Gamete expression of TALE class HD genes activates the diploid sporophyte program in Marchantia polymorpha
Figures
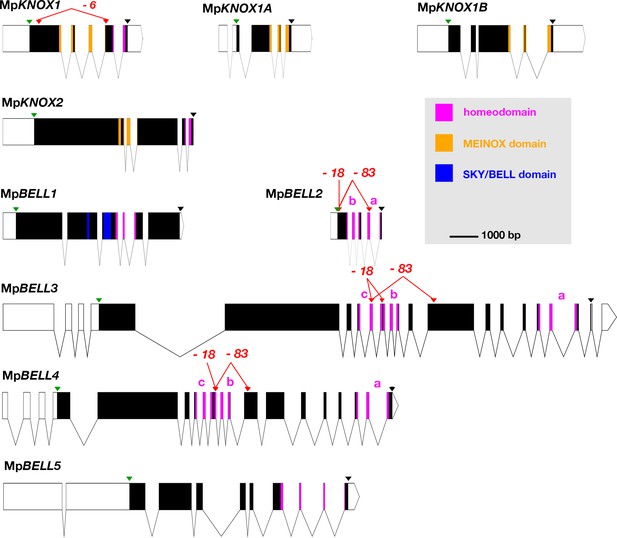
Schematic representations of the nine TALE-HD loci of Marchantia polymorpha.
White, 5' and 3' untranslated regions (UTRs); black, coding exons; thin lines, introns; green triangles, start codon; black triangles, stop codon; red triangles, guide RNA-targeted positions. The mutant alleles generated via CRISPR-Cas9 are described in more detail in table 1; molecular lesions of some alleles can be found in Figure 1—source data 1. All protein annotation models are based on the Marchantia genome assembly of v5.1 except for MpKNOX2. The MpKNOX2 model is based on sequences derived from reverse transcription-polymerase chain reaction (RT-PCR). In genes with multiple homeodomains, they are denoted a, b, and c. Gene models were assembled using wormweb (http://wormweb.org/exonintron).
-
Figure 1—source data 1
Molecular lesions of alleles listed in Figure 1 and Table 1.
- https://cdn.elifesciences.org/articles/57088/elife-57088-fig1-data1-v3.pdf
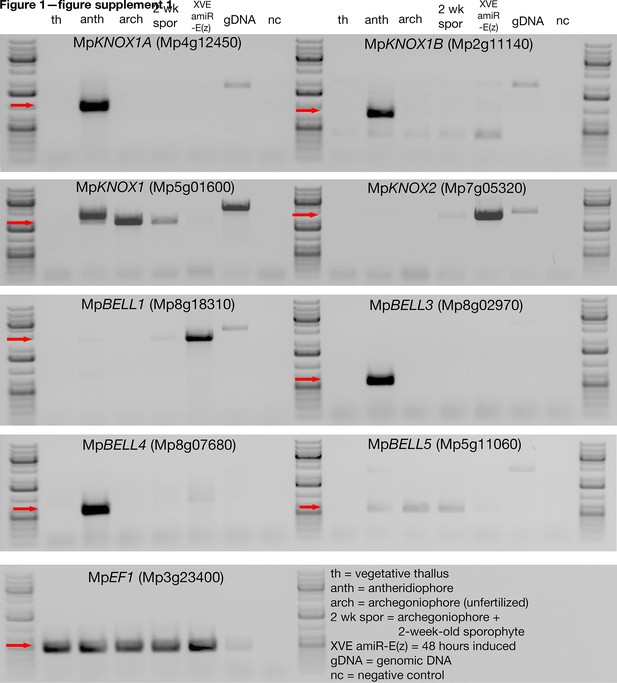
Expression of Marchantia polymorpha TALE-HD genes.
RNA extracted from various tissues of Marchantia polymorpha ssp ruderalis, ecotype BoGa, were subjected to semiquantitative reverse transcription-polymerase chain reaction (sqRT-PCR) (30 cycles, except MpKNOX1 and MpKNOX2 at 32 cycles; 55°C anneal; 1 min extension, except MpKNOX1, MpKNOX2, and MpBELL1 at 2 min). The expected size band from complementary DNA (cDNA) is indicated by a red arrow at the left of each panel. We were unable to amplify a product from MpBELL2. See (Supplementary file 1) for primer sequences.
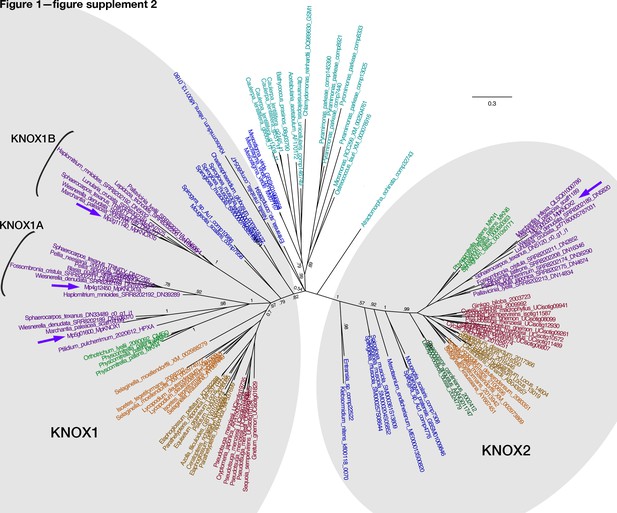
Unrooted Bayesian phylogram of Viridiplantae KNOX-TALE class homeodomain genes.
Tree constructed using nucleotide alignment of the homeodomain and MEINOX and ELK domains. The four Marchantia polymorpha KNOX genes are highlighted (purple arrows). The tree topology suggests a single KNOX gene in the common ancestor of the Viridiplantae, and possibly the Streptophyta, with a gene duplication in an ancestral charophycean alga after the divergence of Mesostigma, producing ancestral KNOX1 and KNOX2 genes (Sakakibara, 2016; Joo et al., 2018; Frangedakis et al., 2017). The KNOX2 genes, including charophyte sequences, form a well-supported monophyletic clade (posterior probability = 1). The KNOX1 clade is less well-supported with regard to the early diverging charophyte sequences; however, land plant KNOX1 genes, including MpKNOX1, share a conserved MEINOX domain (see also Figure 3—figure supplement 3). The ancestral land plant likely had single KNOX1 and KNOX2 orthologs, a condition retained in M. polymorpha. MpKNOX1A and MpKNOX1B orthologs are found throughout liverworts, suggesting their origins in the ancestral liverwort. Chlorophyte, light blue; charophyte, dark blue; liverwort, purple; moss, green; hornwort, dark green; lycophyte, brown; fern, orange; seed plant, red. Numbers indicate posterior probability values >50%.
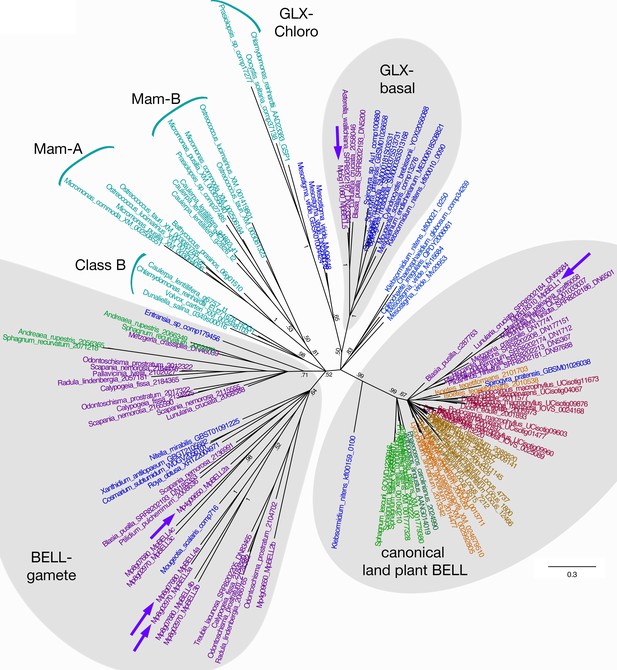
Unrooted Bayesian phylogram of Viridiplantae BELL-TALE class homeodomain genes.
Tree was constructed using a nucleotide alignment of the homeodomain. The five Marchantia polymorpha BELL genes are highlighted (purple arrows). A clade of genes representing previously identified ‘canonical’ land plant BELL genes is recovered with high support. This clade includes MpBELL1 and charophycean algal sequences dating as far back as prior to the divergence of Klebsormidium. These canonical land plant BELL genes also share significant sequence similarity outside the homeodomain, including the SKY and BELL domains. The remaining BELL genes, including all chlorophyte sequences, sequences from charophycean algae, and the other four M. polymorpha sequences, reside in either other well-supported clades or in polytomies. Several subclades (labeled) previously identified are evident (Joo et al., 2018). MpBELL2/3/4 resides in a polytomy composed of liverwort and charophyte sequences, with other liverwort, moss, and charophyte sequences residing in a second clade; these are labeled ‘BELL-gamete’. The multiple homeodomains of MpBELL2, MpBELL3 and MpBELL4 are designated a, b, and c (see Figure 1). MpBELL5 resides, with other liverwort and charophyte sequences, in the GLX-basal clade. Chlorophyte, light blue; charophyte, dark blue; liverwort, purple; moss, green; hornwort, dark green; lycophyte, brown; fern, orange; seed plant, red. Numbers at branches indicate posterior probability values >50%; branches explicitly shown have probability values >50%, whereas polytomies represent nodes with probability values <50%; values within subclades are omitted for clarity.
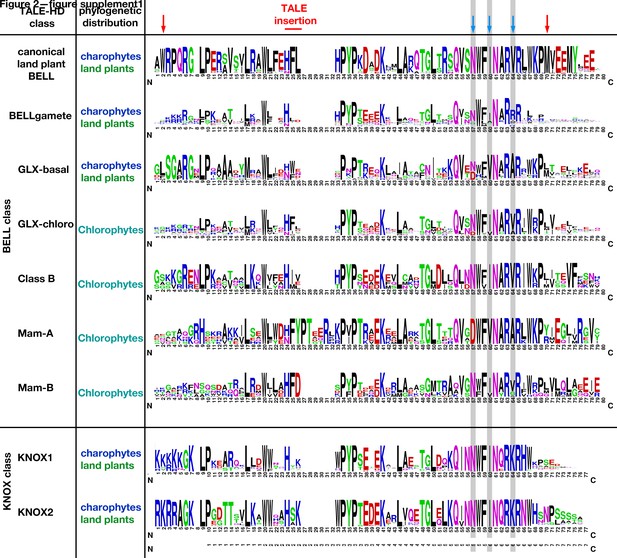
Comparison of Viridiplantae TALE homeodomains.
The alignments used in the construction of phylogenetic trees in Figure 2 and Figure 1—figure supplement 1 were used to generate consensus sequences for nine different classes of Viridiplantae TALE-homeodomain (HD) proteins. Homeodomain is flanked by red arrows. Some TALE-HD classes have insertions relative to others and some subclasses of BELL protein have conserved motifs carboxyl to the HD. The invariant asparagine (N) at position 50 (61 in this figure) within the HD is critical for specific binding to AT-rich sequences, with the amino acids at positions 47, 50, and 54 (57, 60, and 64 in this figure highlighted by blue arrows and gray shading) influencing sequence specificity adjacent to the AT-rich core (Berger et al., 2008; Noyes et al., 2008). Of note is that, based on these critical residues, the three different subclasses of land plant BELL HD proteins are predicted to have distinct DNA-recognition specificities.
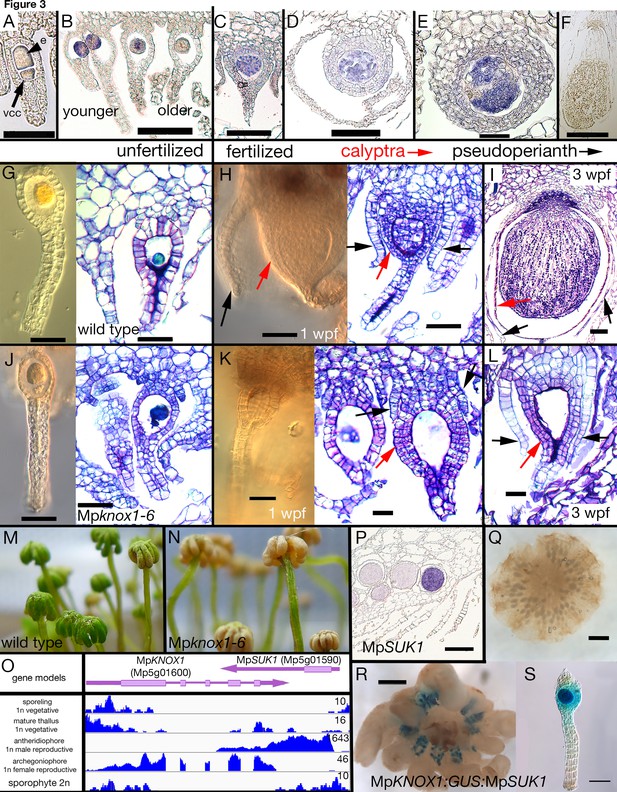
Maternal MpKNOX1 is required for post-zygotic embryo development.
(A–F) MpKNOX1 expression pattern. MpKNOX1 transcripts could be detected via in situ hybridization in the egg (e) cells of unfertilized archegonia (B), but not in the venter canal cell (A; vcc). After fertilization, expression is detected in young developing sporophytes (approximately 1 week post fertilization (wpf); C, D), with expression continuing until foot and seta begin to differentiate (E). MpKNOX1 is not expressed in older sporophytes where sporogenous tissue has differentiated (F). (G–L) Comparison of wild-type and Mpknox1 development; unfertilized (G, J), 1 wpf (H, K), and 3 wpf (I, L). Mpknox1-6ge egg cells appear wild-type-like (unfertilized; G, J). In contrast to wild-type (H, I), the embryo of Mpknox1-6ge mutants does not develop post fertilization (K, L). The zygote fails to undergo cytokinesis, with the nucleus disappearing after 3 weeks, leaving an empty space (approximately 3 wpf; L). The initial outgrowth of the pseudoperianth (black arrows) and calyptra (red arrows) post-fertilization is not affected in Mpknox1-6ge mutants, but their development is arrested as well approximately 1 wpf (K, L). (M, N) Archegoniophores of Mpknox1-6ge mutants begin to senesce approximately 2 weeks after maturation (N), while wild-type archegoniophores of the same age remain green (M). (O) Based on RNA-sequencing (RNA-seq) data and associated gene models, the 3' end of MpKNOX1 overlaps with the 3' untranslated region (UTR) of MpSUK1, which is transcribed from the opposite strand. Predicted full-length MpKNOX1 transcripts, that is, those including exons 2 and 3, are present primarily in the archegoniophore, consistent with the in situ data and semiquantitative reverse transcription-polymerase chain reaction (sqRT-PCR) data in Figure 1—figure supplement 1. FPKM (Fragments Per Kilobase of exon model per Million mapped fragments) scales are set to a standard within each tissue to allow potential weakly expressed transcripts to be visualized. (P) Signal of MpSUK1 in situ hybridization is detected in antheridia of the antheridiophore. (Q, R) Expression of proMpKNOX1:GUS:SUK1 in an archegoniophore (R), an archegonium (S), and an antheridiophore (Q). Scale bar = 50 µm (A, E, G, J, K, L, S); 100 µm (B, C, D, H, I); 200 µm (P); 500 µm (F, R); 1000 µm (Q).
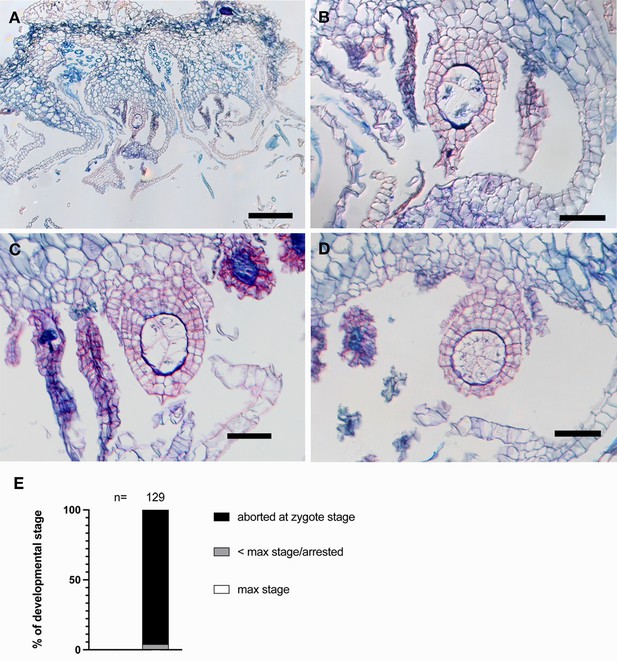
Mpknox1-6ge mutants rarely form embryos on senescing archegoniophores.
(A–D) Cross-sections of Mpknox1-6ge female mutants crossed with wild-type males and observed 2 weeks post fertilization (wpf). Note that some cells are deformed and collapsed, a characteristic of senescing tissue. (B) is the close up of (A). (E) Percentages of developmental stages observed from this cross. Plants were crossed once, and then examined after 1 or 2 wpf. Observed embryos were grouped into the following developmental stages: aborted zygotes, sporophytes that were arrested at a stage younger than the maximum stage possible, and sporophytes that had reached the maximum stage expected. n = total number of observed fertilization events. See Figure 3—figure supplement 1—source data 1 for details. Scale bar = 50 µm (B–D); 200 µm (A).
-
Figure 3—figure supplement 1—source data 1
Developmental staging data of crosses between Mpknox1-6 females and wild-type males.
- https://cdn.elifesciences.org/articles/57088/elife-57088-fig3-figsupp1-data1-v3.xlsx
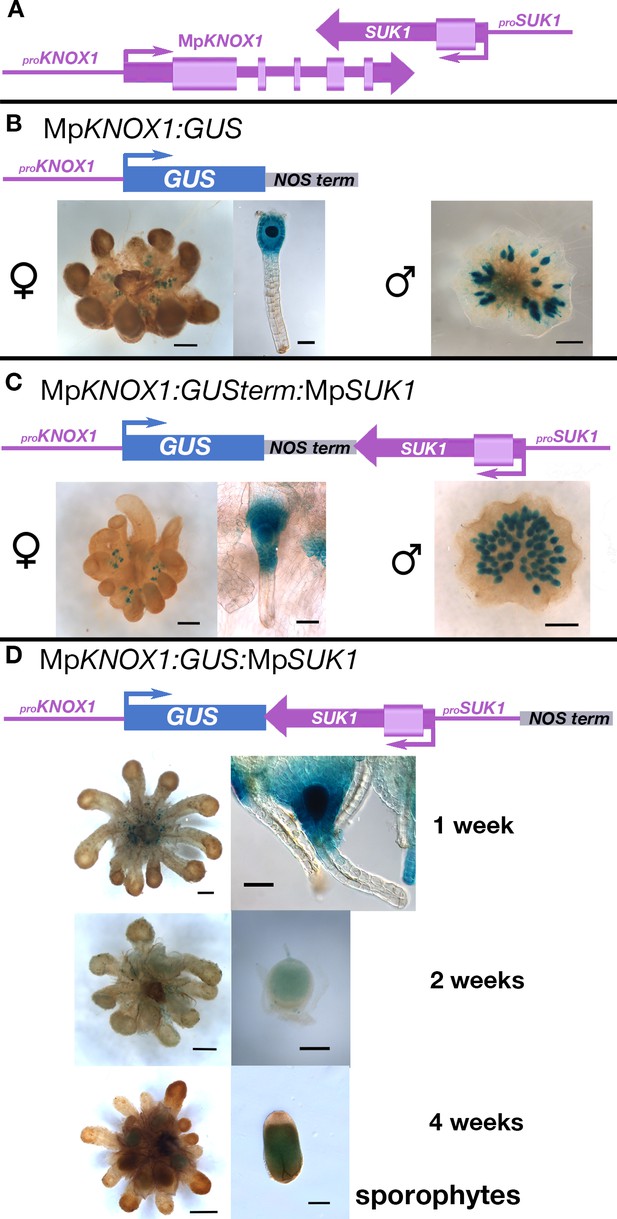
Expression profile of proMpKNOX1 transcriptional β-glucuronidase reporter gene fusions unravels regulation by antisense transcript MpSUK1.
Sequences in purple are from the endogenous locus, while sequences in other colors are from the heterologous locus. (A) Endogenous MpKNOX1/MpSUK1 locus. (B) A construct, proMpKNOX1:GUS, including a 4.6 kb sequence upstream (5') of the MpKNOX1 translational start site fused with the β-glucuronidase (GUS) reporter gene followed by a nopaline synthase (NOS) transcriptional terminator (250 bp), drives expression in both the egg cell (left) and antheridia (right). (C) Inclusion of the MpSUK1 locus, proMpKNOX1:GUSterm:SUK1, including 1.5 kb upstream (5') of the MpSUK1 translational start site, downstream of the NOS terminator, results in the same expression pattern, that is, in both the egg cell and antheridia. (D) Only in constructs featuring an assembly in which proMpKNOX1:GUS and proMpSUK1:MpSUK1 are opposing and not interrupted by a transcriptional terminator (proMpKNOX1:GUS:SUK1) is expression of MpKNOX1 restricted to the female egg cell (Figure 3) and the developing sporophyte, shown here at 1 week post fertilization (wpf), 2 wpf, and 4 wpf. No expression is observed in males (Figure 3). Scale bar = 50 µm (archegonia in B, C and 1-week-old sporophyte in D), 250 µm (2- and 4-week-old sporophytes in D), 500 µm (archegoniophores in B, C), and 1000 µm (antheridiophores in B, C and archegoniophores in D).
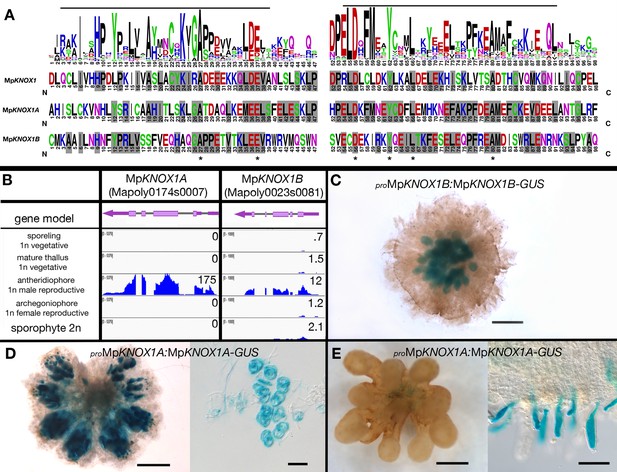
MpKNOX1A and MpKNOX1B lack the homeodomain and are both primarily expressed in the antheridiophores.
(A) Alignment of MpKNOX1 MEINOX domains with land plant KNOX1 proteins. A consensus sequence of the MEINOX domain (Bürglin, 1997; Crooks et al., 2004) of land plant KNOX1 proteins was computed from sequences in Figure 1—figure supplement 1, with the alignment of the three Marchantia polymorpha KNOX1 proteins displayed below. The lines above the sequence highlight the previously defined KNOX subdomains of the MEINOX domain. Gray shading indicates amino acids conserved with the land plant consensus and asterisks highlight amino acids conserved in all three M. polymorpha KNOX1 sequences. (B) Panels as described in Figure 3. Expression of both genes is predominantly detected in antheridiophores. (C–E) Translational fusion reporter line expression containing 1 kb (MpKNOX1A) and 2.3 kb (MpKNOX1B) of the putative 5' regulatory sequences suggests that antheridiophore expression identified by RNA-sequencing (RNAseq) is primarily in the antheridia. Expression of a translational fusion of MpKNOX1A with β-glucuronidase (GUS) was detectable in the sperm (D) and in mucilage papillae of the archegoniophore (E), but not in mucilage papillae in other parts of the thallus (not shown). Scale bar = 200 µm (antheridiophores and archegoniophores), 10 µm (sperm), and 50 µm (mucilage cells).
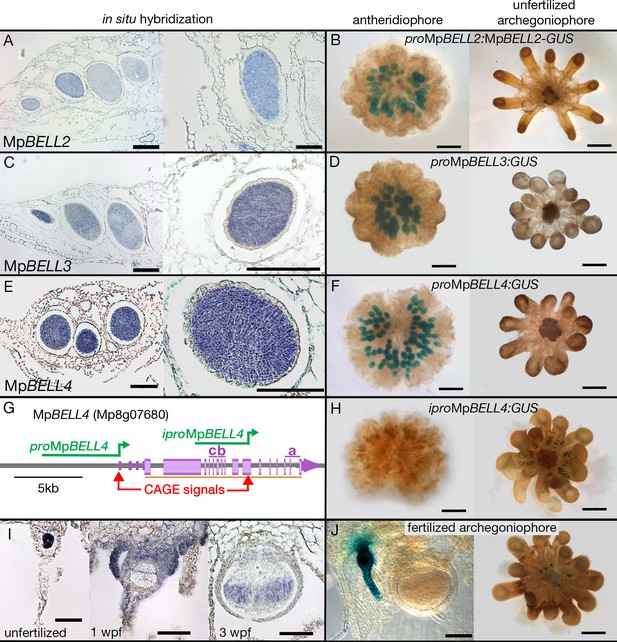
Expression patterns of MpBELL2, MpBELL3, and MpBELL4.
In situ localization of all three MpBELL mRNAs in antheridia as seen in antheridiophore cross-sections; younger antheridia are toward the left (A, C, E). Antheridiophores with translational (B) or transcriptional (D, F) β-glucuronidase reporter gene fusions of MpBELL2 (B), MpBELL3 (D), and MpBELL4 (F). These reporter genes all harbor sequences 5’ of the longest predicted transcript at each of the loci; for example, proMpBELL4 in (G). Signal appears strongest in young- to medium-aged antheridia (stage 3 and stage 4; Higo et al., 2016), with the signal being lost in older antheridia toward the center of the antheridiophore (B, F), possibly due to draining of spermatogenous tissue. Staining was not observed in unfertilized archegonia (B, D, F). (I) In situ hybridization using the full-length MpBELL4 coding sequence exhibits signals in egg cells of unfertilized archegonia, gametophytic tissues surrounding fertilized archegonia at 1 week post fertilization (wpf), as well as sporophytes up to at least 3 wpf. (H, J) Reporter genes constructed using an alternative promoter internal of the MpBELL4 locus (iproMpBELL4) marked with a Cap Analysis of Gene Expression (CAGE) signal (G) exhibit a signal in unfertilized archegonia, but not in antheridia (H). Signal remains visible in developing unfertilized archegonia within the center of the archegoniophore, but no signal was observed in sporophytes at 2 wpf (J). Scale bar = 200 μm for left panels in A, C, E and the right panel in C, panels B, D, F, H, and the right panel in J; 100 μm for right panels in A, E, I and the left panel in J; 50 μm for the left and middle panel in I.
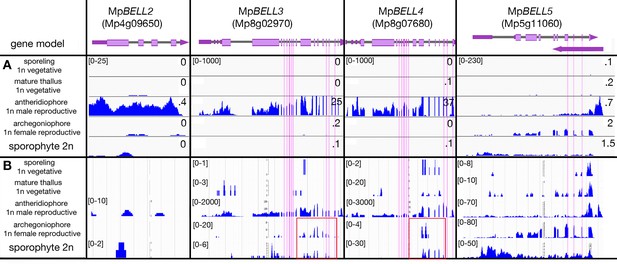
Expression profiles of MpBELL loci.
Panels as described in Figure 3. Vertical pink lines highlight exons encoding the homeodomains, each spanning three exons, in MpBELL3, MpBELL4, and MpBELL5. (A) Expression in different tissues of wild-type Marchantia polymorpha based on RNA-sequencing (RNA-seq) experiments (Bowman et al., 2017). FPKM scales are set to a standard within each outlined box (if less than 10, rounded to.1). (B) Strand-specific expression at each MpBELL locus (marchantia.info). Potential alternative shorter transcripts (highlighted in red boxes) are detected for MpBELL3 and MpBELL4 in archegoniophores and sporophytes. Scales in the strand-specific expression panels vary according to tissue to allow potential weakly expressed transcripts to be visualized.
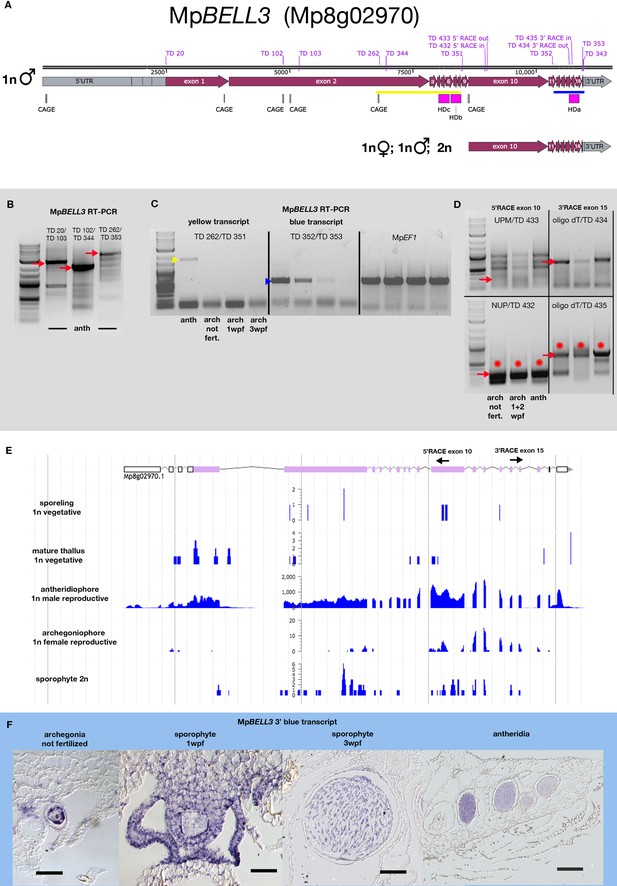
MpBELL3 locus is a source of multiple transcripts in antheridia versus archegonia/sporophytes.
(A) Schematics of MpBELL3 transcripts predicted to be produced in each of the male gametophytes, female gametophytes, and sporophytes are depicted, showing Cap Analysis of Gene Expression (CAGE) signals of transcription start sites (TSS), intron/exon boundaries (from https://marchantia.info), position of homeodomains, and primers used in subsequent experiments. The (B) reverse transcription polymerase chain reaction (RT-PCR) using different primer combinations (as shown in A) suggests a continuous long transcript in antheridia. (C) RT-PCR indicates that transcripts including homeodomains HDb and HDc are only expressed in antheridiophores (yellow), while a shorter 3’ transcript (blue) including HDa is also expressed in archegoniophores. (D) MpBELL3 nested 5’ and 3’ RACE (Rapid Amplification of cDNA Ends) on complementary DNA (cDNA) derived from archegoniophores, archegoniophores harboring 1- + 2-week-old sporophytes, and antheridiophores. Red arrows show the expected amplicon length, while red asterisks indicate that a correct amplicon could be verified via direct sequencing. Both panels show that a truncated 3’ transcript indicated by a CAGE signal around the exon 9/10 boundary including homeodomain HDa is present in all three tissues. (E) Strand-specific expression profile of MpBELL3 in different tissues of wild-type Marchantia polymorpha based on RNA-sequencing (RNA-seq) experiments (Bowman et al., 2017) as in Figure 4—figure supplement 1 (B), highlighting the positions of 5’ and 3’ RACE primers in relation to the transcript signal. The transcriptome signal suggests that some introns may experience alternative splicing. (F) In situ experiments using the truncated MpBELL3 3’ transcript spanning exons 11–16 (blue transcript in A) give signal in archegonia, archegonia harboring 1- and 3-week-old sporophytes as well as antheridia. Scale bar = 50 μm for the two left panels in (F) and 200 μm for the two right panels in (F).
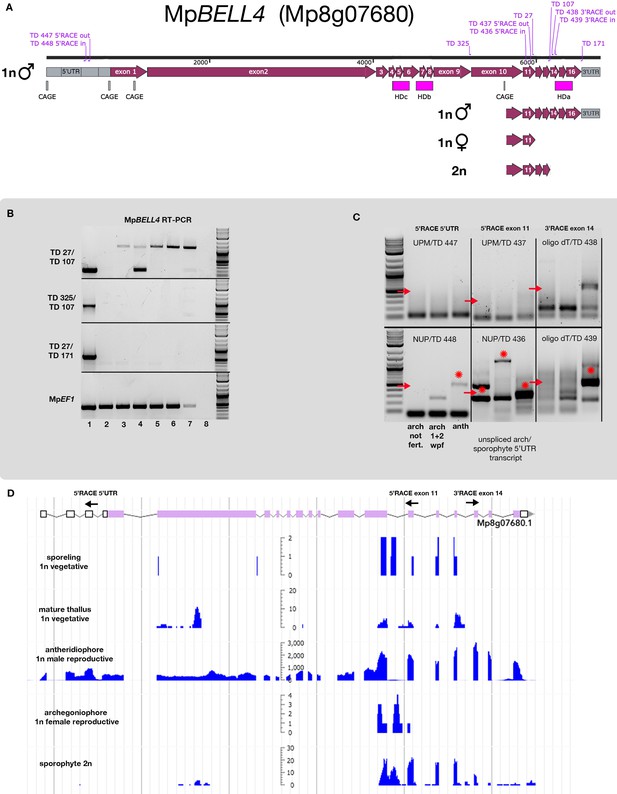
MpBELL4 transcripts in archegonia/sporophytes lack homeodomains.
(A) Schematics of MpBELL4 transcripts predicted to be produced in each of the male gametophytes, female gametophytes, and sporophytes are depicted, showing Cap Analysis of Gene Expression (CAGE) signals, intron/exon boundaries (from https://marchantia.info), position of homeodomains, and primers used in subsequent experiments. (B) Reverse transcription-polymerase chain reaction (RT-PCR) using different primer combinations (as shown in A) suggests that transcripts harboring HDa are only expressed in antheridia, whereas a truncated 3’ transcript is expressed in archegonia harboring sporophytes at 3 weeks post fertilization (wpf). Tissues from which RNA was extracted: 1, wild-type antheridiophores; 2, wild-type archegoniophores (not fertilized); 3, wild-type 1 wpf; 4, wild-type 3 wpf; 5, wild-type female x Mpbell34-4 (1 wpf); 6, wild-type female x Mpbell2/3/4-15 (1 wpf); 7, genomic DNA; 8, no template control. (C) MpBELL4 nested 5’ and 3’ RACE on complementary DNA (cDNA) derived from archegoniophores, archegoniophores harboring 1- + 2-week-old sporophytes, and antheridiophores. Red arrows show the expected amplicon length, while red asterisks indicate that a matching amplicon could be verified via direct sequencing. 5’ RACE and 3’ RACE on untranslated regions (UTRs) (panels 1 and 3) only give signal in antheridia, suggesting that a full-length transcript harboring the carboxy-terminal homeodomain (HDa) is only present in antheridia. A truncated 3’ transcript could be detected by 5’ RACE from exon 11 in all tissue examined (panel 2), although only antheridia-derived transcripts were spliced. (D) Strand-specific expression profile of MpBELL4 in different tissues of wild-type Marchantia polymorpha based on RNA-sequencing (RNA-seq) experiments (Bowman et al., 2017) as used in Figure 4—figure supplement 1 (B), highlighting the positions of 5’ and 3’ RACE primers in relation to the transcript signal. The transcriptome signal suggests that some introns may experience alternative splicing.
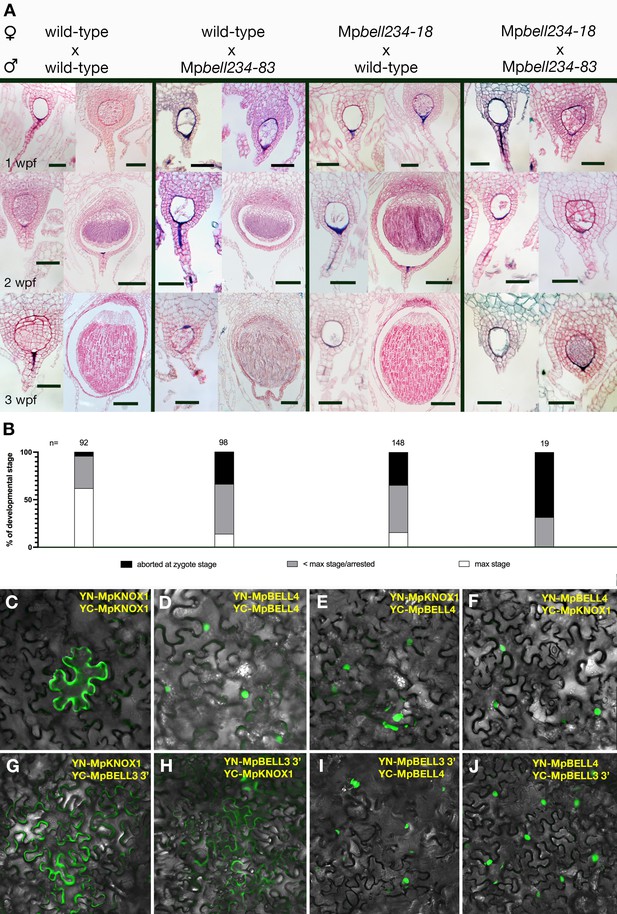
Sporophyte development entails BELL activity in both male and female gametes.
(A) Wild-type females crossed with wild-type males (left column), reciprocal crosses between wild-type and Mpbell mutant alleles (middle two columns), and crosses between Mpbell234 homozygotes (right column) observed after 1, 2, and 3 weeks post fertilization (wpf). All crosses produced aborted embryos or arrested sporophytes (left pictures in each panel). However, crosses between Mpbell234-18ge and Mpbell234-83ge only produced embryos aborted at the zygote stage or arrested at approximately the 1 wpf stage. See Table 1 for details on mutant alleles. (B) Percentages of developmental stages observed for each of the crosses. Plants were crossed once and then examined after 1, 2, or 3 wpf. Observed embryos were grouped into the following developmental stages: aborted zygotes, sporophytes that were arrested at a stage younger than the maximum stage possible, and sporophytes that had reached the maximum stage expected. N = the total number of observed fertilization events. See Figure 5—source data 1 for details. (C–F) BiFC (Bimolecular fluorescence complementation) assay of protein-protein interaction of MpKNOX1 and MpBELL4 in Nicotiana benthamiana leaves. Homodimers of MpKNOX1 show cytoplasmic localization (C), while MpBELL4 homodimers localize to the nucleus (D). Co-expression of both proteins resulted in nuclear localization of heterodimers (E, F). (G–J) BiFC assays of the protein derived from the shorter 3’ transcript of MpBELL3 (Figure 4—figure supplement 2) with MpKNOX1 show weak cytoplasmic interaction (G, H). However, MpBELL4 interacts strongly with shortMpBELL3 and results in nuclear localization (I, J). Scale bar = 50 µm, for all left pictures in (A) panels, all double mutant pictures, and all 1 wpf pictures; 100 µm for right pictures of the 2 wpf panel; 200 µm for all right pictures of the 3 wpf panel.
-
Figure 5—source data 1
Developmental staging data of crosses between wild-type and Mpbell234 mutants.
(a) Developmental staging data of crosses between wild-type females and wild-type males. (b) Developmental staging data of crosses between wild-type females and Mpbell234-83 males. (c) Developmental staging data of crosses between Mpbell234-18 females and wild-type males. (d) Developmental staging data of crosses between Mpbell234-18 females and Mpbell234-83 males. (e) Summary and calculations of developmental staging data.
- https://cdn.elifesciences.org/articles/57088/elife-57088-fig5-data1-v3.xlsx
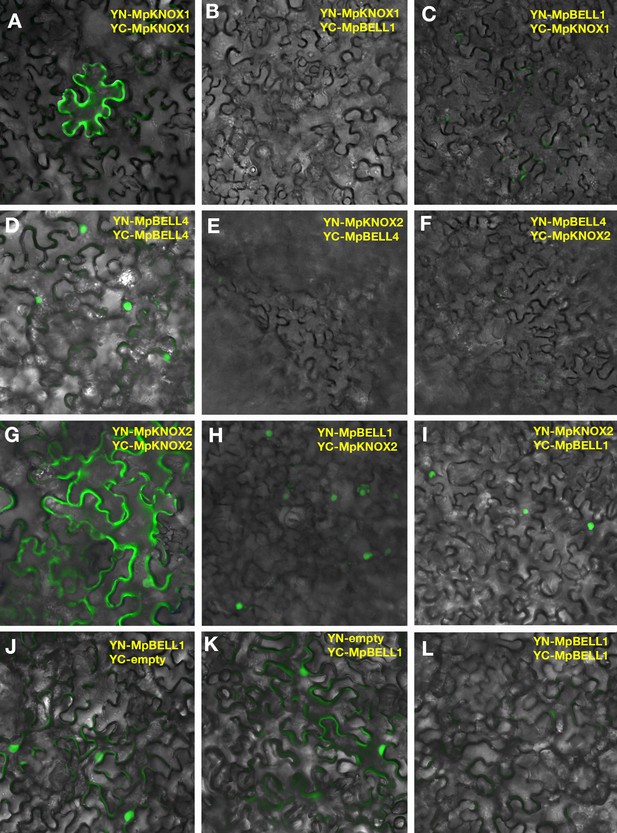
MpKNOX1/MpBELL4 and MpKNOX2/MpBELL1 exhibit specificity in their interactions.
BiFC assay of protein-protein interaction in Nicotiana benthamiana leaves. MpKNOX1 homodimers showed cytoplasmic localization (A), but no interaction could be observed when MpKNOX1 was co-expressed with MpBELL1 (B, C). MpBELL4 homodimers localized to the nucleus (D), but co-expression of both MpBELL4 and MpKNOX2 showed no interaction (E, F). MpKNOX2 homodimers alone showed cytoplasmic localization (G); however, when MpKNOX2 and MpBELL1 were co-expressed, the interaction signal was located to the nucleus (H, I). MpBELL1 did not homodimerize (J–L). Note that (A) and (D) are taken from Figure 5C and D for reference.
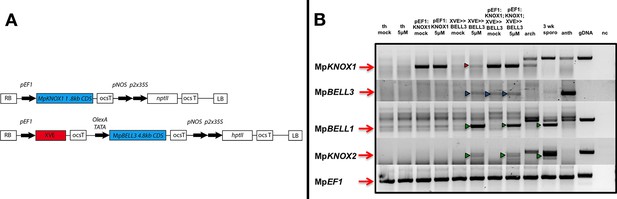
Co-expression strategy to express MpKNOX1 and MpBELL3 simultaneously in the vegetative gametophyte.
(A) Plants were doubly transformed using one construct in cis and one in trans: proEF1:MpKNOX1 constitutively expressing MpKNOX1 CDS and proEF1:XVE>> MpBELL3 expressing a 4.8 kb fragment of the putative 9 kb coding sequence after induction. Note that the pair of constructs features two different selection markers (nptII for selection on G418 and hptII for hygromycin selection) for selection of doube transformants. (B) Semiquantitative reverse transcription-polymerase chain reaction (sqRT-PCR) of mock and 5 μM 17-β-estradiol-treated plants after 72 hr of induction. MpKNOX1 transcription was induced after 72 hr in proEF1:XVE>> MpBELL3 lines (red arrowhead). Antheridia-specific MpBELL3 expression was induced in proEF1:XVE>> MpBELL3 lines, in case of the double transformants also in the un-induced line (blue arrowheads). MpKNOX2 and MpBELL1, both normally expressed primarily in the sporophyte, were de-repressed after induction via MpBELL3 expression (green arrowheads). Th = thallus; arch = archegoniophore; 3 wk sporo = archegoniophore including sporophytes at 3 weeks post fertilization (wpf); anth = antheridiophores; gDNA = genomic DNA; nc = negative control.
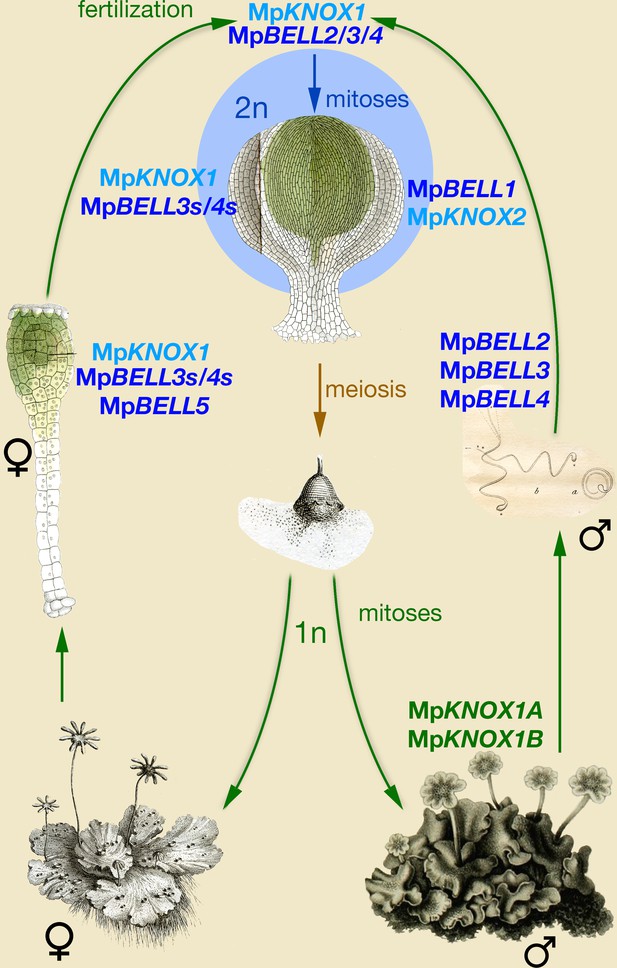
Model of KNOX and BELL function in Marchantia polymorpha.
Three MpBELL genes (MpBELL2/3/4) are expressed in antheridia in the cells that will differentiate into sperm cells. MpKNOX1 is expressed in the egg cell of the archegonia, along with three MpBELL genes (MpBELL5 and MpBELL3/4). In contrast to the antheridia, where full-length transcripts are found, alternative short transcripts of MpBELL3/4 (MpBELL3s and MpBELL4s; see Figure 4—figure supplements 2 and 3) are produced in the egg cell. After fertilization, MpBELL3/4 derived from both sperm and egg cells and MpKNOX1 derived from the egg cell act together to activate the zygotic program. MpBELL1 and MpKNOX2 form a distinct heterodimer that acts at later stages of sporophyte development. The homeodomain lacking MpKNOX1A and MpKNOX1B genes expressed in the antheridia along with MpBELL2/3/4 potentially prevents functionality by sequestering MpBELL2/3/4 proteins. The function of MpBELL5 is unknown. Images are obtained from Marchant, 1713; Mirbel, 1835; Thuret, 1851; and Unger, 1837.
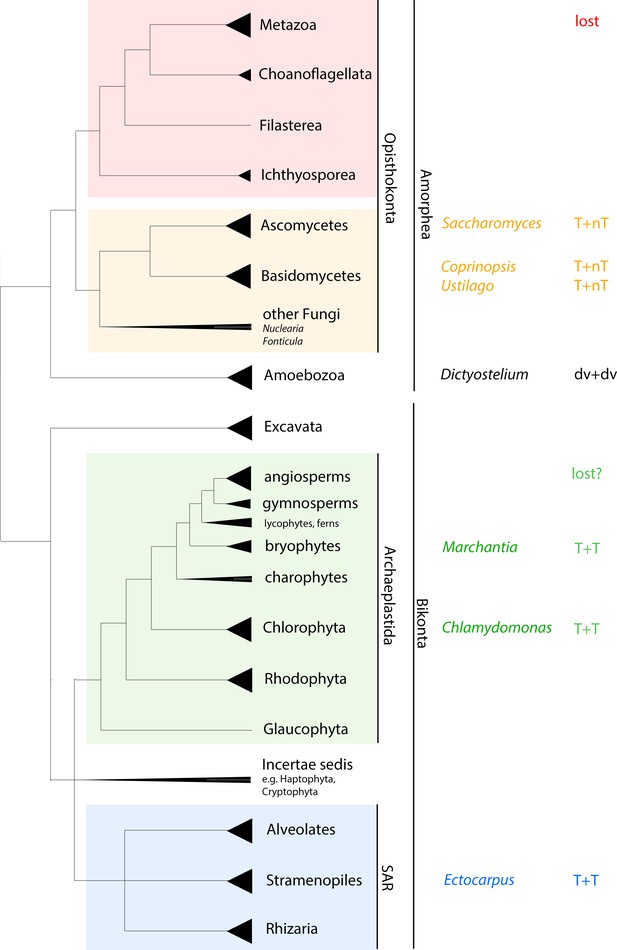
Phylogenetic perspective on HD gene-mediated haploid-to-diploid transitions in eukaryotes.
Unrooted phylogenetic tree adapted from Adl et al., 2012 and Keeling, 2014. Triangles at the end of branches represent monophyletic groups; triangles extending to the node represent paraphyletic groups. Taxa for which functional data are available are listed, as is the mode of homeodomain (HD) involvement if known. While the ancestral eukaryote possessed both TALE- and non-TALE-HD genes, the predicted composition of the heterodimer in the ancestral eukaryote is equivocal. T = TALE-HD; nt = non-TALE-HD; dv = divergent HD.
Tables
Alleles used in this study.
Nomenclature is as outlined previously (Bowman et al., 2016c; Montgomery et al., 2020).
| Name | Gene number(s) | Type | Details | Gender | Putative effect |
|---|---|---|---|---|---|
| Mpknox1-6ge | Mp5g01600 | CRISPR 2 gRNAs | 2360 nt del | Female | Null |
| Mpbell2-1ge | Mp4g09650 | CRISPR 1 gRNAs | 1 nt in | Female | Hypomorph |
| Mpbell2-2ge | Mp4g09650 | CRISPR 1 gRNAs | 10 nt del | Male | Hypomorph |
| Mpbell2-11ge | Mp4g09650 | CRISPR 1 gRNAs | 1 nt del | Male | Hypomorph |
| Mpbell3-4ge | Mp8g02970 | CRISPR 2 gRNAs | 2217 nt del | Male | Null |
| Mpbell3-10ge | Mp8g02970 | CRISPR 2 gRNAs | 2218 nt del | Male | Null |
| Mpbell4-5ge | Mp8g07680 | CRISPR 2 gRNAs | 1135 nt del | Male | Null |
| Mpbell4-24ge | Mp8g07680 | CRISPR 2 gRNAs | 1135 nt del | Male | Null |
| Mpbell34-4ge (Mpbell3-1ge Mpbell4-1ge) | CRISPR 4 gRNAs | Male | |||
| MpBELL2 | Mp4g09650 | wt | Wild type | ||
| Mpbell3-1ge | Mp8g02970 | 6 nt in/1 nt del; 1 nt in | Hypomorph | ||
| Mpbell4-1ge | Mp8g07680 | 1 nt del | Hypomorph | ||
| Mpbell234-15ge (Mpbell2-15ge Mpbell3-15ge Mpbell4-15ge) | CRISPR 4 gRNAs | Male | |||
| Mpbell2-15ge | Mp4g09650 | 23 nt in/10 nt del | Hypomorph | ||
| Mpbell3-15ge | Mp8g02970 | 49 nt in/1 nt del; 1 nt in | Hypomorph | ||
| Mpbell4-15ge | Mp8g07680 | 1 nt del | Hypomorph | ||
| Mpbell234-83ge (Mpbell2-83ge Mpbell3-83ge Mpbell4-83ge) | CRISPR 6 gRNAs | Male | |||
| Mpbell2-83ge | Mp4g09650 | 110 nt in/1401 nt del | Null? | ||
| Mpbell3-83ge | Mp8g02970 | 2217 nt del | Null? | ||
| Mpbell4-83ge | Mp8g07680 | 7 nt in/1150 nt del | Null? | ||
| Mpbell234-7ge (Mpbell2-7ge Mpbell3-7ge Mpbell4-7ge) | CRISPR 4 gRNAs | Female | |||
| Mpbell2-7ge | Mp4g09650 | 16 nt in/8 nt del | Hypomorph | ||
| Mpbell3-7ge | Mp8g02970 | 1 nt in/30 nt del; wt | Hypomorph | ||
| Mpbell4-7ge | Mp8g07680 | 44 nt in | Hypomorph | ||
| Mpbell234-18ge (Mpbell2-18ge Mpbell3-18ge Mpbell4-18ge) | CRISPR 4 gRNAs | Female | |||
| Mpbell2-18ge | Mp4g09650 | 13 nt in/7 nt del | Hypomorph | ||
| Mpbell3-18ge | Mp8g02970 | 97 nt in/304 nt del; 1 nt del | Hypomorph | ||
| Mpbell4-18ge | Mp8g07680 | 1 nt del | Hypomorph | ||
| proEF1:XVE>> amiR-MpE(z)1SkmiR166 | Mp5g18040 | Transgene | Knock-down | Both | Hypomorph |
| proEF1:MpKNOX1 | Mp5g01600 | Transgene | Ectopic overexpression | Both | Hypermorph |
| proEF1:XVE>> MpBELL3 | Mp8g02970 | Transgene | Inducible overexpression | Both | Hypermorph |
| proEF1:MpKNOX1; proEF1:XVE>> MpBELL3 | Mp5g01600 | Transgene | Ectopic overexpression | Both | Hypermorph |
| proMpBELL2:MpBELL2-GUS | Mp4g09650 | Transgene | Translational β-glucuronidase gene fusion | Both | |
| proMpBELL3:GUS | Mp8g02970 | Transgene | Transcriptional β-glucuronidase gene fusion | Both | |
| proMpBELL4:GUS | Mp8g07680 | Transgene | Transcriptional β-glucuronidase gene fusion | Both | |
| internal_proMpBELL4:GUS | Mp8g07680 | Transgene | Transcriptional β-glucuronidase gene fusion | Both | |
| proMpKNOX1:GUS | Mp5g01600 | Transgene | Transcriptional β-glucuronidase gene fusion | Both | |
| proMpKNOX1:GUSterm:SUK1 | Mp5g01600Mp5g01590 | Transgene | Transcriptional β-glucuronidase gene fusion | Both | |
| proMpKNOX1:GUS:SUK1term | Mp5g01600Mp5g01590 | Transgene | Transcriptional β-glucuronidase gene fusion | Both | |
| proMpKNOX1A:MpKNOX1A-GUS | Mp4g12450 | Transgene | Translational β-glucuronidase gene fusion | Both | |
| proMpKNOX1B:MpKNOX1B-GUS | Mp2g11140 | Transgene | Translational β-glucuronidase gene fusion | Both | |
Additional files
-
Supplementary file 1
Primers and guide RNAs used in this study.
Table 1a. Primer name, gene number, sequence, and the corresponding figure used for sqRT-PCR and RACE. Table 1b. Primer name, gene name, and sequence used for in situ probe preparation. Table 1c. Primer name, gene name, and sequence used for overexpression, inducible overexpression, and transcriptional and translational β-glucuronidase reporter constructs. Table 1d. Guide RNA sequences used to generate mutant alleles.
- https://cdn.elifesciences.org/articles/57088/elife-57088-supp1-v3.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57088/elife-57088-transrepform1-v3.pdf





