Harmful DNA:RNA hybrids are formed in cis and in a Rad51-independent manner
Figures
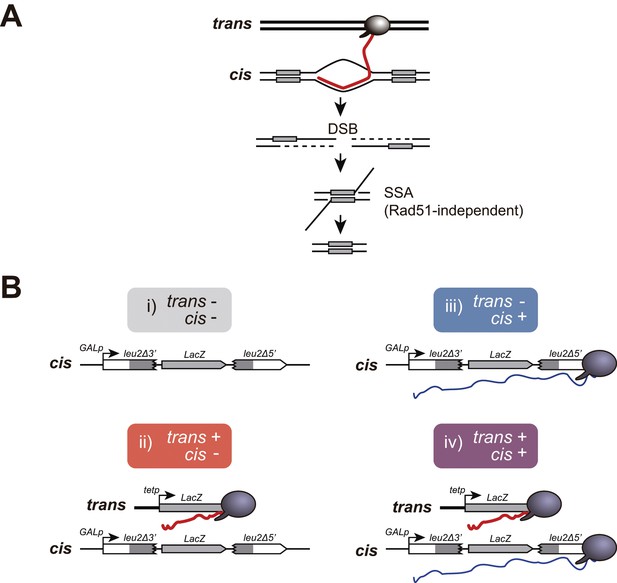
A new genetic assay to detect recombinogenic DNA:RNA hybrids in trans.
(A) DSBs induced in between direct repeats by DNA:RNA hybrids putatively formed with RNA produced in trans would be repaired by Rad51-independent Single-Strand Annealing (SSA) causing the deletion of one of the repeats. A DSB is depicted for simplicity, but other recombinogenic lesions such as nicks or ssDNA gaps cannot be ruled out. (B) Schematic representation of the recombination assay to study the recombinogenic potential RNA produced by transcription (Trx) in cis or in trans. Four combinations were studied: i) no transcription, with GL-LacZ construct turned transcriptionally off (2% glucose) and an empty plasmid; ii) transcription in trans, with GL-LacZ construct turned transcriptionally off (2% glucose) and the tetp:LacZ construct; iii) transcription in cis, with GL-LacZ construct turned transcriptionally on (2% galactose) and an empty plasmid; and iv) transcription in cis and in trans, with GL-LacZ construct turned transcriptionally on (2% galactose) and the tetp:LacZ construct.
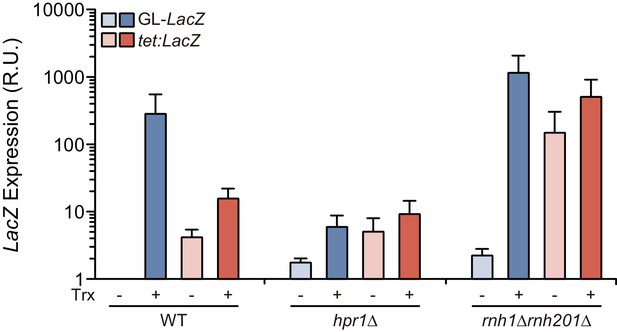
LacZ expression levels in the GL-LacZ and tetp:LacZ constructs.
Relative RNA levels at the LacZ gene from GL-LacZ and tetp:LacZ constructs when transcription was turned either off (Trx -) or on (Trx +) in WT (W303), hpr1Δ (U678.4C) and rnh1Δ rnh201Δ (HRN2.10C) strains transformed with either pRS314-GL-LacZ or pCM179. Transcription at the GL-LacZ construct was turned on or off by growth in media with 2% galactose or 2% glucose, respectively. Transcription at the tetp:LacZ construct was turned on or off by growth in media without or with 5 μg/mL doxycycline, respectively. Average and SEM of three independent experiments are shown.
-
Figure 1—figure supplement 1—source data 1
Relative RNA levels at theLacZgene from GL-LacZandtetp:LacZconstructs.
- https://cdn.elifesciences.org/articles/56674/elife-56674-fig1-figsupp1-data1-v2.xlsx
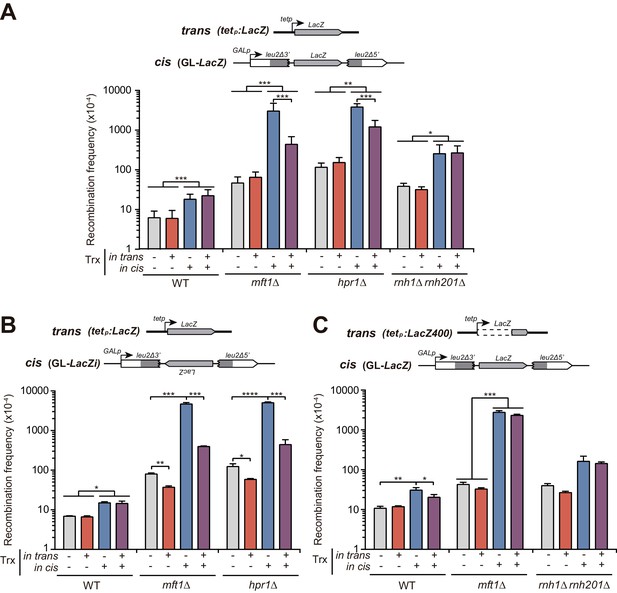
Analysis of the effect on genetic recombination of RNA produced in cis or in trans.
(A) Recombination analysis in WT (W303), rnh1Δ rnh201Δ (HRN2.10C), mft1Δ (WMK.1A) and hpr1Δ (U678.4C) strains carrying GL-LacZ plasmid system (pRS314-GL-LacZ) plus either the pCM190 empty vector or the same vector carrying the LacZ gene (pCM179). (B) Recombination analysis in WT (W303), mft1Δ (WMK.1A) and hpr1Δ (U678.4C) strains carrying GL-LacZi plasmid plus either the pCM190 empty vector or the same vector carrying the sequence of the LacZ gene (pCM179). (C) Recombination analysis in WT (W303), rnh1Δ rnh201Δ (HRN2.10C) and mft1Δ (WMK.1A) strains carrying GL-LacZ plasmid system (pRS314-GL-LacZ) plus either the pCM190 empty vector or the same vector carrying the last 400 bp from the 3’ end of the LacZ gene (pCM190:LacZ400). In all panels, average and SEM of at least three independent experiments consisting in the median value of six independent colonies each are shown. *, p≤0.05; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001 (unpaired Student’s t-test).
-
Figure 2—source data 1
Analysis of the effect on genetic recombination of RNA producedin cisorin trans.
- https://cdn.elifesciences.org/articles/56674/elife-56674-fig2-data1-v2.xlsx
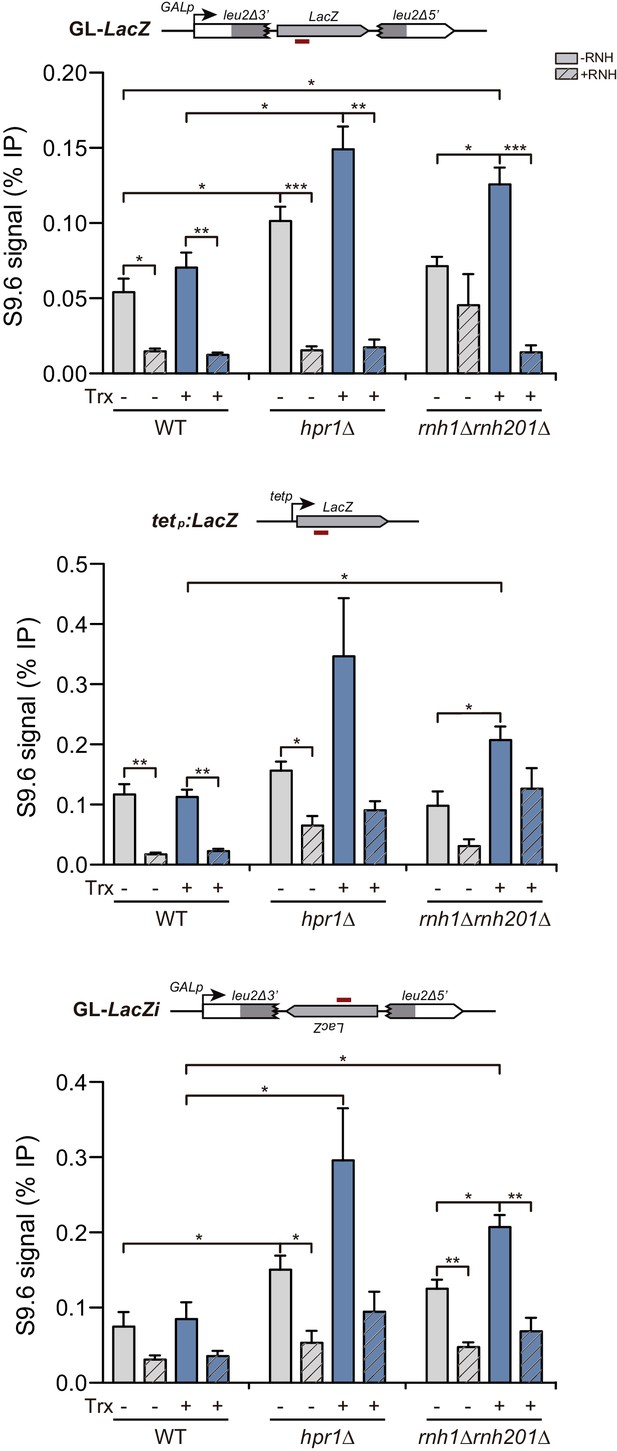
Detection of co-transcriptional DNA:RNA hybrids in hpr1Δ and rnh1Δ rnh201Δ mutants at the LacZ-containing constructs under the GAL1 or tet promoters.
DNA:RNA Immuno-Precipitation (DRIP) with the S9.6 antibody in WT (W303), hpr1Δ (U678.4C) and rnh1Δ rnh201Δ (HRN2.10C) strains in asynchronous cultures treated or not in vitro with RNase H in the GL-LacZ, tetp:LacZ and GL-LacZi constructs turned transcriptionally off (2% glucose or 5 μg/mL doxycycline) or on (2% galactose and in the absence of doxycycline). Average and SEM of three independent experiments are shown *, p≤0.05; **, p≤0.01; ***, p≤0.001 (unpaired Student’s t-test).
-
Figure 3—source data 1
Detection of co-transcriptional DNA:RNA hybrids.
- https://cdn.elifesciences.org/articles/56674/elife-56674-fig3-data1-v2.xlsx
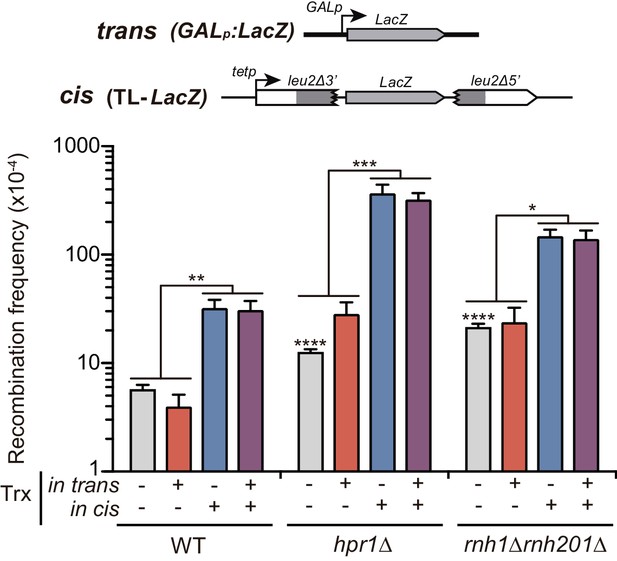
Analysis of the effect on genetic recombination of RNA produced in cis or in trans with different promoters.
Recombination analysis in WT (W303), hpr1Δ (U678.4C) and rnh1Δ rnh201Δ (HRN2.10C) carrying TL-LacZ plasmid system (pCM184-TL-LacZ) plus either the pRS416 empty vector or the same vector carrying the LacZ gene (pRS416-GALLacZ). In this case, the four combinations studied were: i) no transcription, with TL-LacZ construct turned transcriptionally off (5 μg/mL doxycycline) and an empty plasmid; ii) transcription in trans, with TL-LacZ construct turned transcriptionally off (5 μg/mL doxycycline) and the GAL-LacZ construct switched on (2% galactose); iii) transcription in cis, with TL-LacZ construct turned transcriptionally on and an empty plasmid; and iv) transcription in cis and in trans, with TL-LacZ construct turned transcriptionally on and the GAL-LacZ construct switched on (2% galactose). Average and SEM of at least three independent experiments consisting in the median value of six independent colonies each are shown. *, p≤0.05; **, p≤0.01; ***, p≤0.001; ****, p≤0.0001 (unpaired Student’s t-test).
-
Figure 4—source data 1
Analysis of the effect on genetic recombination of RNA producedin cisorin transwith different promoters.
- https://cdn.elifesciences.org/articles/56674/elife-56674-fig4-data1-v2.xlsx
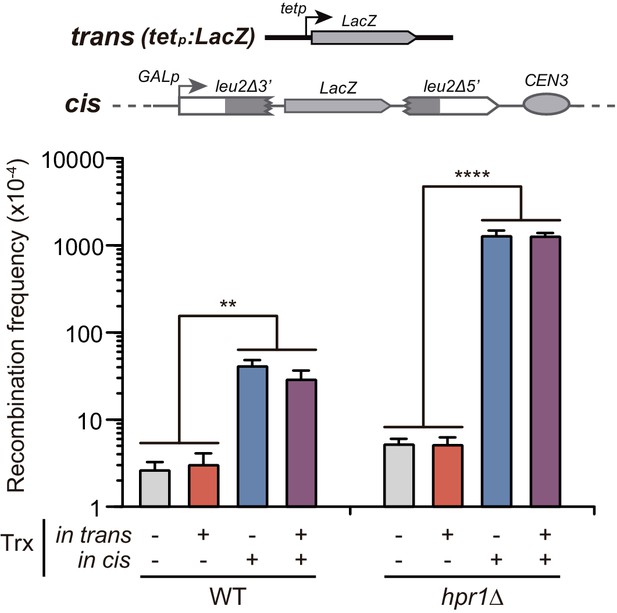
Analysis of the effect on genetic recombination of RNA produced in trans of chromosome III.
Recombination analysis in WT (WGLZN), hpr1Δ (HGLZN) strains carrying the GL-LacZ recombination system integrated in chromosome III. Strains were transformed with empty vector pCM190 or the same vector carrying the LacZ gene (pCM179). Average and SEM of at least three independent experiments are shown consisting in the median value of six independent colonies each. **, p≤0.01; ****, p≤0.0001 (unpaired Student’s t-test).
-
Figure 5—source data 1
Analysis of the effect on genetic recombination of RNA producedin transof chromosome III.
- https://cdn.elifesciences.org/articles/56674/elife-56674-fig5-data1-v2.xlsx
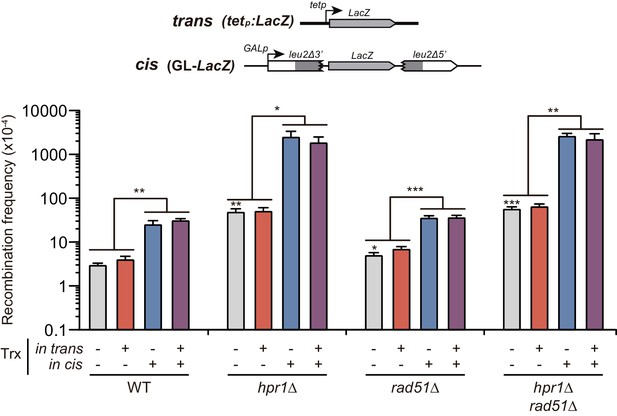
Analysis of the effect on genetic recombination of RNA produced in trans with or without Rad51.
Recombination analysis in WT (W303), hpr1Δ (U678.1C), rad51Δ (WSR51.4A) and hpr1Δ rad51Δ (HPR51.15A) strains carrying GL-LacZ plasmid system (pRS314-GL-LacZ) plus either the pCM190 empty vector or the same vector carrying the LacZ gene (pCM179). Average and SEM of at least three independent experiments consisting in the median value of six independent colonies each are shown. *, p≤0.05; **, p≤0.01, ***, p≤0.001 (unpaired Student’s t-test).
-
Figure 6—source data 1
Analysis of the effect on genetic recombination of RNA producedin transwith or without Rad51.
- https://cdn.elifesciences.org/articles/56674/elife-56674-fig6-data1-v2.xlsx
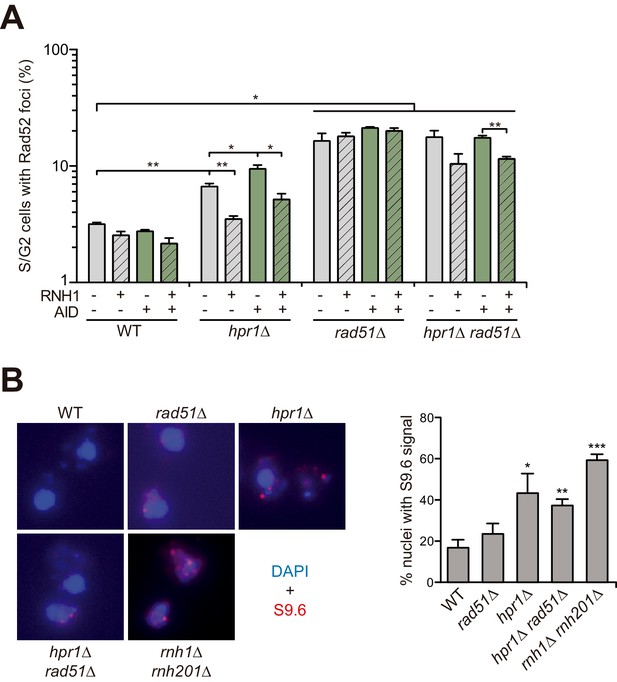
The increased genetic instability and DNA:RNA hybrids of hpr1∆ are independent on Rad51.
(A) Spontaneous Rad52-YFP foci formation in WT (W303), hpr1Δ (U678.1C), rad51Δ (WSR51.4A) and hpr1Δ rad51Δ (HPR51.15A) strains carrying the empty vectors pCM184 and pCM189, or a combination of both carrying the RNH1 or AID genes as indicated in the legend. (B) Representative images and value of the percent of the total nuclei scored that stained positively for DNA:RNA hybrids in chromatin spreads stained with the S9.6 antibody in WT (W303), hpr1Δ (U678.1C), rad51Δ (WSR51.4A), hpr1Δ rad51Δ (HPR51.15A) and RNH-R (rnh1Δ rnh201Δ) strains. In both panels, average and SEM of at least three independent experiments performed with more than 100 cells are shown. *, p≤0.05; **, p≤0.01, ***, p≤0.001 (unpaired Student’s t-test).
-
Figure 7—source data 1
Genetic instability and DNA:RNA hybrids in the absence of Rad51.
- https://cdn.elifesciences.org/articles/56674/elife-56674-fig7-data1-v2.xlsx
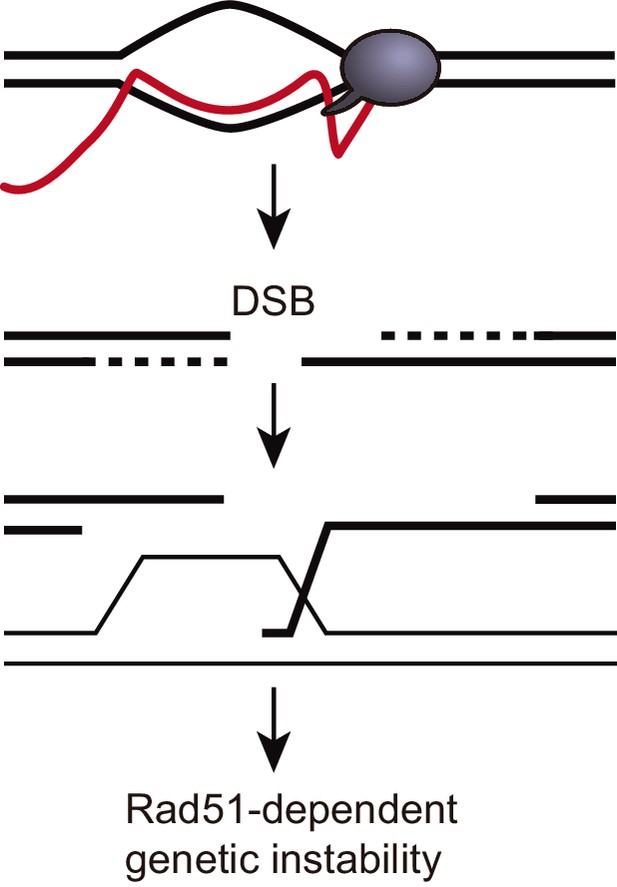
A model to explain how DNA:RNA hybrids could induce Rad51-dependent genetic instability in trans.
DNA:RNA hybrid produced in cis can induce a DSB in the same sequence. The 3’ end of such a DSB could invade an ectopic homologous sequence and destabilize it. This DNA strand invasion event would require Rad51. In this model, genetic instability caused by hybrids in trans would be Rad51-dependent without the need of invoking a Rad51-mediated DNA:RNA hybrid formation in trans. A DSB is depicted for simplicity, but other recombinogenic lesions such as nicks or ssDNA gaps cannot be ruled out.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent Saccharomyces cerevisiae | W303 background strains with different gene deletions | various | (See Materials and methods section) | |
| Recombinant DNA reagent | Yeast expression plasmids and recombination systems | various | (See Materials and methods section) | |
| Sequence-based reagent | Primers for DRIP and RT-PCR | Condalab | (See Materials and methods section) | |
| Antibody | Cy3 conjugated anti-mouse (goat monoclonal) | Jackson laboratories | Cat# 115-165-003, RRID:AB_2338680 | IF (1:1000) |
| Antibody | S9.6 anti DNA:RNA hybrids (mouse monoclonal) | ATCC Hybridoma cell line | Cat# HB-8730, RRID:CVCL_G144 | DRIP (1 mg/ml) and IF (1:1000) |
| Commercial assay, kit | Macherey-Nagel DNA purification | Macherey- Nagel | Cat# 740588.250 | |
| Commercial assay, kit | Qiagen’s RNeasy | Quiagen | Cat# 75162 | |
| Commercial assay, kit | Reverse Transcription kit | Qiagen | Cat # 205311 | |
| Peptide, recombinant protein | Zymolyase 20T | US Biological | Z1001 | (50 mg/ml) |
| Chemical compound, drug | Doxycyclin hyclate | Sigma-Aldrich | D9891 | (5 mg/ml) |
| Peptide, recombinant protein | Proteinase K (PCR grade) | Roche | Cat # 03508811103 | |
| Peptide, recombinant protein | Rnase A | Roche | Cat # 10154105103 | |
| Software, algorithm | GraphPad Prism V8.4.2 | GraphPad Software, La Jolla, CA, USA | RRID:SCR_002798 | |
| Other | iTaq Universal SYBR Green | Bio-RAD | Cat # 1725120 | |
| Other | DAPI stain | Invitrogen | D1306 | 1 µg/mL |





