Neuronal octopamine signaling regulates mating-induced germline stem cell increase in female Drosophila melanogaster
Figures
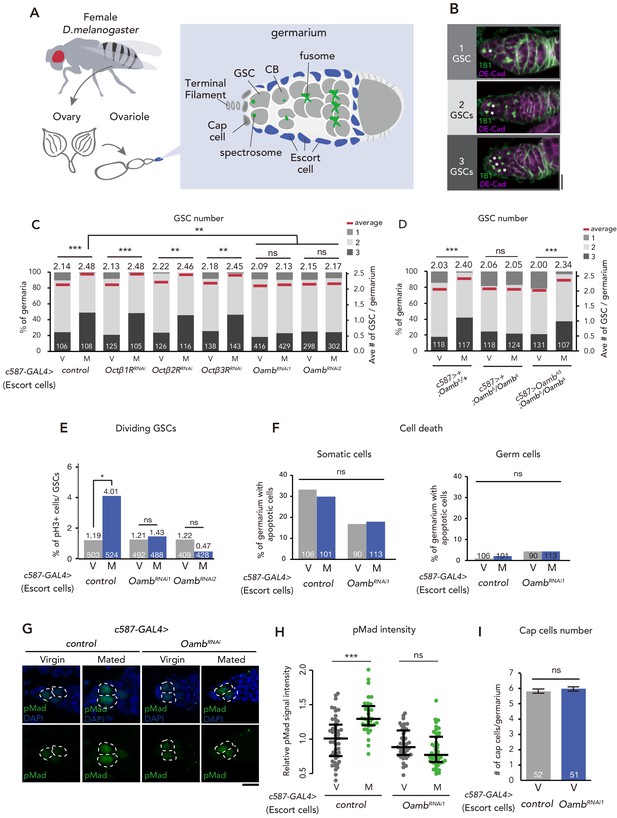
Post-mating GSC increase requires Oamb in the escort cells.
(A) A schematic representation of Drosophila germarium. GSCs reside in a niche consisting of somatic cells such as cap cells, terminal filament cells, and escort cells and are identifiable by their stereotypical spectrosome morphology and location (adjacent to cap cells). GSC division produces one self-renewing daughter and one cystoblast (CB) that differentiates into a germline cyst. (B) Representative images of wild-type (w1118) female adult germariums, containing 1, 2 and 3 GSCs from top to bottom. The samples were stained with monoclonal antibody 1B1 (green) and anti-DE-cadherin (magenta), which stain the spectrosome and overall cell membranes, respectively. GSCs are indicated by asterisk. Scale bar, 20 µm. (C–D) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control in D. (E) The ratio of pH3+ GSCs per total GSCs. (F) The ratio of apoptotic (Dcp-1+) somatic cells and germ cells per germarium. c587>+ flies were used as the control. (G) Representative images of adult female germaria immunostained with anti-pMad antibody (green) and DAPI (blue) are shown. GSCs are outlined with dotted lines. Scale bar, 10 µm. (H) Quantification of relative pMad intensity levels in the GSCs (i.e. virgin (V), mated (M)) as normalized to the pMad intensity in CBs. Each sample number was at least 25. The three horizontal lines for each sample indicate lower, median, and upper quartiles. (I) The number of cap cells per germarium in the control and Oamb RNAi driven by c587-GAL4. Values on the y-axis are presented as the mean with standard error of the mean. c587>+ flies were used as the control. For C-F, and I the number of germaria analyzed is indicated inside the bars. Wilcoxon rank sum test with Holm’s correction was used for C, D, H, and I. Fisher’s exact test with Holm’s correction was used for E and F. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, nonsignificant (p>0.05). All source data are available in Source data 1 and 2.
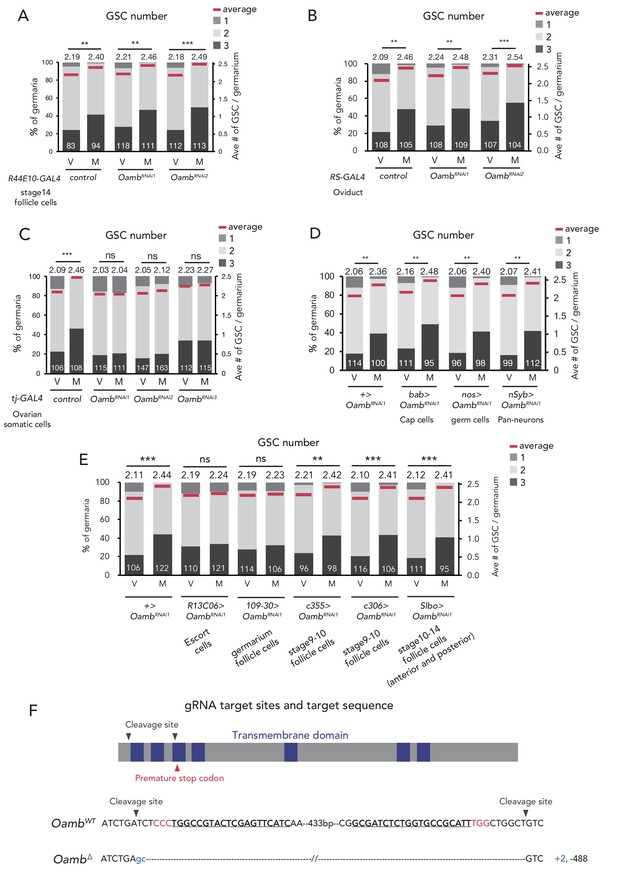
Oamb acts in the escort cells for post-mating GSC increase.
(A–E) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies in (A) Oamb RNAi in mature follicle cells by (R44E10-GAL4); (B) Oamb RNAi in the oviduct (by RS-GAL4); (C) Oamb RNAi by tj-GAL4; (D) Oamb RNAi in cap cells (by bab-GAL4), nervous system (by nSyb-GAL4), and germ cells (by nos-GAL4); and (E) Oamb RNAi in escort cells (by R13C06-GAL4), follicle cells in germarium (by 109–30 GAL4), and stage 9–10 follicle cells (by c355-GAL4 and c306-GAL4), late stage follicle cells and border cells (slbo-GAL4); The number of germaria analyzed is indicated inside the bars. (F) A schematic representation of gRNA target sites (cleavage sites: gray arrowhead) and premature stop codon (red arrowhead) in coding sequences of Oamb genes. Regions of the putative transmembrane domains of Oamb are highlighted in blue. The target locus in Cas9-induced mutant was PCR-amplified and sequenced. The WT sequence is shown on the top of sequences as reference. The Cas9-gRNA target sequence is underlined with the PAM indicated in red. Inserted nucleotides are indicated in light blue lowercase letters. The indel size is shown next to the sequence. The indel mutation results in a premature stop codon. Wilcoxon rank sum test was used for A-E. ***p≤0.001 and **p≤0.01; NS, non-significant (p>0.05). All source data are available in Source data 1.
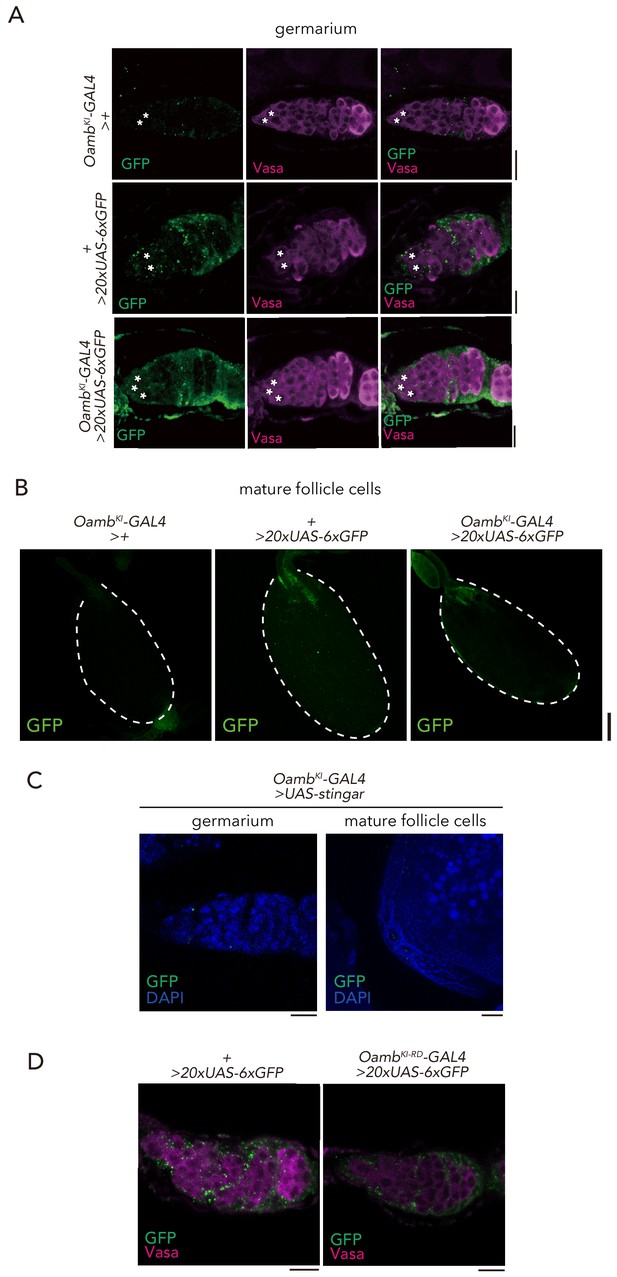
Expression of Oamb knock-in GAL4.
(A–B) Immunofluorescence of germarium in adult female flies expressing 20xUAS-6xGFP reporter under OambKI-T2A-GAL4. The GSCs are indicated by asterisk. Note that 20xUAS-6xGFP has leak signal in the germarium even in the control (+ > 20xUAS-6xGFP). Scale bar, 20 µm. (B) Immunofluorescence of stage 14 egg chamber expressing 20xUAS-6xGFP reporter under OambKI-T2A-GAL4. Note that GFP expression was not observed in the stage 14 egg chamber. Scale bar, 100 µm. (C) Immunofluorescence of germarium (left) and posterior follicle cells of stage 14 egg chamber (right) in adult female flies expressing UAS-Stinger reporter under OambKI-T2A-GAL4. Note that GFP signal is not detected in the germarium and stage 14 egg chamber. Scale bar, 20 µm. (D) Immunofluorescence of germarium in adult female flies expressing 20xUAS-6xGFP reporter under OambKI-RD-T2A-GAL4. Scale bar, 20 µm.
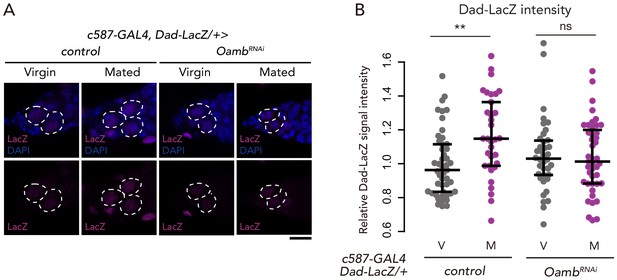
Oamb in the escort cells is necessary on mating-induced BMP signaling increase.
(A) Representative images of adult female germaria immunostained with anti-LacZ antibody (magenta) and DAPI (blue) are shown. GSCs are outlined with dotted lines. Scale bar, 10 µm. (B) Quantification of relative Dad-LacZ intensity levels in the GSCs (i.e. virgin (V), mated (M)) as normalized to the Dad-LacZ intensity in CBs. Each sample number was at least 25. The three horizontal lines for each sample indicate lower, median, and upper quartiles. Wilcoxon rank sum test was used for B. **p≤0.01; NS, non-significant (p>0.05). All source data are available in Source data 2.
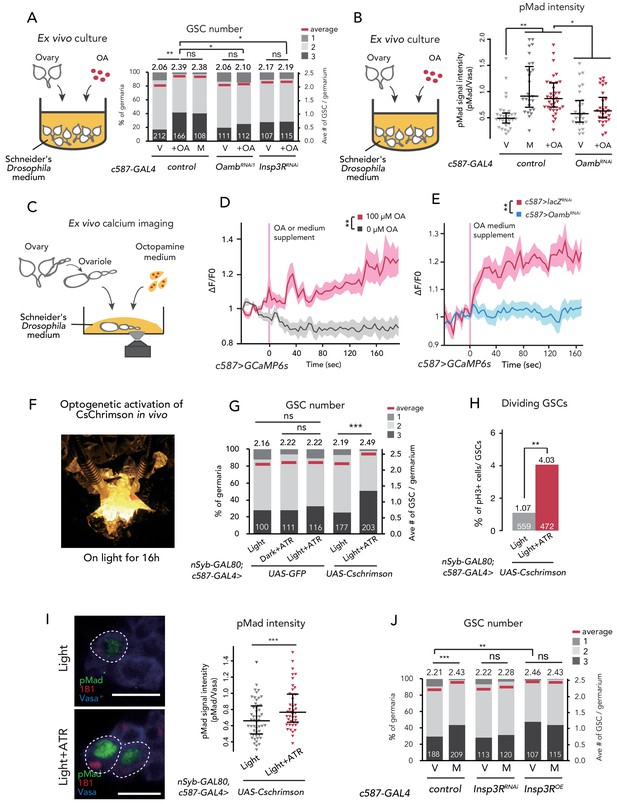
Ca2+ signaling is necessary for mating-induced GSC increase.
(A) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis). The ovaries were dissected from virgin (V), mated (M), and virgin ovaries cultured with OA (+OA). c587>+ flies were used as the control. The number of germaria analyzed is indicated inside the bars. (B) Quantification of relative pMad intensity levels in the GSCs of ex vivo cultured ovaries (i.e. virgin (V), mated (M), and virgin cultured with OA (+OA)) as normalized to the pMad intensity in CBs. For the quantification of pMad intensity, the cell boundaries of GSCs and CBs were determined using anti-Vasa staining. Each sample number was at least 25. The three horizontal lines for each sample indicate lower, median, and upper quartiles. (C) A schematic representation of ex vivo calcium imaging. The dissected ovariole was incubated in Schneider’s Drosophila medium with or without OA. (D) Changes in the relative fluorescence intensity of GCaMP6s after 200 s without stimulation (n = 8) or with stimulation (n = 10) with 100 μM OA, and (E) with 100 μM OA as control (c587 >LacZRNAi, n = 8) and c587 >OambRNAi (n = 8) female ovaries. Note that OA significantly increased the calcium response in escort cells, but OambRNAi impaired the calcium response. Statistical analysis was done at 120 s. (F) Equipment setup for optogenetic activation of ChR. Flies were placed under the light for 16 hr before dissection. (G) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) with light, with light and all trans-retinal (ATR) or with dark and ATR. Germarium was dissected from virgin females. nSyb-GAL80; c587 >GFP flies were used as control. The number of germaria analyzed is indicated inside the bars. (H) The ratio of pH3+ GSCs and total GSCs. The number of GSCs analyzed is indicated inside the bars. (I, left) Representative images of adult female germaria immunostained with anti-pMad antibody (green), anti-1B1 antibody (red), and anti-Vasa antibody (germ cell marker; blue) are shown. GSCs are outlined with dotted lines. (I, right) Quantification of the relative pMad intensity in GSCs, which was normalized to that in CBs. For the quantification of pMad intensity, the cell boundaries of GSCs and CBs were determined using anti-Vasa staining. Each sample number is at least 30. The three horizontal lines for each data sample indicate lower, median, and upper quartiles. (J) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control. The number of germaria analyzed is indicated inside the bars. Wilcoxon rank sum test with Holm’s correction was used for A, B, D, E, G, I, and J. Fisher’s exact test was used for H. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, non-significant (p>0.05). All source data are available in Source data 1, 2, and 4.
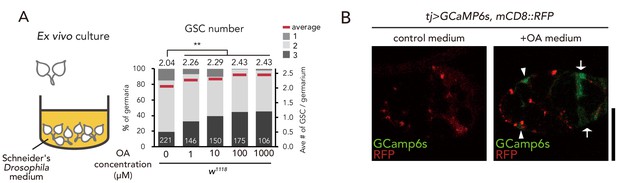
OA treatment induces GSC increase.
(A) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin female flies. The addition of OA to the medium is sufficient to induce GSC increase. (B) Representative images of adult female germaria in response to OA in tj >GCaMP6s; mCD8::RFP. Note that calcium response was observed in the escort cells (arrowheads) and follicle cells (arrow) of the germarium. Scale bar, 10 µm. Wilcoxon rank sum test with Holm’s correction was used for statistical analysis. ***p≤0.001. All source data are available in Source data 1.
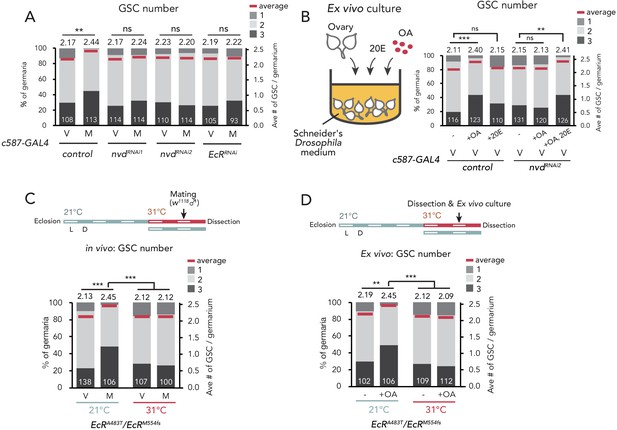
Ecdysteroid signaling is necessary for OA-mediated GSC increase.
(A–D) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control. The number of germaria analyzed is indicated inside the bars. (A) GSC number of nvd and EcR RNAi flies in vivo. (B) Virgin ovaries were cultured ex vivo with or without OA and 20E (+OA, +20E, −), and then the GSC number was determined. (C–D) Experiments using a temperature-sensitive allele EcRA483T. 21°C and 31°C were used as the permissive and restrictive temperatures, respectively. Flies were cultured at 21°C and transferred to 31°C 1 d prior to the assays (L; light, D; dark). (C) GSC number in vivo. (D) Virgin ovaries were cultured ex vivo with or without OA (+OA, −). The number of germaria analyzed is indicated inside the bars. Wilcoxon rank sum test with Holm’s correction was used for statistical analysis. ***p≤0.001 and **p≤0.01; NS, non-significant (p>0.05). All source data are available in Source data 1.
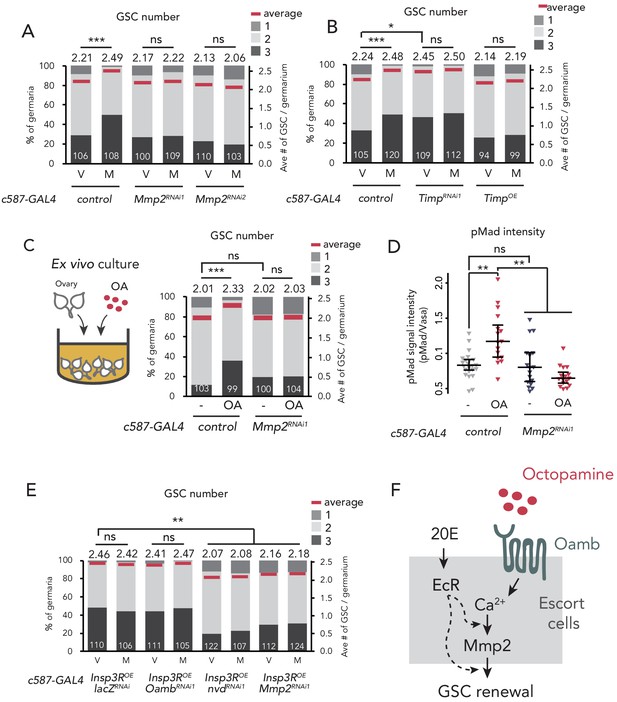
Mmp2 is necessary for OA-mediated GSC increase.
(A–C, E) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control. The number of germaria analyzed is indicated inside the bars. (A) Mmp2 RNAi by c587-GAL4 driver. (B) RNAi and the overexpression of Timp by c587-GAL4 driver. (C) Ex vivo culture experiment using c587 >Mmp2 RNAi. OA was added into the ex vivo culture medium. Cultured with or without OA (+OA, -, respectively) is indicated under each bar. (D) Quantification of the relative pMad intensity in GSCs of the ex vivo cultured ovaries normalized to pMad intensity in CBs. Cultured with or without OA (+OA, −) is indicated under each bar. For the quantification of pMad intensity, the cell boundaries of GSCs and CBs were determined using anti-Vasa staining (n > 15). The three horizontal lines for each data sample indicate lower, median, and upper quartiles. (E) Oamb, nvd, or Mmp2 RNAi in the genetic background of c587 >Insp3R overexpression. (F) A model of signaling in the escort cell to induce the mating-induced GSC increase. Oamb in the escort cells receives OA, and induce [Ca2+]i in the cells. The [Ca2+]i induces GSC increase via Mmp2. Ecdysteroid signaling is also involved in this process. Wilcoxon rank sum test with Holm’s correction was used. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, non-significant (p>0.05). All source data are available in Source data 1 and 2.
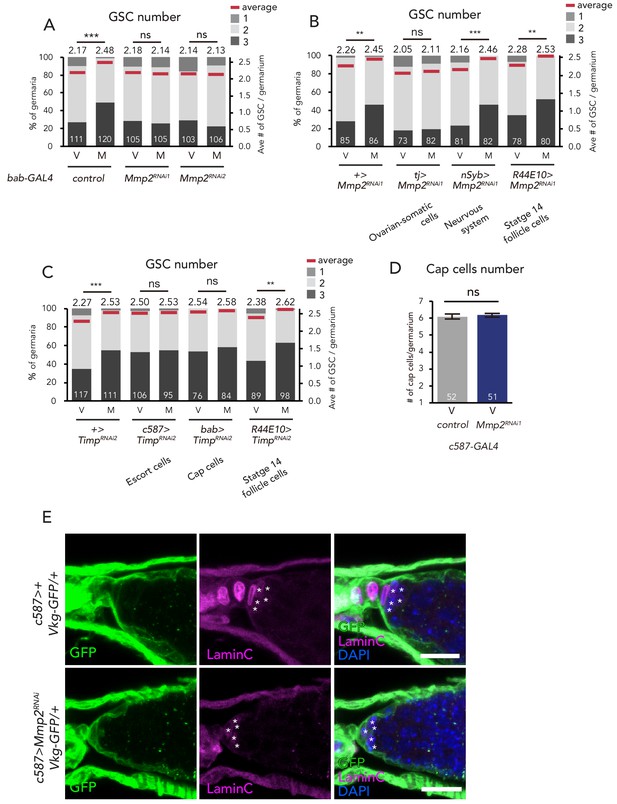
Mmp2 is necessary in the escort cells to induce GSC increase.
(A–C) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) female flies. Mmp2 RNAi in the (A) cap cells by bab-GAL4 driver and in the (B) nervous system by nSyb-GAL4, mature follicle cells by R44E10-GAL4, and ovarian-somatic cells by tj-GAL4. (C) Timp RNAi in the escort cells by c587-GAL4, cap cells by bab-GAL4, and mature follicle cells by R44E10-GAL4. (D) The number of cap cells in the control and Mmp2 RNAi driven by c587-GAL4. Values on y-axis are presented as the mean with standard error of the mean. (E) Representative images of Vkg::GFP adult female germaria immunostained with anti-GFP antibody (green), anti-Lamin C antibody (red; cap cells, asterisk), and DAPI. Note that the Vkg::GFP signal around cap cells is not affected even in Mmp2 RNAi flies. Scale bar, 10 µm. Wilcoxon rank sum test with Holm’s correction was used for A, B, C and D. ***p≤0.001 and **p≤0.01; NS, non-significant (p>0.05). All source data are available in Source data 1.
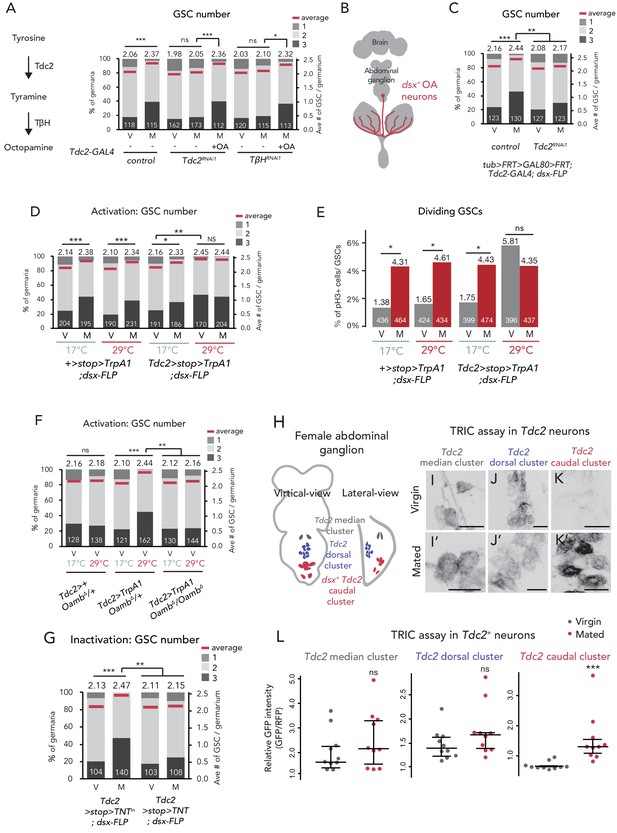
Ovary-projecting OA neurons control the GSC increase.
(A, C–D, F–G) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. The number of germaria analyzed is indicated inside the bars. (A) RNAi of Tdc2 and TβH by Tdc2-GAL4. OA was added into the standard food. (B) A schematic drawing of Drosophila central nervous system and the ovary-projecting OA neurons with the dsx+ OA neurons projecting to the ovary. (C) Tdc2 RNAi in dsx+ Tdc2+ neurons with the genotype indicated. (D–E) TrpA1-mediated activation of dsx+ Tdc2+ neurons. 17°C and 29°C were used as the permissive and restrictive temperatures, respectively, of TrpA1 channel. (D) GSC number. (E) The ratio of pH3+ GSCs and total GSCs. (F) The activation of Tdc2+ neurons with OambΔ genetic background. (G) The inactivation of dsx+ Tdc2+ neurons. (H) Illustration showing the location of three clusters of Tdc2+ neurons in the caudal part of the abdominal ganglion (I–K, I’–K’). Negative images of TRIC labeling (anti-GFP) in the abdominal ganglions of virgin (I–K) and mated females (I’–K’) of TRIC (Tdc2 >UAS-mCD8::RFP, UAS-p65AD::CaM LexAop2-mCD8::GFP; nSyb-MKII::nlsLexADBDo;UAS-p65AD::CaM) flies, indicating intracellular Ca2+ transients. Scale bars, 20 μm. (L) The GFP intensities from the Tdc2+ median cluster, Tdc2+ dorsal cluster, and dsx+ Tdc2+ cluster of TRIC females show Ca2+ activity in virgin (gray) and mated females (red). Wilcoxon rank sum test was used for A, C, D, F, G, and L. Fisher’s exact test with Holm’s correction was used for E. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, non-significant (p>0.05). All source data are available in Source data 1 and 3.
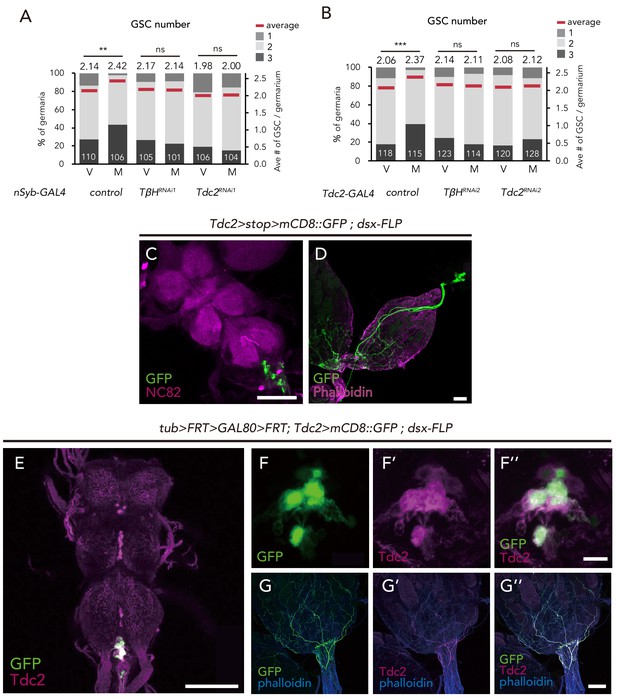
dsx+ Tdc2+ neurons control GSC increase.
(A–B) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies using nSyb-GAL4 (A) and Tdc2-GAL4 (B). (C–D) Images of dsx+ Tdc2+ neurons expressing the UAS > stop >mCD8::GFP reporter under Tdc2-GAL4 with dsx-FLP. GFP expression was detected only in the abdominal ganglion neurons projecting to the ovary. Images of the abdominal ganglion and the reproductive system are shown in C and D, respectively. (E–G) Immunofluorescence of dsx+ Tdc2+ neurons expressing UAS >mCD8::GFP reporter under Tdc2-GAL4 with tub >FRT >GAL80>FRT. GFP expression was only observed in the anti-Tdc2 positive neurons in the abdominal ganglion that projected to the ovary (E). Scale bars, 100 µm in C, D, E, and G; 10 µm in F. Wilcoxon rank sum test with Holm’s correction was used. ***p≤0.001; NS, non-significant (p>0.05). All source data are available in Source data 1.
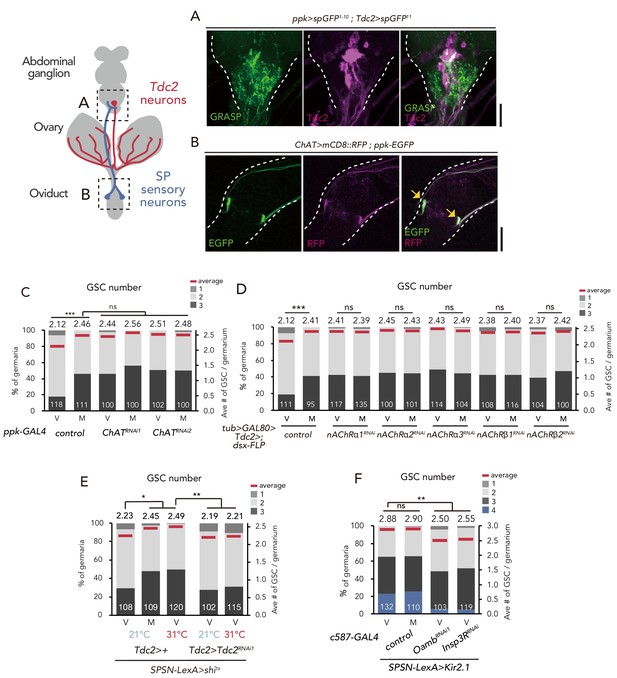
SPSNs control GSC increase through OA neurons.
(A) Neuronal proximity of SPSNs and Tdc2+ neurons in the abdominal ganglion of female flies stained with anti-Tdc2 (magenta). Note that reconstituted GFP (GRASP) signal was detected in the caudal part of the abdominal ganglion surrounded by broken white lines. Scale bar, 25 µm. (B) Cell bodies of SPSNs (yellow arrows) of ChAT-GAL4; UAS-mCD8::RFP; ppk-EGFP virgin females. Note that mCD8::RFP and EGFP signals overlapped in the cell bodies (yellow arrow) of SPSNs. White broken lines outline the oviduct. Scale bar, 25 µm. (C–F) Frequencies of germaria containing 1, 2, 3, and 4 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. The number of germaria analyzed is indicated inside the bars. (C) ChaT RNAi by ppk-GAL4. (D) RNAi of nAChRs in dsx+ Tdc2+ neurons. (E) RNAi of Tdc2 by Tdc2-GAL4 along with the silencing of SPSNs. 21 and 31°C were used as the permissive and restrictive temperatures, respectively, of shibirets (shits). (F) RNAi of Oamb and Insp3R by c587-GAL4 along with the silencing of SPSNs. Kir2.1 was used in this experiment. Note that frequencies of germaria containing 4 GSCs increased. Wilcoxon rank sum test with Holm’s correction was used for C, D, E and F. ***p≤0.001 and **p≤0.01; NS, non-significant (p>0.05). All source data are available in Source data 1.
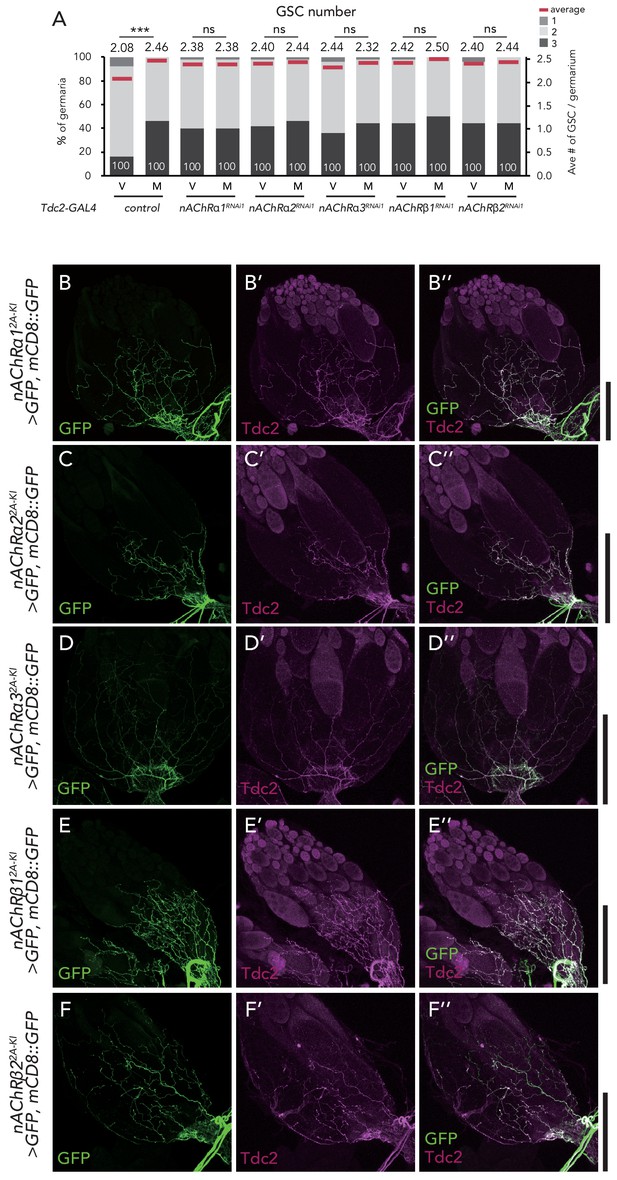
nAChRs are expressed in the ovary-projecting Tdc2 neurons.
(A) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (v) female flies. (B–F) Representative images of the ovaries stained by anti-GFP (B–F) and anti-Tdc2 (B’–F’). Both signals merged on the surface of the ovary (B’’–F’’). Scale bar, 50 µm. Wilcoxon rank sum test was used. ***p≤0.001; NS, non-significant (p>0.05).
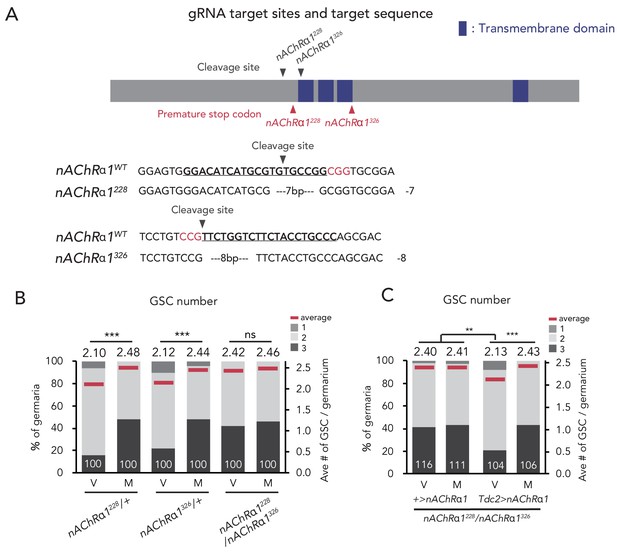
nAChRα1 in the Tdc2 neurons regulates GSC increase.
(A) Schematic representation of gRNA target sites (cleavage sites: gray arrowhead) and premature stop codon (red arrowhead) in coding sequences of nAChRα1 genes. Regions of the putative transmembrane domains of nAChRα1 are highlighted in blue. The target locus in Cas9-induced mutant was PCR-amplified and sequenced. The WT sequence (nAChRα1WT) is shown as reference. The Cas9-gRNA target sequence is underlined with the PAM indicated in red. Inserted nucleotides are indicated in light blue lowercase letters. The indel size is shown next to the sequence. The indel mutation results in a premature stop codon at the 228th and 326th amino acid sequence. (B–C) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. (B) GSC numbers in nAChRα1 genetic mutants. (C) nAChRα1 overexpression in the Tdc2 neurons was sufficient to restore increased GSC in nAChRα1 mutants. Wilcoxon rank sum test with Holm’s correction was used. ***p≤0.001 and **p≤0.01; NS, non-significant (p>0.05). All source data are available in Source data 1.
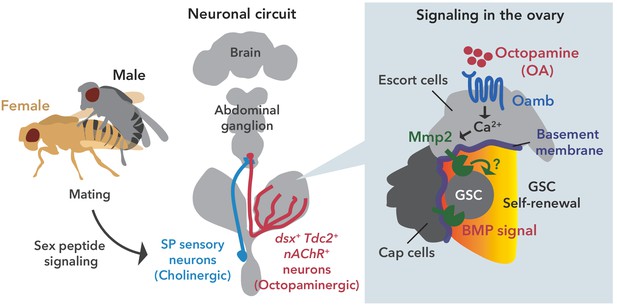
Neuronal octopamine signaling, followed by Oamb-Ca2+-Mmp2 signaling, regulates the mating-induced GSC increase.
The illustration is the proposed working model from our findings here. SP signaling and SP sensory neurons activate dsx+ Tdc2 neurons via acetylcholine signaling. The octopamine released from dsx+ Tdc2 neurons is received by the Oamb in escort cells and then activates intracellular Ca2+ flux. The OA-mediated signaling increases pMad levels in GSCs to evoke mating-induced GSC increase via Mmp2.
Videos
A video image of the GCaMP6 signal in the ex vivo-cultured germarium without OA administration.
A genotype of the germarium was Tj-GAL4 >UAS-GCaMP6s UAS-mCD8::RFP.
A video image of the GCaMP6 signal in the ex vivo-cultured germarium with 100 mM OA administration.
A genotype of the germarium was Tj-GAL4 >UAS-GCaMP6s UAS-mCD8::RFP.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | c587-GAL4 | Manseau et al., 1997 | FBal0150629 RRID:BDSC_67747 | A gift from Hiroko Sano, Kurume University, Japan |
| Genetic reagent (D. melanogaster) | OambΔ | This paper | Detail described in Figure 1—figure supplement 1F | |
| Genetic reagent (D. melanogaster) | nAChRa1228 | This paper | Detail described in Figure 6—figure supplement 2A | |
| Genetic reagent (D. melanogaster) | nAChRa1326 | This paper | Detail described in Figure 6—figure supplement 2A | |
| Genetic reagent (D. melanogaster) | EcRA483T | Bloomington Drosophila Stock Center | BDSC: #5799 RRID:BDSC_5799 FBal0083501 | |
| Genetic reagent (D. melanogaster) | EcRM554fs | Bloomington Drosophila Stock Center | BDSC: #4894 RRID:BDSC_4894 FBal0083490 | |
| Genetic reagent (D. melanogaster) | Vkg::GFP | KYOTO stock center | DGRC #110692 RRID:DGGR_110692 FBal0286156 | |
| Genetic reagent (D. melanogaster) | Dad-LacZ | Tsuneizumi et al., 1997 | FBal0065787 RRID:DGGR_118114 | A gift from Yoshiki Hayashi, University of Tsukuba, Japan |
| Genetic reagent (D. melanogaster) | R44E10-GAL4 | Deady and Sun, 2015 | FBal0252601 PMID:26473732 | A gift from Jianjun Sun, University of Connecticut, USA |
| Genetic reagent (D. melanogaster) | RS-GAL4 | Lee et al., 2009 | FBal0263794 PMID:19262750 | A gift from Kyung-An Han, Pennsylvania State University, USA |
| Genetic reagent (D. melanogaster) | nSyb-GAL4 | Bloomington Drosophila Stock Center | BDSC: #51941 RRID:BDSC_51941 FBti0154973 | FACS (5 ul per test) |
| Genetic reagent (D. melanogaster) | nSyb-GAL80 | Harris et al., 2015 | PMID:26193122 | A gift from James W. Truman, Janelia Research Campus, USA |
| Genetic reagent (D. melanogaster) | tj-GAL4 | KYOTO stock center | DGRC: #104055 RRID:DGGR_104055 FBti0034540 | |
| Genetic reagent (D. melanogaster) | R13C06-GAL4 | Bloomington Drosophila Stock Center | BDSC: #47860 RRID:BDSC_47860 FBal0249828 | |
| Genetic reagent (D. melanogaster) | 109–30 GAL4 | Bloomington Drosophila Stock Center | BDSC: #7023 RRID:BDSC_7023 FBti0027548 | |
| Genetic reagent (D. melanogaster) | c355-GAL4 | Bloomington Drosophila Stock Center | BDSC: #3750 RRID:BDSC_3750 FBti0002591 | |
| Genetic reagent (D. melanogaster) | c306-GAL4 | Bloomington Drosophila Stock Center | BDSC: #3743 RRID:BDSC_3743 FBal0048787 | |
| Genetic reagent (D. melanogaster) | slbo-GAL4 | Bloomington Drosophila Stock Center | BDSC: #6458 RRID:BDSC_6458 FBst0006458 | |
| Genetic reagent (D. melanogaster) | bab1-GAL4 | Bolívar et al., 2006 | FBal0242654 PMID:17013875 | A gift from Satoru Kobayashi, University of Tsukuba, Japan |
| Genetic reagent (D. melanogaster) | nos-GAL4 | KYOTO stock center | DGRC: #107748 RRID:DGGR_107748 FBst0306396 | |
| Genetic reagent (D. melanogaster) | tub > FRT > GAL80>FRT | Bloomington Drosophila Stock Center | BDSC: #38879 RRID:BDSC_38879 FBti0147580 | |
| Genetic reagent (D. melanogaster) | OambKI-RD-GAL4 | Deng et al., 2019 Bloomington Drosophila Stock Center | BDSC: #84677 RRID:BDSC_84677 FBti0209942 | |
| Genetic reagent (D. melanogaster) | Oamb-KI-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRα1-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRα2-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRα3-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRβ1-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRβ2-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | ChaT-GAL4 | Bloomington Drosophila Stock Center | BDSC: #6793 RRID:BDSC_6793 FBst0006793 | |
| Genetic reagent (D. melanogaster) | ppk-GAL4 | Grueber et al., 2007 | FBtp0039691 PMID:17164414 | A gift from Hiroko Sano, Kurume University, Japan |
| Genetic reagent (D. melanogaster) | SPSNs-LexA | Feng et al., 2014 | FBtp0110869 PMID:24991958 | A gift from Young-Joon Kim, Gwangju Institute of Science and Technology, South Korea |
| Genetic reagent (D. melanogaster) | 20xUAS-6xGFP | Bloomington Drosophila Stock Center | BDSC: #52261 RRID:BDSC_52261 FBst0052261 | |
| Genetic reagent (D. melanogaster) | UAS-GFP;UAS-mCD8::GFP | Ito et al., 1997; Lee and Luo, 1999 | FBtp0002652 PMID:9043058 PMID:10457015 | A gift from Kei Ito, University of Cologne, Germany |
| Genetic reagent (D. melanogaster) | UAS-Stinger | Bloomington Drosophila Stock Center | BDSC: #84277 RRID:BDSC_84277 | |
| Genetic reagent (D. melanogaster) | UAS-mCD8::RFP | Bloomington Drosophila Stock Center | BDSC: #32219 RRID:BDSC_32219 FBti0131967 | |
| Genetic reagent (D. melanogaster) | UAS-CsChrimson | Bloomington Drosophila Stock Center | BDSC: #55134 RRID:BDSC_55134 FBti0160571 | |
| Genetic reagent (D. melanogaster) | UAS-Insp3R | Bloomington Drosophila Stock Center | BDSC: #30742 RRID:BDSC_30742 FBti0129829 | |
| Genetic reagent (D. melanogaster) | UAS-OambK3 | Lee et al., 2009 | FBtp0069415 PMID:19262750 | A gift from Kyung-An Han, Pennsylvania State University, USA |
| Genetic reagent (D. melanogaster) | UAS-Timp | Bloomington Drosophila Stock Center | BDSC: #58708 RRID:BDSC_58708 FBti0164930 | A gift from Andrea Page-McCaw, Vanderbilt University, USA |
| Genetic reagent (D. melanogaster) | UAS-nAChRα1 | This paper | Detail described in Material and method | |
| Genetic reagent (D. melanogaster) | UAS > stop > dTrpA1mcherry | von Philipsborn et al., 2011 | FBtp0064577 PMID:21315261 | A gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan |
| Genetic reagent (D. melanogaster) | UAS > stop > TNT | von Philipsborn et al., 2011 | FBtp0020863 PMID:21315261 | A gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan |
| Genetic reagent (D. melanogaster) | UAS > stop > TNTin | von Philipsborn et al., 2011 | FBtp0020863 PMID:21315261 | A gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan |
| Genetic reagent (D. melanogaster) | dsx-FLP | Rezával et al., 2014 | FBal0296301 PMID:24631243 | A gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan |
| Genetic reagent (D. melanogaster) | TRiC; UAS-mCD8::RFP, LexAop2-mCD8::GFP;nSyb-MKII::nlsLexADBDo;UAS-p65AD::CaM | Bloomington Drosophila Stock Center | BDSC: #61679 RRID:BDSC_61679 FBst0061679 | |
| Genetic reagent (D. melanogaster) | ppk-eGFP | Grueber et al., 2003 | FBtp0041053 PMID:12699617 | A gift from Tadashi Uemura, Kyoto University, Japan |
| Genetic reagent (D. melanogaster) | LexAop-Kir2.1 | Feng et al., 2014 | FBtp0110870 PMID:24991958 | A gift from Young-Joon Kim, Gwangju Institute of Science and Technology, South Korea |
| Genetic reagent (D. melanogaster) | UAS-LacZRNAi | Kennerdell and Carthew, 2000 | FBtp0016505 PMID:10932163 | A gift from Masayuki Miura, The University of Tokyo, Japan |
| Genetic reagent (D. melanogaster) | UAS-OambRNAi1 | Bloomington Drosophila Stock Center | BDSC: #31171 RRID:BDSC_31171 FBst0031171 | |
| Genetic reagent (D. melanogaster) | UAS-OambRNAi2 | Bloomington Drosophila Stock Center | BDSC: #31233 RRID:BDSC_31233 FBst0031233 | |
| Genetic reagent (D. melanogaster) | UAS-OambRNAi3 | Vienna Drosophila Resource Center | VDRC: #106511 FBst0478335 | |
| Genetic reagent (D. melanogaster) | UAS-Octβ1RRNAi | Vienna Drosophila Resource Center | VDRC: #110537 FBst0482104 | |
| Genetic reagent (D. melanogaster) | UAS-Octβ2RRNAi | Vienna Drosophila Resource Center | VDRC: #104524 FBst0476382 | |
| Genetic reagent (D. melanogaster) | UAS-Octβ3RRNAi | Vienna Drosophila Resource Center | VDRC: #101189 FBst0473062 | |
| Genetic reagent (D. melanogaster) | UAS-Insp3RRNAi | Bloomington Drosophila Stock Center | BDSC: #25937 FBst0025937 | |
| Genetic reagent (D. melanogaster) | UAS-EcRRNAi | Vienna Drosophila Resource Center | VDRC: #37059 FBst0461818 | |
| Genetic reagent (D. melanogaster) | UAS-Mmp2RNAi1 | Bloomington Drosophila Stock Center | BDSC: #31371 RRID:BDSC_31371 FBst0031371 | |
| Genetic reagent (D. melanogaster) | UAS-Mmp2RNAi2 | Vienna Drosophila Resource Center | VDRC: #330203 FBst0490996 | |
| Genetic reagent (D. melanogaster) | UAS-TimpRNAi1 | Bloomington Drosophila Stock Center | BDSC: #61294 RRID:BDSC_61294 FBst0061294 | |
| Genetic reagent (D. melanogaster) | UAS-TimpRNAi2 | Vienna Drosophila Resource Center | VDRC: #109427 FBst0481116 | |
| Genetic reagent (D. melanogaster) | UAS-Tdc2RNAi1 | Vienna Drosophila Resource Center | VDRC: #330541 FBst0492256 | |
| Genetic reagent (D. melanogaster) | UAS-Tdc2RNAi2 | Bloomington Drosophila Stock Center | BDSC: #25871 RRID:BDSC_25871 FBst0025871 | |
| Genetic reagent (D. melanogaster) | UAS-TβhRNAi1 | Vienna Drosophila Resource Center | VDRC: #107070 FBst0478893 | |
| Genetic reagent (D. melanogaster) | UAS-TβhRNAi2 | Bloomington Drosophila Stock Center | BDSC: #67968 RRID:BDSC_67968 FBst0067968 | |
| Genetic reagent (D. melanogaster) | UAS-ChATRNAi1 | Vienna Drosophila Resource Center | VDRC: #330291 FBst0490951 | |
| Genetic reagent (D. melanogaster) | UAS-ChATRNAi2 | Bloomington Drosophila Stock Center | BDSC: #25856 RRID:BDSC_25856 FBst0025856 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRα1RNAi | Vienna Drosophila Resource Center | VDRC #48159 FBst0467755 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRα2RNAi | Vienna Drosophila Resource Center | VDRC: #101760 FBst0473633 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRα3RNAi | Vienna Drosophila Resource Center | VDRC: #101806 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRβ1RNAi | Vienna Drosophila Resource Center | VDRC: #106570 FBst0478394 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRβ2RNAi | Vienna Drosophila Resource Center | VDRC: #109450 FBst0481138 | |
| Genetic reagent (D. melanogaster) | UAS-nvdRNAi1 | Yoshiyama et al., 2006 | FBal0193613 PMID:16763204 | |
| Genetic reagent (D. melanogaster) | UAS-nvdRNAi2 | Yoshiyama et al., 2006 | FBal0193614 PMID:16763204 | |
| Chemical, compound, drug | Octopamine | Sigma-Aldrich | #O0250 | |
| Chemical, compound, drug | Schneider’s Drosophila medium | Thermo Fisher Scientific | #21720024 | |
| Chemical, compound, drug | 20-hydroxyecdysone | Enzo Life Sciences | ALX-370–012 | |
| Antibody | anti-GFP (chicken polyclonal) | Abcam | #ab13970 | 1:4000 dilution |
| Antibody | anti-RFP (rabbit polyclonal) | Medical and Biological Laboratories | #PM005 | 1:2000 dilution |
| Antibody | anti-Hts 1B1 (mouse monoclonal) | Developmental Studies Hybridoma Bank | 1:50 dilution | |
| Antibody | anti-DE-cadherin DCAD2 (rat monoclonal) | Developmental Studies Hybridoma Bank | 1:50 dilution | |
| Antibody | anti-pH3 (rabbit polyclonal) | Merck Millipore | #06–570 | 1:2000 dilution |
| Antibody | anti-pMad (rabbit polyclonal) | Abcam | #ab52903 | 1:2000 dilution |
| Antibody | anti-Lamin C LC28.26 (mouse monoclonal) | Developmental Studies Hybridoma Bank | 1:10 dilution | |
| Antibody | anti-cleaved Dcp-1 (rabbit polyclonal) | Cell Signaling Technology | #9578 | 1:1000 dilution |
| Antibody | anti-Vasa (rat monoclonal) | Developmental Studies Hybridoma Bank | 1:50 dilution | |
| Antibody | anti-LacZ 40-1a (mouse monoclonal) | Developmental Studies Hybridoma Bank | 1:50 dilution | |
| Antibody | anti-Tdc2 (rabbit polyclonal) | Abcam | #ab128225 | 1:2000 dilution |
| Antibody | Alexa Fluor 546 phalloidin | Thermo Fisher Scientific | #A22283 | 1:200 dilution |
| Antibody | Alexa Fluor 633 phalloidin | Thermo Fisher Scientific | #A22284 | 1:200 dilution |
| Chemical, compound, drug | FluorSave reagent | Merck Millipore | #345789 | |
| Chemical, compound, drug | all trans-Retinal | Sigma-Aldrich | #R2500 | |
| Software, algorithm | ImageJ | https://imagej.nih.gov/ij; RRID:SCR_003070 PMID:22930834 | ||
| Software, algorithm | R | RRID:SCR_001905 |
Additional files
-
Source data 1
Source data of numbers of GSC and other cells.
Raw data of numbers of GSC and other cells for Figure 1, Figure 1—figure supplement 1, Figure 1—figure supplement 3, Figure 2, Figure 2—figure supplement 1, Figure 3, Figure 4, Figure 4—figure supplement 1, Figure 5, Figure 5—figure supplement 1, Figure 6, and Figure 6—figure supplement 2.
- https://cdn.elifesciences.org/articles/57101/elife-57101-data1-v1.xlsx
-
Source data 2
Source data of pMad and Dad-lacZ signal intensity.
Raw data of pMad and Dad-LacZ signal intensity for Figure 1, Figure 1—figure supplement 3, Figure 2, and Figure 4.
- https://cdn.elifesciences.org/articles/57101/elife-57101-data2-v1.xlsx
-
Source data 3
Source data of the TRIC signal intensity.
Raw data of the TRIC signal intensity for Figure 5.
- https://cdn.elifesciences.org/articles/57101/elife-57101-data3-v1.xlsx
-
Source data 4
Source data of the GCaMPs signal intensity.
Raw data of the GCaMPs signal intensity for Figure 2.
- https://cdn.elifesciences.org/articles/57101/elife-57101-data4-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57101/elife-57101-transrepform-v1.docx





