1—source data 1
The numerical data that are represented as a graph in Figure 3—figure supplement 1B.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig3-figsupp1-data1-v2.xlsx
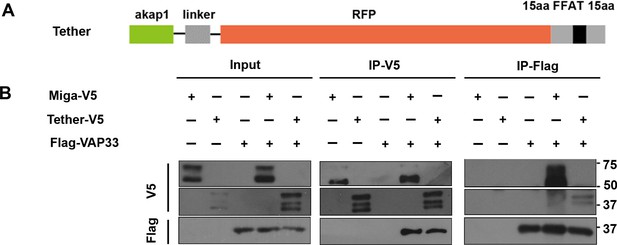
The artificial ER-mitochondria tether interacts with Vap33.
(A) A diagram shows the design of the ER- mitochondrial tether. (B) IP experiments indicated that the tether could interact with VAP33. V5 tagged Miga was used as a control.
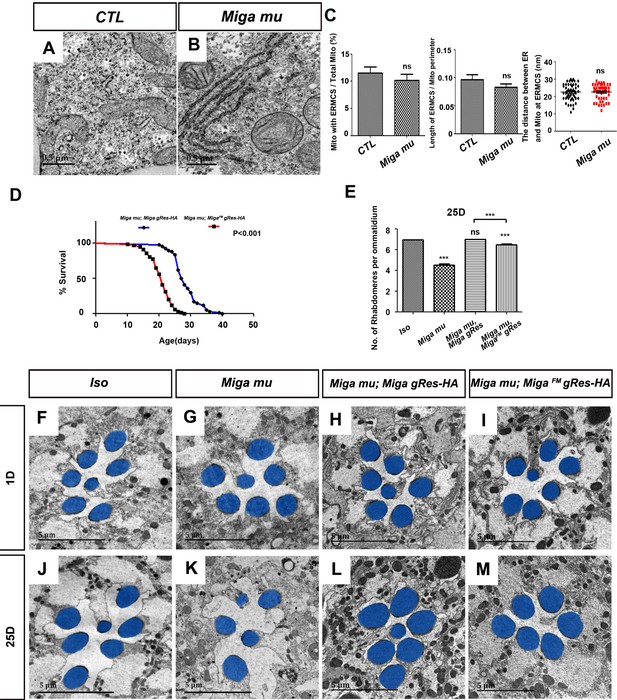
FFAT motif is important for the physiological functions of Miga.
(A, B) TEM analysis was performed for the fat body thin sections of the early third instar larvae with indicated genotypes. The loss of Miga did not affect MERCS significantly in fat body tissues. (C) Quantification of the ERMCSs in the fat bodies of the indicated genotypes. ns, not significant; two-tailed unpaired t-test. (D) The genomic fragment of Miga with FFAT motif mutated (MigaFMgRes-HA) could rescue the fatality of Miga mutants (Miga mu), but the rescued flies (Miga mu; MigaFM gRes-HA) have reduced life span compared with Miga mutants rescued with wildtype Miga genomic fragment (Miga mu; Miga gRes-HA). The male flies were analyzed. p<0.001, log rank test; n = 100 flies. (E) Quantification of the rhabdomere numbers per ommatidia of the fly eyes with indicated genotypes n = 50 for each genotype, data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (F–M) TEM analysis was performed for the retina thin sections of 1-day and 25-day-old flies with indicated genotypes. The seventh/eighth photoreceptor cells often degenerated in the 25-day-old Miga mu; MigaFM gRes-HA flies. The rhabdomeres were highlighted with blue pseudo-color.
-
Figure 4—source data 1
The numerical data that are represented as a graph in Figure 4C.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig4-data1-v2.xlsx
-
Figure 4—source data 2
The numerical data that are represented as a graph in Figure 4D.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig4-data2-v2.xlsx
-
Figure 4—source data 3
The numerical data that are represented as a graph in Figure 4E.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig4-data3-v2.xlsx
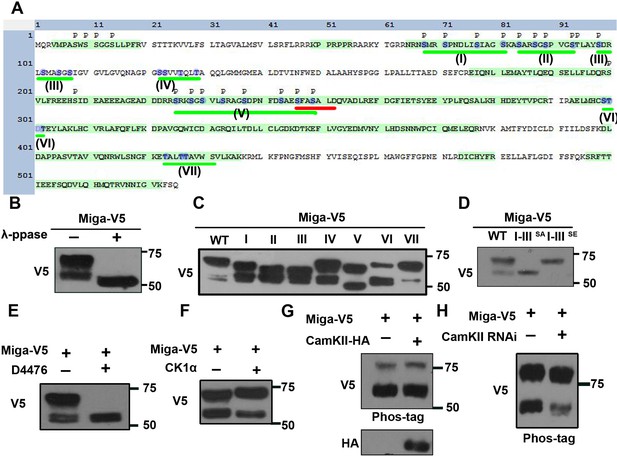
Miga was phosphorylated at multiple clusters.
(A) A diagram showed the protein sequence of Miga. The sequences highlighted with green color were the peptides identified in the mass spectrometry analysis. The phosphorylation sites identified in the mass spectrometry analysis were labeled with ‘p’. The Ser/Thr clusters were underlined with green lines and labeled with I-VII sequentially. The blue color highlighted the amino acid residues were mutated to Ala or Glu in our study. The FFAT motif was underlined with red line. (B) λ-phosphatase treatment abolished the upper shift of V5-tagged Miga protein in the western blot assay. (C) V5-tagged Miga proteins with mutated Ser/Thr in cluster I, II, III, IV, V, VI or VII were analyzed by western blot and the mobility shift of the mutated proteins were compared with the wildtype Miga. (D) Miga proteins with Ser/Thr residues in cluster I-III mutated to Ala (I-IIISA) or Glu (I-IIISE) were analyzed by western blot the mobility shifts of the mutated proteins were compared with the wildtype Miga (WT). (E) CKI inhibitor D4476 inhibited the shift of the upper band of Miga. (F) Coexpressing CKIα with Miga increased the proportion of the upper band vs lower bands. (G) Overexpressed CAMKII together with Miga led to up-shift of both lower and upper bands of Miga in a phospho-tag gel. (H) Phospho-tag gel analysis together with western blot indicated that a slight reduction of band shift when CAMKII was knocked down.
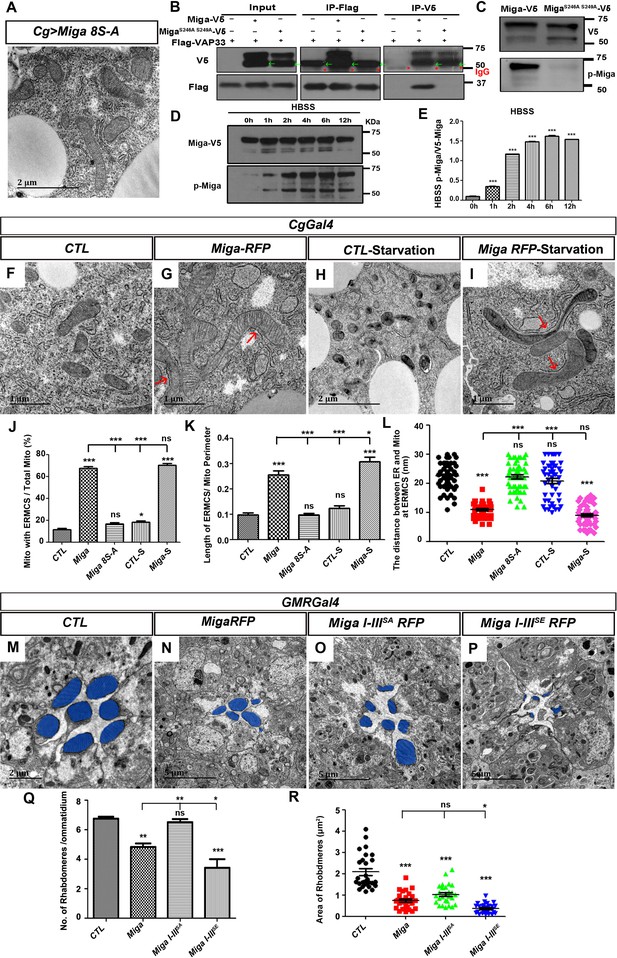
Hyper-phosphorylation regulates ERMCS formation and fine-tuned activity of Miga.
(A) TEM analysis was performed for the fat body thin sections of the early third instar larvae. Overexpression of the mutant form of Miga with Ser residues in the V cluster mutated to Ala (Miga 8S-A) did not affect ERMCSs in fat body tissues. (B) Miga with Ser 246 and Ser 249 mutated to Ala (MigaS246A, S249A) fail to bind to Vap33. MigaS246A, S249A-V5 showed two bands in most of the blots. The lower bands have a molecular weight close to 50 KD (indicated with green arrows), which were often merged with the IgG heavy chain (indicated with red *) in the blots after IP experiments. (C) A phospho-specific antibody of Miga (p-Miga) recognize overexpressed wildtype Miga-V5 but not MigaS246A, S249A-V5. Western blot with anti-V5 antibody indicated that both proteins were expressed at comparable levels. (D, E) HBSS treatment increased the phosphorylation on the Ser246 and Ser249 residues. (E) was the quantification of the ratios between p-Miga and total V5 tagged Miga when the cells were treated with HBSS. n = 3. Data are represented as mean + SD. ns, not significant; *, p<0.05, **, p<0.01, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (F–I) TEM analysis was performed for the thin sections of fly early third instar larval fat body tissues with indicated genotypes and treatments. Starvation was performed by treating the dissected fat body tissues with HBSS for 6 hr. (J–L) Quantification of the ERMCSs in the fat body tissues with indicated genotypes and treatments. Data are represented as mean + SD. ns, not significant; *, p<0.05, **, p<0.01, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. Starvation slightly increased the proportion of mitochondria with ERMCSs in the control group (CTL-S). When Miga was overexpressed, starvation increased the length of ERMCSs per mitochondria. (M–P) TEM analysis was performed for the retina thin sections of 1-day-old flies with indicated genotypes. Overexpression of Miga I-IIISE led to more severe eye defects than the overexpression of wild type Miga or Miga I-IIISA. Overexpression Miga I-IIISA had the weakest eye defects when compared with the overexpression of the wildtype Miga or the overexpression of Miga I-IIISE. The rhabdomeres were highlighted with blue pseudo-color. (Q) Quantification of the rhabdomere numbers per ommatidia in the fly eyes with indicated genotypes. n = 12 for each genotype, data are represented as mean + SD. ns, not significant; *, p<0.05, **, p<0.01, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (R) Quantification of the rhabdomere size in the fly eyes with indicated genotypes. n = 27 for each genotype, data are represented as mean + SD. ns, not significant; *, p<0.05, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test.
-
Figure 6—source data 1
The numerical data that are represented as a graph in Figure 6E.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-data1-v2.xlsx
-
Figure 6—source data 2
The numerical data that are represented as a graph in Figure 6J.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-data2-v2.xlsx
-
Figure 6—source data 3
The numerical data that are represented as a graph in Figure 6K.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-data3-v2.xlsx
-
Figure 6—source data 4
The numerical data that are represented as a graph in Figure 6L.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-data4-v2.xlsx
-
Figure 6—source data 5
The numerical data that are represented as a graph in Figure 6Q.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-data5-v2.xlsx
-
Figure 6—source data 6
The numerical data that are represented as a graph in Figure 6R.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-data6-v2.xlsx
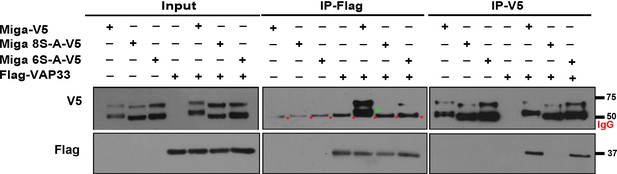
The phosphorylation on the cluster V Ser residues are critical for the binding between Miga and Vap33.
V5-tagged wildtype Miga or indicated mutant forms of Miga co-expressed with Flag-tagged Vap33 in S2 cells. Miga 8SA is the mutant form of Miga with 8 Ser in the cluster V mutated to Ala. Miga 6SA is the mutant form of Miga with 6 Ser in the cluster V mutated to Ala and two Ser residues in the FFAT motif are intact. The IP expreiments were performed by IP with V5 or IP with Flag. The input and the pulldown products were detected by western blot with indicated antibodies. The lower bands of Miga have a molecular weight close to 50 KD (indicated with green arrows), which were often merged with the IgG heavy chain (indicated with red *) in the blots after IP experiments.
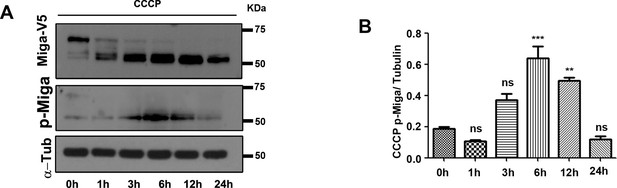
The phosphorylation of Miga was regulated upon stimulation.
(A) The phosphorylation of Ser246 and Ser249 of Miga is increased with short time exposure to CCCP (6 hr). But the hyperphosphorylation of Miga was reduced upon CCCP treatment. (B) was the quantification of the ratios between p-Miga and α-tubulin when the cells as treated with CCCP with indicated time period. n = 3. Data are represented as mean + SD. ns, not significant; *, p<0.05, **, p<0.01, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test.
-
Figure 6—figure supplement 2—source data 1
The numerical data that are represented as a graph in Figure 6—figure supplement 2B.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-figsupp2-data1-v2.xlsx
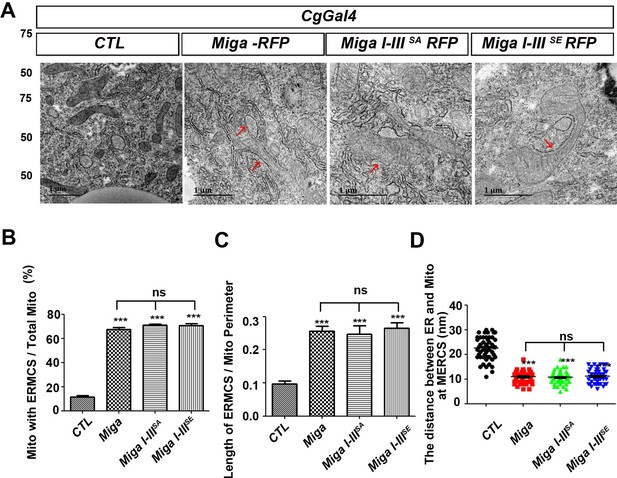
The phosphorylation of Miga on cluster I-III did not affect its ability to mediate ERMCSs.
(A) TEM analysis was performed for the fat body thin sections of the early third instar larvae. Miga I-IIISA or Miga I-IIISE overexpression increased ERMCSs in a similar manner as the wildtype Miga overexpression did. (B) Quantification of the proportion of mitochondria with ERMCSs. n = 6 images for each genotype. Data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (C) Quantification of ratio between the length of ERMCSs and the mitochondrial perimeter. n = 17 for each genotype. Data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (D) Quantification of the distance between ER and mitochondria at ERMCSs. n = 50 for each genotype. Data are represented as mean + SD. ns, not significant; ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test.
-
Figure 6—figure supplement 3—source data 1
The numerical data that are represented as a graph in Figure 6—figure supplement 3B.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-figsupp3-data1-v2.xlsx
-
Figure 6—figure supplement 3—source data 2
The numerical data that are represented as a graph in Figure 6—figure supplement 3C.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-figsupp3-data2-v2.xlsx
-
Figure 6—figure supplement 3—source data 3
The numerical data that are represented as a graph in Figure 6—figure supplement 3D.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-figsupp3-data3-v2.xlsx
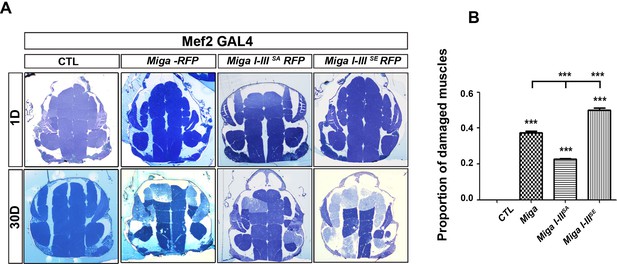
The phosphorylation on the I-III clusters on Miga increased Miga activity.
(A) The thick sections of adult fly flight muscles in the young (1 Day) and old (30 Day) flies with indicated genotypes. Mef2-Gal4 driven UAS-Miga-RFP overexpression induced severe muscle degeneration in the 30-day-old flies. Miga I-IIISE overexpression caused more severe muscle degeneration in 30-day-old flies than the wildtype Miga overexpression. Miga I-IIISA overexpression led to less severe muscle degeneration in the aged flies than the wildtype Miga overexpression. (B) The quantification of the proportion of damaged muscle fragments in the thick sections of the 30 day old flies with indicated genotype. n = 3 images for each genotype. Data are represented as mean + SD. ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test.
-
Figure 6—figure supplement 4—source data 1
The numerical data that are represented as a graph in Figure 6—figure supplement 4B.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig6-figsupp4-data1-v2.xlsx
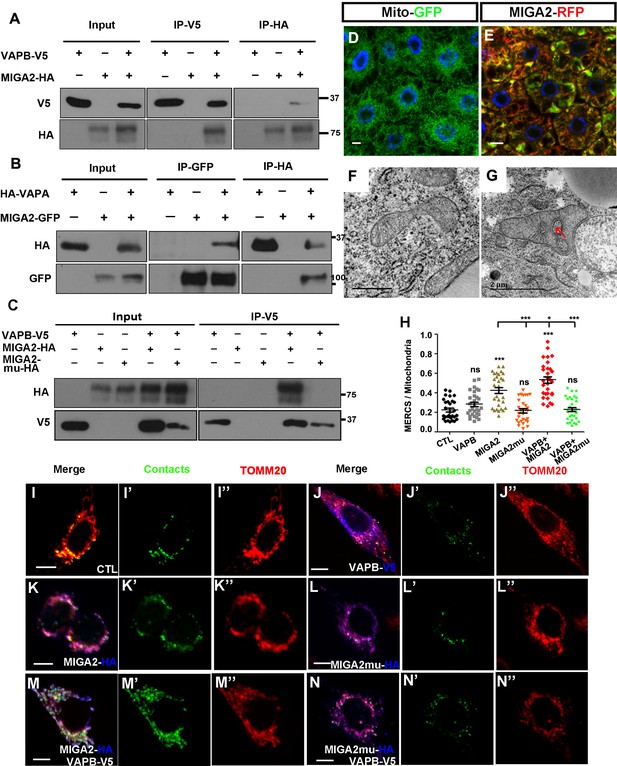
The mammalian homolog of Miga, MIGA2, had conserved function in ERMCS formation.
(A) MIGA2-HA and VAPB-V5 could pull down each other in both directions in the IP assay. (B) MIGA2-GFP and VAPA-HA could pull down each other in both directions in the IP assay. (C) Mutating the FFAT motif in MIGA2 (MIGA2-mu) abolished the interaction between VAPB and MIGA2 in the IP assay. (D and E) Human MIGA2RFP (red) ectopically overexpressed in fly fat body tissues led to the change of MitoGFP (green) patterns. (F and G) TEM analysis were performed for the early third instar larval fat body tissues with MitoGFP (F) or MIGA2RFP together with MitoGFP overexpressed (G). MIGA2RFP overexpression increased ERMCS. (H–N”) MIGA2 overexpression but not MIGA2-mu overexpression increased the signals of MERCSs. VAPB co-expression with MIGA2 further increased MERCSs. (H) Quantification of the ratio between contacts and the total mitochondrial signals of the cells with indicated genotypes. n = 30 for each genotype, ns, not significant; *, p<0.05, ***, p<0.001; one-way ANOVA/Bonferroni’s multiple comparisons test. (I–N’’) A genetic encoded split-GFP based MERCS reporter (green) stably expressed in U2OS cells was used to indicate MERCS. Mitochondria were labeled with anti-TOMM20 staining (red). The expression of VAPB, MIGA2 or MIGA2-mu were indicated by anti-V5 or anti-HA staining (blue).
-
Figure 7—source data 1
The numerical data that are represented as a graph in Figure 7M.
- https://cdn.elifesciences.org/articles/56584/elife-56584-fig7-data1-v2.xlsx
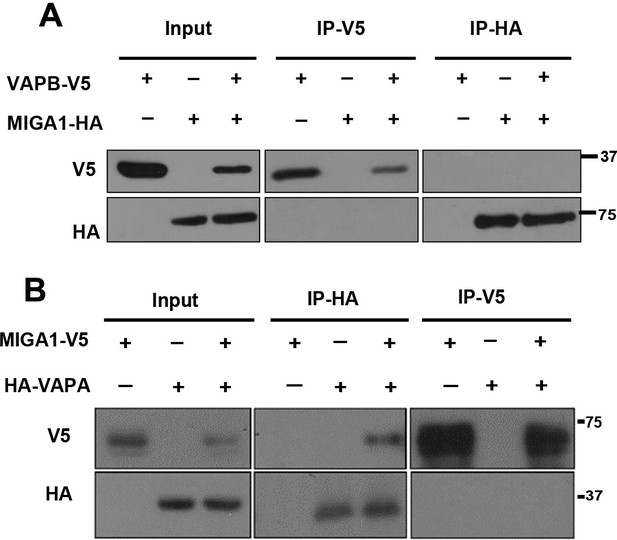
MIGA1 binds to VAPA weakly in HeLa cells when overexpressed.
(A) HA-tagged MIGA1 and V5-tagged VAPB were co-expressed in HeLa cells. MIGA1 and VAPB failed to pulldown each other in the IP experiments. (B) V5-tagged MIGA1 and HA-tagged VAPA were co-expressed in Hela cells. VAPA could pulldown MIGA1, but MIGA1 fail to pulldown VAPA in the IP experiments.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Drosophila melanogaster) | Miga | GenBank | FLYB:FBgn0030037 | |
| Gene (Drosophila melanogaster) | marf | GenBank | FLYB:FBgn0029870 | |
| Gene (Drosophila melanogaster) | MitoPLD | GenBank | FLYB:FBgn0261266 | |
| Gene (Drosophila melanogaster) | Vap33 | GenBank | FLYB:FBgn0029687 | |
| Gene (Homo- sapiens ) | Miga1 | GenBank | HGNC:24741 | |
| Gene (Homo- sapiens ) | PTPIP51 | GenBank | HGNC:25550 | |
| Gene (Homo- sapiens ) | Miga2 | GenBank | HGNC:23621 | |
| Gene (Homo- sapiens ) | VAPA | GenBank | HGNC:12648 | |
| Gene (Homo- sapiens ) | VAPB | GenBank | HGNC:12649 | |
| Cell line (Drosophila melanogaster) | S2 | This paper | FLYB:FBtc0000181; RRID:CVCL_Z992 | |
| Cell line (Homo sapiens) | U2OS | This paper | CLS Cat# 300364/p489_U-2_OS, RRID:CVCL_0042 | |
| Cell line (Homo sapiens) | Hela | This paper | CLS Cat# 300194/p772_HeLa, RRID:CVCL_0030 | |
| Antibody | anti-HA (Rabbit monoclonal) | Cell Signaling | Cell Signaling Technology Cat# 3724, RRID:AB_1549585 | WB (1:1000) |
| Antibody | anti-GFP (Rabbit polyclonal) | MBL | MBL International Cat# 598, RRID:AB_591819 | WB (1:1000) |
| antibody | anti-Flag (mouse monoclonal) | Sigma | Sigma-Aldrich Cat# F3165, RRID:AB_259529 | WB (1:1000) |
| Antibody | anti-V5 (mouse monoclonal) | Invitrogen | Invitrogen: R96025 | WB(1:5000) IF(1:500) |
| Antibody | P-Miga (Rabbit monoclonal) | GL BioChem Ltd | WB (1:5000-1:10000) | |
| Chemical compound, drug | Phos-tag acrylamide | Boppard | Boppard: 300–93523 | 100 μM |
| Chemical compound, drug | D4470 | Selleck | Selleck:S7642 | 80 μM |
| Chemical compound, drug | CCCP | Sigma | Sigma:C2759 | 10 μM |
| Chemical compound, drug | HBSS | ThermoFisher | ThermoFisher: 14025076 | |
| Chemical compound, drug | PhosSTOP | Sigma | Sigma: 4906837001 | |
| chemical compound, drug | FLAG peptide | APExBIO | APExBIO: A6001 | |
| Chemical compound, drug | Paraformaldehyde | Electron Microscopy Sciences | Electron Microscopy Sciences: 15711 | 4% |
| Chemical compound, drug | Cacodylic acid | Electron Microscopy Sciences | Electron Microscopy Sciences: 12201 | 1.4% |
| Chemical compound, drug | Glutaraldehyde | Electron Microscopy Sciences | Electron Microscopy Sciences: 16020 | 1% |
| Chemical compound, drug | Osmium tetroxide | Electron Microscopy Sciences | Electron Microscopy Sciences: 19152 | 2% |
| Chemical compound, drug | Propylene oxide | Sigma | Sigma:82320 | |
| Chemical compound, drug | Embed 812 | Electron Microscopy Sciences | Electron Microscopy Sciences: 14900 | |
| Chemical compound, drug | DDSA | Electron Microscopy Sciences | Electron Microscopy Sciences: 13710 | |
| Chemical compound, drug | NMA | Electron Microscopy Sciences | Electron Microscopy Sciences:19000 | |
| Chemical compound, drug | DMP-30 | Electron Microscopy Sciences | Electron Microscopy Sciences:13600 | |
| Chemical compound, drug | Uranyl acetate | Electron Microscopy Sciences | Electron Microscopy Sciences: 22400 | 4% |
| Chemical compound, drug | Lead nitrate | Electron Microscopy Sciences | Electron Microscopy Sciences: 17800 | 2.5% |
| Chemical compound, drug | Toluidine blue | Electron Microscopy Sciences | Electron Microscopy Sciences: 22050 | |
| Other | HA beads | Sigma | Sigma:E6779 | |
| Other | Flag beads | Sigma | Sigma, A2220 | |
| Other | Protein A Sepharose 4 Fast Flow beads | GE Healthcare | GE Healthcare: 17-5280-01 | |
| Sequence-based reagent | CaMKII_F | This paper | dsRNA primers | TAATACGACTCACTATAGGGGCAAAGTCCGCTTATTCTCGTTCTT |
| Sequence-based reagent | CaMKII_R | This paper | dsRNA primers | TAATACGACTCACTATAGGGAATTCTTTGGCTCCCCTCATGC |
| Sequence-based reagent | CaMKII_F | This paper | Real-time PCR primers | ATCCCAACATAGTGCGGCTACATGA’ |
| Sequence-based reagent | CaMKII_R | This paper | Real-time PCR primers | AAGTCAGCGAGTTTCACTGCTGCA |
Additional files
-
Supplementary file 1
The genotypes of the fly strains used in this study.
- https://cdn.elifesciences.org/articles/56584/elife-56584-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/56584/elife-56584-transrepform-v2.docx





