Neuronal octopamine signaling regulates mating-induced germline stem cell increase in female Drosophila melanogaster
Abstract
Stem cells fuel the development and maintenance of tissues. Many studies have addressed how local signals from neighboring niche cells regulate stem cell identity and their proliferative potential. However, the regulation of stem cells by tissue-extrinsic signals in response to environmental cues remains poorly understood. Here we report that efferent octopaminergic neurons projecting to the ovary are essential for germline stem cell (GSC) increase in response to mating in female Drosophila. The neuronal activity of the octopaminergic neurons is required for mating-induced GSC increase as they relay the mating signal from sex peptide receptor-positive cholinergic neurons. Octopamine and its receptor Oamb are also required for mating-induced GSC increase via intracellular Ca2+ signaling. Moreover, we identified Matrix metalloproteinase-2 as a downstream component of the octopamine-Ca2+ signaling to induce GSC increase. Our study provides a mechanism describing how neuronal system couples stem cell behavior to environmental cues through stem cell niche signaling.
eLife digest
Stem cells have the unique ability to mature into the various, specialized groups of cells required for organisms to work properly. Local signals released by the tissues immediately surrounding stem cells usually trigger this specialization process. However, recent studies have revealed that external signals, such as hormones or neurotransmitters (the chemicals used by nerve cells to communicate), can also control the fate of stem cells. This is particularly the case during development, or in response to events such as injury.
In the right conditions, germline stem cells can specialize into the egg or sperm required for many animals to reproduce. In fruit flies for example, the semen contains proteins that activate a cascade of molecular events in the female nervous system, ultimately resulting in female germline stem cells multiplying in the ovaries after mating. Yet, exactly how this process takes place was still unclear. To investigate this question, Yoshinari et al. focused on nerve cells in the fruit fly ovary which produce a neurotransmitter called octopamine.
The experiments assessed changes in the ovaries of female fruit flies after mating, piecing together the sequence of events that activate germline stem cells. This showed that first, mating triggers the release of octopamine from the nerve cells. In turn, this activates a protein called Oamb, which is studded through the membrane of cells present around germline stem cells. Turning on Oamb prompts a cascade of molecular events which include an enzyme called Matrix metalloproteinase 2 regulating the signal sent from the local environment to germline stem cells.
As mammals use a neurotransmitter similar to octopamine, future fruit fly studies could shed light on how neurotransmitters activate stem cells in other animals. Ultimately, unravelling the way external signals trigger the specialization process may offer insight into how diseases arise from uncontrolled stem cell activity.
Introduction
Animal tissues are built from cells originally derived from stem cells (Spradling et al., 2001). During normal development and physiology, this robust stem cell system is precisely regulated (Drummond-Barbosa, 2008). Conversely, the dysregulation of these cells can result in abnormal tissue integrity and lead to deleterious diseases. Previous studies have revealed that many types of stem cells reside in a specialized microenvironment, or niche, where they are exposed to local signals required for their function and identity (Morrison and Spradling, 2008; Spradling et al., 2001). Recently, researchers have demonstrated how stem cell activity is regulated by tissue-extrinsic signals, such as hormones and neurotransmitters. For instance, in mammals, hematopoietic stem cells, mammary stem cells, muscle stem cells, and neural stem cells are influenced by sex hormones such as estrogen (Asselin-Labat et al., 2010; Bramble et al., 2019; Kim et al., 2016; Nakada et al., 2014). Retinoic acid and thyroid hormone play essential roles in the differentiation of testicular stem cells and neural stem cells, respectively (Gothié et al., 2017; Ikami et al., 2015). In addition, mesenchymal stem cell proliferation is stimulated by adrenaline (Wu et al., 2014). However, the when, how, and why these humoral factors are produced, circulated, and received during stem cell regulation remain to be elucidated.
The ovaries of the fruit fly Drosophila melanogaster are an excellent model system on how stem cell lineages are shaped by both local niche signals and tissue-extrinsic signals (Drummond-Barbosa, 2019). D. melanogaster ovary is composed of 16–20 chains of developing egg chambers called ovarioles. The anterior-most region of which, known as the germarium, contains germline stem cells (GSCs) that give rise to the eggs (Figure 1A and B). GSCs are adjacent to the somatic niche cells, which comprises cap cells, escort cells, and terminal filament cells (Figure 1A). After GSC divides, one daughter cell that remains attached to the niche cells retains its GSC identity, whereas the remaining daughter cells are displaced away from the niche cells and differentiate into cystoblast (CB). Each CB then undergoes differentiation into 15 nurse cells and one oocyte in each egg chamber, which is surrounded by somatic follicle cells.
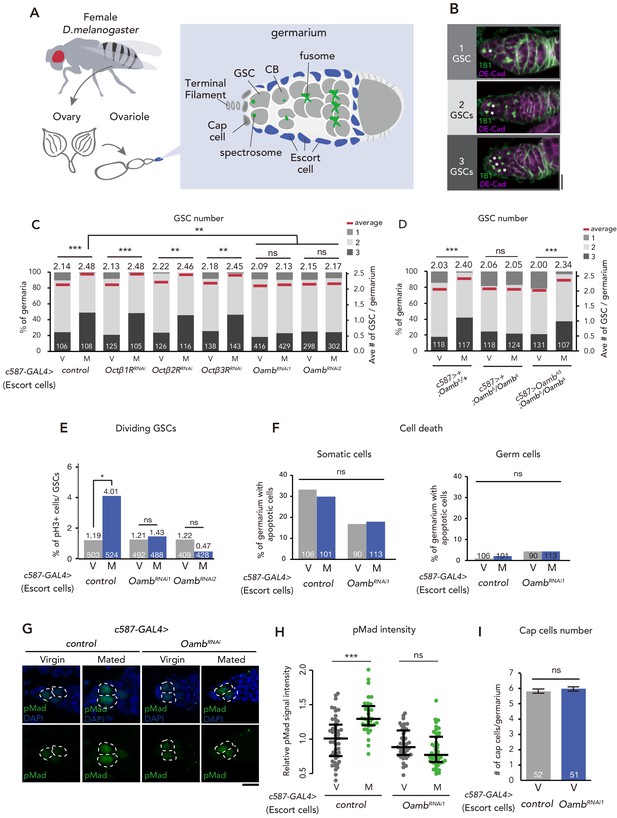
Post-mating GSC increase requires Oamb in the escort cells.
(A) A schematic representation of Drosophila germarium. GSCs reside in a niche consisting of somatic cells such as cap cells, terminal filament cells, and escort cells and are identifiable by their stereotypical spectrosome morphology and location (adjacent to cap cells). GSC division produces one self-renewing daughter and one cystoblast (CB) that differentiates into a germline cyst. (B) Representative images of wild-type (w1118) female adult germariums, containing 1, 2 and 3 GSCs from top to bottom. The samples were stained with monoclonal antibody 1B1 (green) and anti-DE-cadherin (magenta), which stain the spectrosome and overall cell membranes, respectively. GSCs are indicated by asterisk. Scale bar, 20 µm. (C–D) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control in D. (E) The ratio of pH3+ GSCs per total GSCs. (F) The ratio of apoptotic (Dcp-1+) somatic cells and germ cells per germarium. c587>+ flies were used as the control. (G) Representative images of adult female germaria immunostained with anti-pMad antibody (green) and DAPI (blue) are shown. GSCs are outlined with dotted lines. Scale bar, 10 µm. (H) Quantification of relative pMad intensity levels in the GSCs (i.e. virgin (V), mated (M)) as normalized to the pMad intensity in CBs. Each sample number was at least 25. The three horizontal lines for each sample indicate lower, median, and upper quartiles. (I) The number of cap cells per germarium in the control and Oamb RNAi driven by c587-GAL4. Values on the y-axis are presented as the mean with standard error of the mean. c587>+ flies were used as the control. For C-F, and I the number of germaria analyzed is indicated inside the bars. Wilcoxon rank sum test with Holm’s correction was used for C, D, H, and I. Fisher’s exact test with Holm’s correction was used for E and F. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, nonsignificant (p>0.05). All source data are available in Source data 1 and 2.
GSC niche produces and secretes several local niche signals that regulate the balance between GSC self-renewal and differentiation (Hayashi et al., 2020; Kirilly and Xie, 2007; Spradling et al., 2011). For example, bone morphogenetic protein (BMP) ligands Decapentaplegic (Dpp) and Glass bottom boat (Gbb) are produced from the niche cells and directly activate BMP receptors in GSCs, leading to the repression of the differentiation inducer, bag-of-marbles (bam) (Morrison and Spradling, 2008; Zhang and Cai, 2020). Recent D. melanogaster GSC studies have also contributed to understanding of the systemic regulation of stem cell proliferation and maintenance in response to external environmental cues (Ables and Drummond-Barbosa, 2017; Drummond-Barbosa, 2019; Lin and Hsu, 2020; Yoshinari et al., 2019). For example, protein restriction results in a reduction in GSC division, which is mediated by Drosophila insulin-like peptides (DILPs) (LaFever, 2005). In addition, nutrients influence GSC maintenance via the adipocyte metabolic pathway (Armstrong and Drummond-Barbosa, 2018; Matsuoka et al., 2017).
Besides nutrients, we have recently found out that mating is another external cue that significantly affects D. melanogaster GSC increase. Mated females show a dramatic increase in egg production, as well as GSC, which is induced by a male-derived peptide from the seminal fluid called sex peptide (SP) (Kubli, 2003; Yoshinari et al., 2019). SP is received by its specific receptor, sex peptide receptor (SPR), in a small subset of SPR-positive sensory neurons (SPSNs), which are located in the uterine lumen and send afferent axons into the tip of the abdominal ganglion (Häsemeyer et al., 2009; Yapici et al., 2008). The SP-SPR signaling in the SPSNs stimulates the biosynthesis of the ovarian insect steroid hormones (ecdysteroids), which play an essential role in mating-induced GSC increase (Ameku et al., 2017; Ameku and Niwa, 2016; Uryu et al., 2015). Because SPSNs do not directly innervate into the ovary, it is hypothesized that a signal and its signaling pathway are involved in bridging the gap between SPSNs and GSCs. However, it is still unclear how mating information is transmitted from SPSNs to GSCs at the molecular and cellular levels.
Here, we present a series of new findings that reveal a novel and fundamental neuronal mechanism connecting SPSNs and GSCs to regulate mating-induced GSC increase. We demonstrate that a small subset of neurons directly innervating into the ovary plays an indispensable role in regulating mating-induced GSC increase. These neurons produce the monoamine neurotransmitter, octopamine (OA), the insect equivalent of noradrenaline (Roeder, 2005). We also show that the neuronal activity of the OA-producing neurons is required for mating-induced GSC increase. Moreover, we find that the OA directory activates GSC increase through its receptor, octopamine receptor in mushroom body (Oamb), followed by Ca2+ signaling in the ovarian escort cells. Furthermore, OA/Oamb signaling requires Matrix metalloproteinase 2 (Mmp2) to activate GSC increase in the ovarian escort cells. Finally, we show that SPSNs relay the mating signal to the ovary-projecting OA neurons via nicotinic acetylcholine receptor signaling. Taken together, we propose a novel efferent neuronal pathway that transmits mating stimulus to the GSC to control stem cell number. Our study provides a mechanism describing how neuronal system couples stem cell behavior to environmental cues, such as mating, through stem cell niche signaling.
Results
Mating-induced GSC increase requires the octopamine receptor Oamb in ovarian escort cells
As a candidate signal that bridges between SPSNs and GSC increase, we focused on the biogenic amine, OA, because a part of the octopaminergic neurons innervate to the ovary and the oviduct (Heifetz et al., 2014; Rezával et al., 2014). Moreover, it has been reported that OA and Oamb signaling regulate ovulation process and ovarian-muscle contraction (Deady and Sun, 2015; Monastirioti, 2003; Rezával et al., 2014). We first conducted transgenic RNAi screen against 4 OA receptor genes with c587-GAL4, which is active in the ovarian-somatic cells, including the escort cells of the germarium (Manseau et al., 1997). In control females, the mated ones exhibited an increase in GSC number as we have reported previously (Ameku and Niwa, 2016; Figure 1C). In contrast, c587-GAL4–mediated Oamb knock-down (c587 >OambRNAi) showed significantly impaired mating-induced GSC increase (Figure 1C). This phenotype was observed with two independent UAS-Oamb-RNAi strains (OambRNAi1 and OambRNAi2) (Figure 1C), each of which targeted a different region in the Oamb mRNA. The specificity of Oamb was also confirmed by the fact that the c587-GAL4–driven transgenic RNAi of other octopamine receptor genes (Octβ1R, Octβ2R and Octβ3R) (Ohhara et al., 2012) had no significant effect on the GSC number between virgin and mated females (Figure 1C). Therefore, Oamb has a pivotal role in mating-induced GSC increase.
We next examined in which ovarian-somatic cells Oamb regulates mating-induced GSC increase with several GAL4 lines that are active in the specific ovarian-somatic cells. Previous studies have demonstrated that Oamb in mature follicle cells and in the oviduct has a significant role in ovulation (Deady and Sun, 2015; Lee et al., 2009; Lee et al., 2003). Therefore, it is possible that Oamb may indirectly induce GSC increase via Oamb-mediated ovulation processes. However, mating-induced GSC increase was not impaired by Oamb RNAi in the stage-14 follicle cells by R44E10-GAL4 (Deady and Sun, 2015) (R44E10 >OambRNAi), in the stage 9–10 follicle cells by c355-GAL4, c306-GAL4, and slbo-GAL4 (Barth et al., 2012), or in the common oviduct by RS-GAL4 (Lee et al., 2003) (RS >OambRNAi) (Figure 1—figure supplement 1A,B and E). These data suggest that mating-induced GSC increase is independent from the ovulation process.
Consistent with the observation using c587-GAL4, Oamb RNAi by Traffic jam (Tj)-GAL4 (Olivieri et al., 2010) (Tj >OambRNAi), R13C06-GAL4, and 109–30-GAL4 (Sahai-Hernandez and Nystul, 2013), which are active in the pan-ovarian-somatic cells, the escort cells, and the germarium follicle cells, respectively, also resulted in the failure of mating-induced GSC increase (Figure 1—figure supplement 1C and E). On the other hand, Oamb RNAi in the cap cells (bab >OambRNAi) and germ cells (nos >OambRNAi) had no effect on GSC increase (Figure 1—figure supplement 1D). These results suggest that Oamb in the escort cells or the follicle cells of the germarium plays an essential role in mating-induced GSC increase.
It must be noted that c587-GAL4 and Tj-GAL4 are expressed not only in the ovarian-somatic cells but also in the nervous system (Ameku et al., 2018). Moreover, Oamb is expressed in the nervous system (Han et al., 1998). However, Oamb RNAi in the nervous system (nSyb >OambRNAi) did not affect the mating-induced GSC increase (Figure 1—figure supplement 1D), suggesting that the impairment of GSC increase of c587 >OambRNAi or tj >OambRNAi is not due to gene knock-down in neuronal cells but rather in the ovarian-somatic cells. These data also support our idea that Oamb in the ovarian-somatic cells regulate mating-induced GSC increase.
To confirm the role of Oamb in mating-induced GSC increase, we generated a Oamb complete loss-of-function genetic allele by Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) technology (Kondo and Ueda, 2013; Figure 1—figure supplement 1F). Similar to Oamb RNAi females, Oamb homozygous mutant females (c587>+; OambΔ/OambΔ) did not exhibit mating-induced GSC increase (Figure 1D). In addition, the GSC increase of OambΔ/OambΔ was restored by overexpression of Oamb in the escort cells (c587 >OambAS; OambΔ/OambΔ). These findings are all consistent with the idea that Oamb in escort cells modulates GSC increase after mating.
We also examined Oamb expression in the ovarian-somatic cells by two Oamb-knock-in GAL4 lines (Deng et al., 2019; Kondo et al., 2020). However, we could not detect any reliable signals in the germarium or the mature follicle cells via these GAL4 lines with UAS-GFP and UAS-Stinger lines (Figure 1—figure supplement 2A,B,C and D). We speculate that this may be due to lower amounts of Oamb transcript in the germarium.
Oamb in escort cells is required for GSC increase after mating
Because mating-induced GSC increase is accompanied by GSC division (Ameku and Niwa, 2016), We next examined whether Oamb in the escort cells is involved in GSC division after mating. We determined the number of GSCs during the M phase by staining using anti-phospho-Histone H3 (pH3) in control and c587 >OambRNAi females. In control female flies, mating increased the frequency of GSCs in the M phase (Figure 1E), whereas in c587 >OambRNAi flies, this was not observed. We also monitored the fraction of apoptotic cells in the germarium by staining with anti-cleaved death caspase-1 (Dcp-1), a marker for apoptotic cells (Song et al., 1997). The number of apoptotic cells in the germarium did not change in c587 >OambRNAi female flies compared with controls (Figure 1F), suggesting that Oamb activates GSC increase by pushing the cell cycle of GSCs and that the lack of mating-induced GSC increase in Oamb RNAi is not due to the enhancement of cell death.
Our previous studies revealed that mating-induced GSC increase is mediated by GSC niche signals (Ameku et al., 2018; Ameku and Niwa, 2016). In particular, Decapentaplegic (Dpp), the fly counterpart to bone morphogenetic protein (BMP), is the essential niche signal (Spradling et al., 2011; Xie and Spradling, 1998). We therefore examined whether Oamb knock-down affects Dpp signaling in GSCs by measuring the level of phosphorylated Mad (pMad), a readout of Dpp signaling activation (Chen and McKearin, 2003; Raftery and Sutherland, 1999). We confirmed that mating induced the increase in pMad level in GSCs, whereas mating did not increase pMad levels in c587 >OambRNAi animals (Figure 1G and H). We also confirmed that mating increased the signal intensity of Daughters against dpp (Dad)-LacZ, a reporter gene that reflect an expression of the BMP target gene Dad, while Oamb knock-down in the escort cells impaired the increase of Dad-LacZ signal after mating (Figure 1—figure supplement 3A and B). These results suggest that mating activates the BMP signal in GSCs through Oamb in the escort cells, thereby resulting in the increase in GSCs.
Further, we determined the number of cap cells, which are critical components of the GSC niche (Xie and Spradling, 1998). c587 >OambRNAi did not change the number of cap cells in virgin nor mated female flies, suggesting that Oamb knock-down does not affect the overall architecture of the niche (Figure 1I). Overall, Oamb in the escort cells plays a pivotal role in mating-induced GSC increase.
OA administration is sufficient to induce GSC increase and BMP signaling in GSCs in an Oamb-dependent manner
To examine whether OA is received in the ovary but not in other organs to induce GSC increase, we cultured dissected virgin ovaries ex vivo with or without purified OA in the culture medium. After incubation for 12 hr, the ovaries cultured with OA had more GSCs as compared to those without OA (Figure 2—figure supplement 1A). Whereas the minimal OA concentration to induce ex vivo GSC increase was 1 μM, hereafter we used 100 μM OA because the GSC number plateaued with this concentration (Figure 2—figure supplement 1A). This OA-mediated ex vivo GSC increase was not observed in c587 >OambRNAi virgin ovaries (Figure 2A).
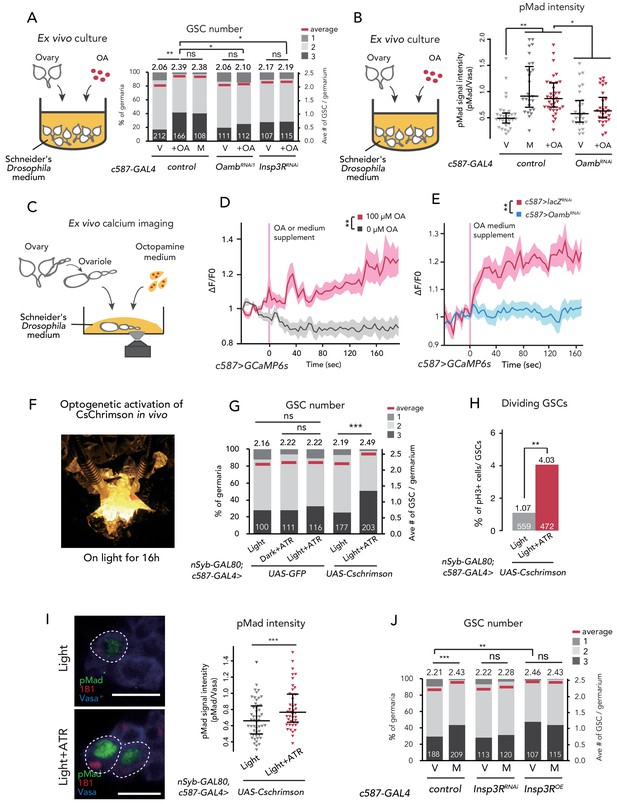
Ca2+ signaling is necessary for mating-induced GSC increase.
(A) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis). The ovaries were dissected from virgin (V), mated (M), and virgin ovaries cultured with OA (+OA). c587>+ flies were used as the control. The number of germaria analyzed is indicated inside the bars. (B) Quantification of relative pMad intensity levels in the GSCs of ex vivo cultured ovaries (i.e. virgin (V), mated (M), and virgin cultured with OA (+OA)) as normalized to the pMad intensity in CBs. For the quantification of pMad intensity, the cell boundaries of GSCs and CBs were determined using anti-Vasa staining. Each sample number was at least 25. The three horizontal lines for each sample indicate lower, median, and upper quartiles. (C) A schematic representation of ex vivo calcium imaging. The dissected ovariole was incubated in Schneider’s Drosophila medium with or without OA. (D) Changes in the relative fluorescence intensity of GCaMP6s after 200 s without stimulation (n = 8) or with stimulation (n = 10) with 100 μM OA, and (E) with 100 μM OA as control (c587 >LacZRNAi, n = 8) and c587 >OambRNAi (n = 8) female ovaries. Note that OA significantly increased the calcium response in escort cells, but OambRNAi impaired the calcium response. Statistical analysis was done at 120 s. (F) Equipment setup for optogenetic activation of ChR. Flies were placed under the light for 16 hr before dissection. (G) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) with light, with light and all trans-retinal (ATR) or with dark and ATR. Germarium was dissected from virgin females. nSyb-GAL80; c587 >GFP flies were used as control. The number of germaria analyzed is indicated inside the bars. (H) The ratio of pH3+ GSCs and total GSCs. The number of GSCs analyzed is indicated inside the bars. (I, left) Representative images of adult female germaria immunostained with anti-pMad antibody (green), anti-1B1 antibody (red), and anti-Vasa antibody (germ cell marker; blue) are shown. GSCs are outlined with dotted lines. (I, right) Quantification of the relative pMad intensity in GSCs, which was normalized to that in CBs. For the quantification of pMad intensity, the cell boundaries of GSCs and CBs were determined using anti-Vasa staining. Each sample number is at least 30. The three horizontal lines for each data sample indicate lower, median, and upper quartiles. (J) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control. The number of germaria analyzed is indicated inside the bars. Wilcoxon rank sum test with Holm’s correction was used for A, B, D, E, G, I, and J. Fisher’s exact test was used for H. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, non-significant (p>0.05). All source data are available in Source data 1, 2, and 4.
We also examined whether OA treatment affects Dpp signaling in GSCs ex vivo by measuring the level of pMad. We confirmed that OA treatment was sufficient to induce the increase in pMad level in GSCs, even in the ex vivo culture system (Figure 2B). On the other hand, OA treatment did not increase pMad levels in c587 >OambRNAi ovaries (Figure 2B). These results suggest that OA activates the BMP signal in GSCs through Oamb in the escort cells.
Ca2+ signaling in escort cells is essential for OA-dependent GSC increase
Upon OA binding, Oamb evokes Ca2+ release from the endoplasmic reticulum (ER) into the cytosol, leading to a transient increase in intercellular Ca2+ concentration ([Ca2+]i) (Han et al., 1998). To determine whether OA induces GSC increase via affecting [Ca2+]i in escort cells, we first monitored [Ca2+]i using a genetically encoded calcium sensor, GCaMP6s (Nakai et al., 2001; Ohkura et al., 2012). We dissected the virgin ovaries, in which GCaMP6s transgene was expressed driven by Tj-GAL4 or c587-GAL4, cultured them ex vivo, and then observed GCaMP6s fluorescence (Figure 2C). We found that 100 μM of OA treatment evoked an increase in GCaMP6s fluorescence in the escort cells and follicle cells, whereas the control medium treatment (0 μM of OA) did not show any increase in fluorescence (Figure 2D and Figure 2—figure supplement 1B; also see Videos 1 and 2). In addition, the OA-mediated increase in GCaMP6s fluorescence intensity was not observed in the germarium of Oamb RNAi flies (Figure 2E), suggesting that the OA-dependent increase in [Ca2+]i in the germarium is required for Oamb. Notably, the timeline of signal increase was very slow as this fluorescence increases progressively. This response to OA treatment was similar to a report featuring mature follicle cells of stage-14 oocytes (Deady and Sun, 2015).
A video image of the GCaMP6 signal in the ex vivo-cultured germarium without OA administration.
A genotype of the germarium was Tj-GAL4 >UAS-GCaMP6s UAS-mCD8::RFP.
A video image of the GCaMP6 signal in the ex vivo-cultured germarium with 100 mM OA administration.
A genotype of the germarium was Tj-GAL4 >UAS-GCaMP6s UAS-mCD8::RFP.
We also employed another approach utilizing the light-gated cation channel, CsChrimsonn (Klapoetke et al., 2014). We prepared c587-GAL4 >CsChrimson flies in combination with nSyb-GAL80 (Harris et al., 2015), allowing the expression of CsChrimson gene only in the germarium but not in the nervous system. Because CsChrimson requires all trans-retinal (ATR) to form its proper protein conformation (Wang et al., 2012), we utilized the flies fed with and without ATR supplement as experimental and control groups, respectively. When we irradiated an orange-light to c587-GAL4, nSyb-GAL80 >CsChrimson flies to induce Ca2+ flux in the germarium (Figure 2F), GSC increased in the virgin females (Figure 2G). In addition, the ratio of GSC in the M phase increased by CsChrimson activation (Figure 2H). Moreover, the pMad level in GSCs was increased by CsChrimson activation, suggesting that the forced Ca2+ flux is sufficient to induce GSC increase through the upregulation of BMP signaling (Figure 2I).
We next confirmed whether the downstream component of Ca2+ signaling is involved in the mating-induced GSC increase. The knock-down of Inositol 3-receptor (c587 >Insp3RRNAi) encoding a protein that releases the stored Ca2+ from ER suppressed GSC increase in mated females (Figure 2J). Conversely, the overexpression of Insp3R in the escort cells increased the GSC number in virgin females (Figure 2J). Furthermore, OA-mediated ex vivo GSC increase was not observed in c587 >Insp3RRNAi virgin ovaries (Figure 2A). Overall, we demonstrated that OA signaling regulates the mating-induced GSC increase by controlling Ca2+ signaling in escort cells, which is thereby necessary and sufficient to induce GSC increase.
Ovarian ecdysteroid signaling is required in the OA-Oamb-Ca2+-dependent GSC increase
The biosynthesis and signaling of ecdysteroid in the ovary are required for the mating-induced GSC increase (Ameku et al., 2017; Ameku and Niwa, 2016). Therefore, we next examined whether ecdysteroid signaling has a pivotal role in the OA-Oamb-Ca2+-dependent GSC increase. As we have previously reported, the RNAi of neverland (nvd), which encodes an ecdysteroidogenic enzyme (Yoshiyama-Yanagawa et al., 2011; Yoshiyama et al., 2006) in the escort cells, suppressed the mating-induced GSC increase (Figure 3A; Ameku et al., 2017; Ameku and Niwa, 2016). We also found a similar phenotype in the RNAi of ecdysone receptor (EcR) in the escort cells (Figure 3A). To assess the requirement of ecdysteroid biosynthesis and signaling in OA-induced GSC increase, we employed an ex vivo experiment. Interestingly, the OA-mediated GSC increase was not observed in nvd RNAi ovaries (c587 >nvdRNAi) (Figure 3B). Moreover, this impairment of GSC increase in c587 >nvdRNAi was restored with the administration of 20-hydroxyecdysone (20E) in the culture media. On the other hand, 20E treatment without OA did not induce GSC increase in the control ovaries (Figure 3B). This observation is consistent with our previous study showing that wild-type virgin females fed with 20E did not exhibit any increase in GSCs (Ameku and Niwa, 2016). Therefore, 20E is required for the OA-mediated increase in GSCs; however, by itself, 20E is not sufficient to induce GSC increase.
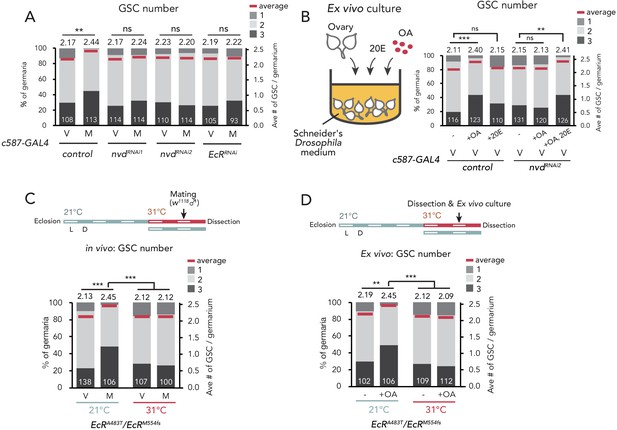
Ecdysteroid signaling is necessary for OA-mediated GSC increase.
(A–D) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control. The number of germaria analyzed is indicated inside the bars. (A) GSC number of nvd and EcR RNAi flies in vivo. (B) Virgin ovaries were cultured ex vivo with or without OA and 20E (+OA, +20E, −), and then the GSC number was determined. (C–D) Experiments using a temperature-sensitive allele EcRA483T. 21°C and 31°C were used as the permissive and restrictive temperatures, respectively. Flies were cultured at 21°C and transferred to 31°C 1 d prior to the assays (L; light, D; dark). (C) GSC number in vivo. (D) Virgin ovaries were cultured ex vivo with or without OA (+OA, −). The number of germaria analyzed is indicated inside the bars. Wilcoxon rank sum test with Holm’s correction was used for statistical analysis. ***p≤0.001 and **p≤0.01; NS, non-significant (p>0.05). All source data are available in Source data 1.
To assess the role of EcR in downstream OA signaling, we utilized the temperature-sensitive and null alleles of EcR (EcRA483T and EcRM54fs, respectively) (Bender et al., 1997). At a restrictive temperature of 31°C, the mating-induced GSC increase was suppressed in EcRA483T/EcRM54fs flies, consistent with our previous report (Ameku and Niwa, 2016; Figure 3C). In the ex vivo experiment, the OA-dependent GSC increase was also suppressed in EcRA483T/EcRM54fs ovaries (Figure 3D). These results suggest that OA-Oamb-Ca2+ signaling requires ovarian ecdysteroid signaling.
Matrix metalloproteinase two acts downstream of OA-Oamb-Ca2+ signaling
So far, we have identified four components with indispensable roles in mating-induced GSC increase, namely OA, Oamb, Ca2+, and ecdysteroids. Interestingly, recent studies have reported that they are also essential in the follicle rupture in D. melanogaster ovary (Deady and Sun, 2015; Knapp and Sun, 2017). In this process, Matrix metalloproteinase 2 (Mmp2), a membrane-conjugated proteinase, acts downstream of the OA-Oamb-Ca2+ signaling pathway in mature follicle cells (Deady et al., 2015). Because recent studies have indicated the expression of Mmp2 in niche cells, including escort cells (Pearson et al., 2016; Wang and Page-McCaw, 2014), we examined Mmp2 function in the escort cells using RNAi of Mmp2 with c587-GAL4. Similar to c587 >OambRNAi, c587 >Mmp2RNAi impaired mating-induced GSC increase (Figure 4A). Notably, Mmp2 knock-down in cap cells by bab-GAL4 also suppressed GSC increase, suggesting that Mmp2 acts in both escort cells and cap cells to induce GSC increase (Figure 4—figure supplement 1A). We also found that Mmp2 RNAi in the ovarian-somatic cells by another GAL4 (Tj >Mmp2RNAi1) resulted in the failure of mating-induced GSC increase, whereas Mmp2 RNAi in the nervous system (nSyb >Mmp2RNAi1) and in the follicle cells of stage 14 oocytes (R44E10 >Mmp2RNAi1) had no effect on GSC increase (Figure 4—figure supplement 1B). These results suggest that Mmp2 in the escort cells and cap cells are necessary to induce post-mating GSC increase.
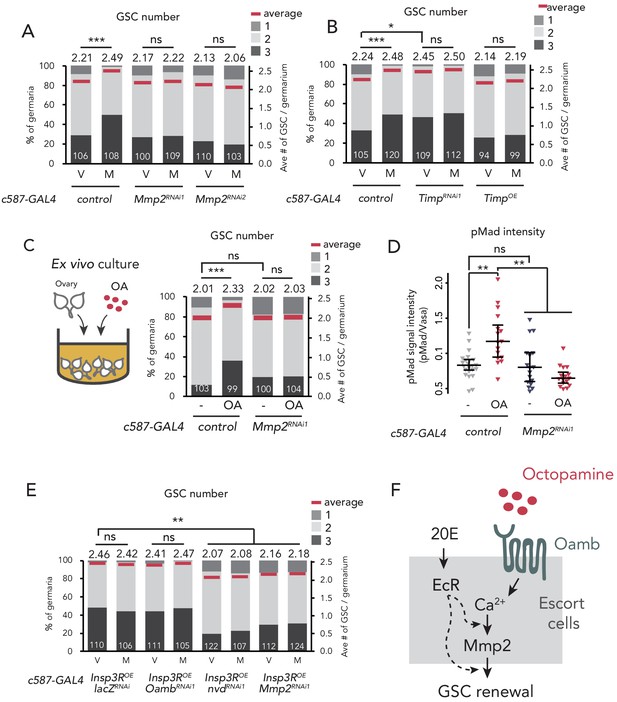
Mmp2 is necessary for OA-mediated GSC increase.
(A–C, E) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. c587>+ flies were used as the control. The number of germaria analyzed is indicated inside the bars. (A) Mmp2 RNAi by c587-GAL4 driver. (B) RNAi and the overexpression of Timp by c587-GAL4 driver. (C) Ex vivo culture experiment using c587 >Mmp2 RNAi. OA was added into the ex vivo culture medium. Cultured with or without OA (+OA, -, respectively) is indicated under each bar. (D) Quantification of the relative pMad intensity in GSCs of the ex vivo cultured ovaries normalized to pMad intensity in CBs. Cultured with or without OA (+OA, −) is indicated under each bar. For the quantification of pMad intensity, the cell boundaries of GSCs and CBs were determined using anti-Vasa staining (n > 15). The three horizontal lines for each data sample indicate lower, median, and upper quartiles. (E) Oamb, nvd, or Mmp2 RNAi in the genetic background of c587 >Insp3R overexpression. (F) A model of signaling in the escort cell to induce the mating-induced GSC increase. Oamb in the escort cells receives OA, and induce [Ca2+]i in the cells. The [Ca2+]i induces GSC increase via Mmp2. Ecdysteroid signaling is also involved in this process. Wilcoxon rank sum test with Holm’s correction was used. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, non-significant (p>0.05). All source data are available in Source data 1 and 2.
In the GSC niche cells in D. melanogaster, Tissue inhibitors of metalloproteinases (Timp) gene encoding an endogenous proteinase inhibitor of Mmp2 is expressed (Gomis-Rüth et al., 1997; Page-McCaw et al., 2003; Pearson et al., 2016). The knock-down of Timp in escort cells induced GSC increase even in virgin females, whereas its overexpression suppressed mating-induced GSC increase (Figure 4B). Consistent with Mmp2 knock-down, Timp knock-down in cap cells (bab >TimpRNAiRNAi2) increased the GSC number in virgin females, whereas its knock-down in the follicle cells of stage 14 oocyte (R44E10 >TimpRNAiRNAi2) had no effect (Figure 4—figure supplement 1C). These data suggest that Mmp2 activity in the GSC niche cells is necessary for mating-induced GSC increase.
We next examined whether Mmp2 is necessary for OA-induced GSC increase. Our ex vivo culture experiment revealed that OA-induced GSC increase was suppressed in c587 >Mmp2 RNAi flies, suggesting that Mmp2 acts downstream of OA signaling (Figure 4C). Moreover, the OA-dependent upregulation of pMad level in GSCs was suppressed in c587 >Mmp2 RNAi flies (Figure 4D). Notably, in virgin females, Mmp2 RNAi affected neither the GSC number nor the cap cell number, indicating that Mmp2 RNAi does not influence the overall niche architecture (Figure 4—figure supplement 1D). Mmp2 in the mature follicle cells cleaves and downregulates collagen VI, also known as Viking (Vkg) and is the major component of the basement membrane (Deady et al., 2017; Wang et al., 2008). However, Mmp2 RNAi had no effect on Vkg::GFP level around the cap cells (Figure 4—figure supplement 1E). Therefore, the suppression of mating-induced GSC increase in Mmp2 RNAi does not likely depend on the collagen VI level.
To examine the epistasis of OA/Oamb-Ca2+ signaling, ecdysteroid signaling, and Mmp2, we knocked down Oamb, nvd, or Mmp2 in c587 >Insp3ROE genetic background, where Ca2+ signaling was forcedly upregulated. Whereas the Oamb RNAi did not suppress GSC increase in virgin female (c587 >Insp3ROE, OambRNAi1), the RNAi of nvd or Mmp2 suppressed GSC increase even when Ca2+ signaling were activated (c587 >Insp3ROE, nvdRNAi or c587 >Insp3ROE, Mmp2RNAi1) (Figure 4E). These results suggest that ecdysteroid signaling and Mmp2 act downstream of Ca2+ signaling in OA-induced GSC increase. Taken together, both the OA-Oamb signaling and the downstream Ca2+ signaling regulate the mating-induced GSC increase via Mmp2 and ecdysteroid signaling (Figure 4F).
Octopamine from dsx+ Tdc2+ neurons regulates the mating-induced GSC increase
To examine the in vivo role of OA in mating-induced GSC increase, we silenced the expression of Tyrosine decarboxylase 2 (Tdc2) and Tyramine β hydroxylase (TβH) genes, which code for enzymes responsible for OA biosynthesis (Cole et al., 2005; Monastirioti et al., 1996; Figure 5A), with Tdc2-GAL4 driver-mediated RNAi (Tdc2 >Tdc2RNAi1, Tdc2 >TbHRNAiRNAi1). Similar to the phenotype of Oamb RNAi, Tdc2 or TbH RNAi with Tdc2-GAL4 or nSyb-GAL4 impaired the mating-induced GSC increase (Figure 5A and Figure 5—figure supplement 1A–B). Moreover, the impairment of GSC increase was restored when Tdc2 or TbH RNAi flies were fed with food supplemented with OA (Figure 5A), supporting our hypothesis that OA is responsible for the mating-induced GSC increase in vivo.
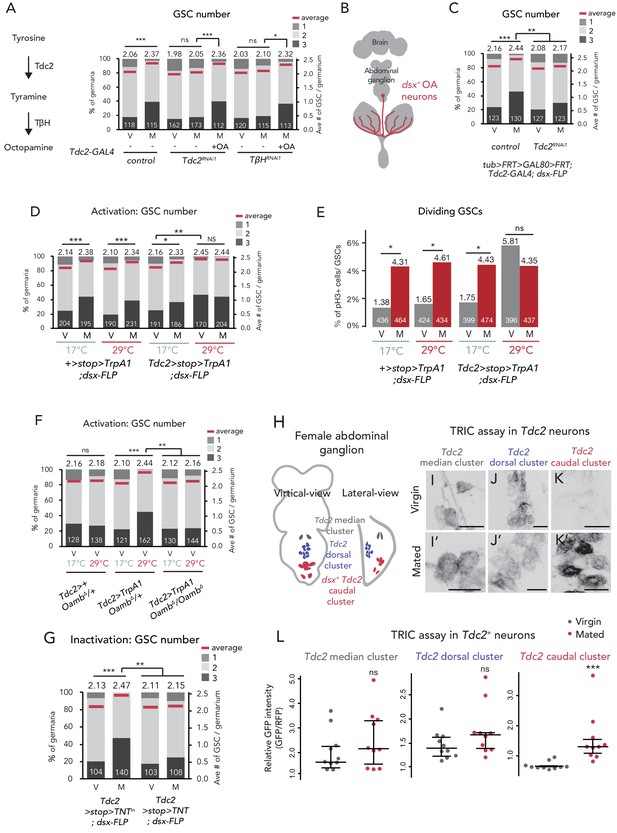
Ovary-projecting OA neurons control the GSC increase.
(A, C–D, F–G) Frequencies of germaria containing 1, 2, and 3 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. The number of germaria analyzed is indicated inside the bars. (A) RNAi of Tdc2 and TβH by Tdc2-GAL4. OA was added into the standard food. (B) A schematic drawing of Drosophila central nervous system and the ovary-projecting OA neurons with the dsx+ OA neurons projecting to the ovary. (C) Tdc2 RNAi in dsx+ Tdc2+ neurons with the genotype indicated. (D–E) TrpA1-mediated activation of dsx+ Tdc2+ neurons. 17°C and 29°C were used as the permissive and restrictive temperatures, respectively, of TrpA1 channel. (D) GSC number. (E) The ratio of pH3+ GSCs and total GSCs. (F) The activation of Tdc2+ neurons with OambΔ genetic background. (G) The inactivation of dsx+ Tdc2+ neurons. (H) Illustration showing the location of three clusters of Tdc2+ neurons in the caudal part of the abdominal ganglion (I–K, I’–K’). Negative images of TRIC labeling (anti-GFP) in the abdominal ganglions of virgin (I–K) and mated females (I’–K’) of TRIC (Tdc2 >UAS-mCD8::RFP, UAS-p65AD::CaM LexAop2-mCD8::GFP; nSyb-MKII::nlsLexADBDo;UAS-p65AD::CaM) flies, indicating intracellular Ca2+ transients. Scale bars, 20 μm. (L) The GFP intensities from the Tdc2+ median cluster, Tdc2+ dorsal cluster, and dsx+ Tdc2+ cluster of TRIC females show Ca2+ activity in virgin (gray) and mated females (red). Wilcoxon rank sum test was used for A, C, D, F, G, and L. Fisher’s exact test with Holm’s correction was used for E. ***p≤0.001, **p≤0.01, and *p≤0.05; NS, non-significant (p>0.05). All source data are available in Source data 1 and 3.
Because Tdc2-GAL4 is active in the nervous system (Busch et al., 2009; Pauls et al., 2018), we then identified which neurons secrete OA to the escort cells. D. melanogaster has more than 70–100 OAergic neurons dispersed throughout the nervous system (Monastirioti, 2003; Schwaerzel et al., 2003; Zhou et al., 2008). Among them, we were particularly interested in a small subset innervating the reproductive system (Figure 5B) as several recent studies have revealed that these neurons regulate mating behavior, egg laying, and ovarian-muscle contraction (Heifetz et al., 2014; Lee et al., 2003; Middleton et al., 2006; Rezával et al., 2014; Rubinstein and Wolfner, 2013). The ovary-projecting OAergic neurons are doublesex (dsx)+ and Tdc2+ double-positive (Rezával et al., 2014; Figure 5B). Therefore, to manipulate the gene expression of dsx+ Tdc2+ neurons only, we implemented a FLP/FRT intersectional strategy using dsx-FLP (Rezával et al., 2014). We could detect GFP expression only in the dsx+ Tdc2+ neurons innervating to the ovary, whose cell bodies are located on a caudal part of the abdominal ganglion (Rezával et al., 2014; Figure 5—figure supplement 1C–E). We next knocked down Tdc2 in dsx+ Tdc2+ neurons by RNAi (tub >GAL80>Tdc2RNAi; dsx-FLP) and found that these RNAi flies failed to increase GSC number after mating. Given the fact that the intersectional strategy using dsx-FLP does not label any neurons in the central nervous system (Rezával et al., 2014), these data suggest that only a small subset of dsx+ Tdc2+ neurons controls the mating-induced GSC increase.
To assess whether the activity of dsx+ Tdc2+ neurons affects the GSC number, we overexpressed TrpA1, a temperature-sensitive cation channel gene, in the dsx+ Tdc2+ neurons only. In Tdc2 >stop >TrpA1;dsx-FLP flies, we can tightly control the TrpA1 expression in the dsx+ Tdc2+ neurons only (Rezával et al., 2014). Both the control flies and TrpA1-overexpressing flies at permissive temperature (17°C) had the normal GSC number in virgin and mated females. On the other hand, at the restrictive temperature (29°C), the TrpA1-overexpressing flies, even the virgin ones, had more GSCs (Figure 5D). We also found that Tdc2 >stop >TrpA1; dsx-FLP virgin females at the restrictive temperature had increased GSC frequency in the M phase (Figure 5E). Importantly, the TrpA1-mediated activation of Tdc2 neurons did not induce the GSC increase in loss-of -Oamb-function females (Tdc2 >TrpA1; OambΔ/OambΔ) (Figure 5F), suggesting that the TrpA1-mediated GSC increase requires Oamb.
Furthermore, we employed the Tetanus toxin light chain (TNT) to inhibit neuronal activity (Sweeney et al., 1995). When we overexpressed TNT in dsx+ Tdc2+ neurons only, the mating-induced GSC increase was suppressed in mated females as compared with the control, whose inactivated TNTin was overexpressed (Figure 5G). Taken together, these findings suggest that the mating-induced GSC increase is mediated by the neuronal activity of dsx+ Tdc2+ neurons innervating to the ovary.
dsx+ Tdc2+ neurons are activated after mating
Because the dsx+ Tdc2+ neuronal activity has a significant role in mating-induced GSC increase, we next examined whether these neurons change their activity before and after mating. We monitored the neuronal activity using an end-point Ca2+ reporting system, the transcriptional reporter of intracellular Ca2+ (TRIC) (Gao et al., 2015). TRIC is designed to increase the GFP expression in proportion to [Ca2+]i. We classified female Tdc2+ neurons in the caudal part of the abdominal ganglion into three clusters based on their location and morphology. We designated the three clusters of these Tdc2+ neurons as the Tdc2+ median, Tdc2+ dorsal, and Tdc2+ caudal clusters (Figure 5H). Among them, the position of first two clusters are not similar to that of dsx+ Tdc2+ neurons, whereas that of the Tdc2+ caudal cluster is similar (Rezával et al., 2014). In virgin females, we detected robust TRIC signals in the Tdc2+ median and Tdc2+ dorsal clusters but not in the Tdc2+ caudal cluster (Figure 5I–L). In contrast, 24 hr after mating, we observed a significant increase in the TRIC signal in the Tdc2+ caudal cluster, whereas those in the Tdc2+ median and Tdc2+ dorsal clusters were not changed in virgin and mated females (Figure 5I–L). This result suggests that the Tdc2+ caudal cluster, which are likely dsx+ Tdc2+ neurons, is significantly activated after mating.
The activity switch of dsx+ Tdc2+ neurons is regulated by the sex peptide sensory neurons via acetylcholine signaling
Our previous study revealed that the mating-induced GSC increase is mediated by the male seminal fluid protein SP (Ameku and Niwa, 2016). SP is received by SPR in a small number of SPSNs, followed by a neural silencing of SPSNs (Häsemeyer et al., 2009; Yapici et al., 2008). Notably, SPSNs project their arbors into a caudal part of the abdominal ganglion, where the cell bodies of the dsx+ Tdc2+ cluster neurons are located (Rezával et al., 2014; Rezával et al., 2012). Therefore, we examined whether SPSNs physically interact with Tdc2+ neurons in the abdominal ganglion by performing the GFP Reconstitution Across Synaptic Partners (GRASP) analysis (Feinberg et al., 2008; Gordon and Scott, 2009), in which two complementary fragments of GFP were expressed in SPSNs and Tdc2+ neurons. GRASP signals were detected in the abdominal ganglion (Figure 6A), suggesting that the axon termini of SPSNs and the cell bodies and/or dendrites of Tdc2+ neurons contact each other likely through synaptic connections.
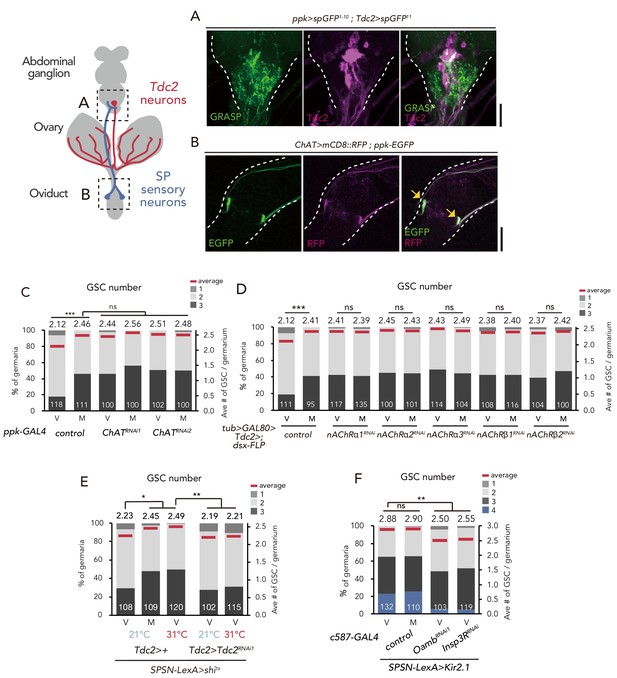
SPSNs control GSC increase through OA neurons.
(A) Neuronal proximity of SPSNs and Tdc2+ neurons in the abdominal ganglion of female flies stained with anti-Tdc2 (magenta). Note that reconstituted GFP (GRASP) signal was detected in the caudal part of the abdominal ganglion surrounded by broken white lines. Scale bar, 25 µm. (B) Cell bodies of SPSNs (yellow arrows) of ChAT-GAL4; UAS-mCD8::RFP; ppk-EGFP virgin females. Note that mCD8::RFP and EGFP signals overlapped in the cell bodies (yellow arrow) of SPSNs. White broken lines outline the oviduct. Scale bar, 25 µm. (C–F) Frequencies of germaria containing 1, 2, 3, and 4 GSCs (left vertical axis) and the average number of GSCs per germarium (right vertical axis) in virgin (V) and mated (M) female flies. The number of germaria analyzed is indicated inside the bars. (C) ChaT RNAi by ppk-GAL4. (D) RNAi of nAChRs in dsx+ Tdc2+ neurons. (E) RNAi of Tdc2 by Tdc2-GAL4 along with the silencing of SPSNs. 21 and 31°C were used as the permissive and restrictive temperatures, respectively, of shibirets (shits). (F) RNAi of Oamb and Insp3R by c587-GAL4 along with the silencing of SPSNs. Kir2.1 was used in this experiment. Note that frequencies of germaria containing 4 GSCs increased. Wilcoxon rank sum test with Holm’s correction was used for C, D, E and F. ***p≤0.001 and **p≤0.01; NS, non-significant (p>0.05). All source data are available in Source data 1.
Because SPSNs have been implied as cholinergic neurons (Rezával et al., 2012), we next examined the expression of Choline acetyltransferase (ChaT)-GAL4 in SPSNs. ChaT encodes an acetylcholine biogenic enzyme (Greenspan, 1980). The SPSNs located on the oviduct, which also co-express pickpocket (ppk) and fruitless (fru), are particularly crucial for inducing the major behavioral changes in female flies after mating (Ameku and Niwa, 2016; Rezával et al., 2012). By using UAS-mCD8::RFP with ChaT-GAL4 alongside ppk-EGFP, we confirmed that the ppk-EGFP–positive population near the oviduct were co-labeled by RFP (Figure 6B), consistent with the speculation that SPSNs are cholinergic.
We next counted the GSC number in ChAT RNAi flies using ppk-GAL4. ppk >ChAT RNAi virgin flies had more GSCs compared with the control (Figure 6C). In addition, mating did not induce GSC increase in ppk >ChATRNAi flies, suggesting that the acetylcholine released from SPSNs is responsible for suppressing the GSC increase.
To further ascertain whether the acetylcholine released from SPSNs is received by dsx+ Tdc2+ neurons to mediate mating-induced GSC increase, we focused on the fast-ionotropic nicotinic acetylcholine receptors (nAChR), which belong to the Cys-loop receptor subfamily of ligand-gated ion channels (Breer and Sattelle, 1987; Gundelfinger and Hess, 1992; Lee and O’Dowd, 1999). In D. melanogaster, 10 genes coding nAChR subunits have been identified. Among these, we focused on nAChRα1, nAChRα2, nAChRα3, nAChRβ1 and nAChRβ2 because the knock-down of these genes in dsx+ Tdc2+ (tub >GAL80>, dsx-FLP; Tdc2-GAL4) or Tdc2 neurons (Tdc2-GAL4) increased the GSC number in virgin females similar to ppk >ChATRNAi (Figure 6D and Figure 6—figure supplement 1A). We then confirmed the expression of these acetylcholine receptor genes in Tdc2 neurons by generating a knock-in T2A-GAL4 line as previously described (Kondo et al., 2020; Ihara et al., 2020) for each 5 nAChR subunits and observed their expression with UAS-mCD8::GFP. All of the five knock-in-GAL4 expressions were detected in anti-Tdc2 positive neurons around the ovary, suggesting that the ovary-projecting dsx+ Tdc2+ neurons expresses these nAChRs (Figure 6—figure supplement 1B–F).
To confirm the role of nAChR in mating-induced GSC increase, we generated nAChRα1 complete loss-of-function genetic alleles by CRISPR/Cas9 technology (Kondo and Ueda, 2013; Figure 6—figure supplement 2A). Similar to nAChRα1 RNAi females, the nAChRα1 transheterozygous mutant virgin females (nAChRα1228/nAChRα326) had more GSCs compared with the controls (Figure 6—figure supplement 2B). In addition, the GSC increase of nAChRα1228/nAChRα326 was restored by the overexpression of nAChRa1 in Tdc2+ neurons (Tdc2 >nAChRα1; nAChRα1228/nAChRα326) (Figure 6—figure supplement 2C). These data support our hypothesis that acetylcholine signaling in Tdc2 neurons has a negative role in mating-induced GSC increase.
We next assessed relationship between SPSNs, Tdc2+ neurons, and OA-Oamb-Ca2+ signaling in ovarian cells. The silencing of SPSNs neuronal activity (SPSNs-LexA and LexAop-shits) increased the GSC number in virgin females (Figure 6E), consistent with our previous study (Ameku and Niwa, 2016). Upon SPSNs silencing, Tdc2 RNAi by Tdc2-GAL4 reduced the GSC number (Figure 6E), suggesting that Tdc2+ neurons act downstream of SPSNs. In addition, the GSC increase through the silencing of SPSNs (SPSNs-LexA >LexAop-kir2.1) was suppressed by Oamb or Insp3R RNAi in the escort cells (Figure 6F), suggesting that OA-Oamb-Ca2+ signaling in ovarian cells acts downstream of SPSNs. Overall, our findings revealed a novel neuronal relay in response to mating that regulates the female GSC increase in the ovary before/after mating (Figure 7).
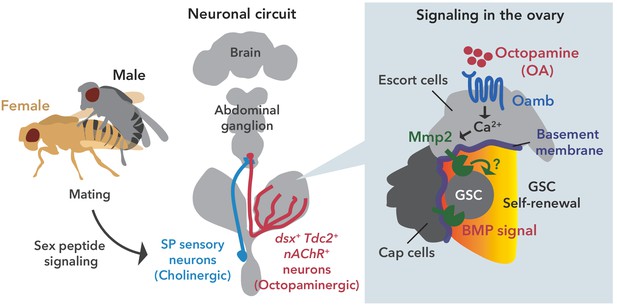
Neuronal octopamine signaling, followed by Oamb-Ca2+-Mmp2 signaling, regulates the mating-induced GSC increase.
The illustration is the proposed working model from our findings here. SP signaling and SP sensory neurons activate dsx+ Tdc2 neurons via acetylcholine signaling. The octopamine released from dsx+ Tdc2 neurons is received by the Oamb in escort cells and then activates intracellular Ca2+ flux. The OA-mediated signaling increases pMad levels in GSCs to evoke mating-induced GSC increase via Mmp2.
Discussion
In this study, we report that the mating-induced GSC increase in female D. melanogaster is regulated by OAergic neurons directly projecting to the ovary. From our in vivo and ex vivo experiments, we propose the following model to explain the mating-induced GSC increase. After mating, the male seminal fluid SP is transferred into the female uterus, stimulating SPR-positive neurons. As the liganded SPR silences the neuronal activity of SPSNs (Häsemeyer et al., 2009), the acetylcholine released from SPSNs is suppressed. As SPSNs and dsx+ Tdc2+ neurons are directly connected, this suppression directly modulates dsx+ Tdc2+ neuronal activity. Because we have shown that nAChRs in dsx+ Tdc2+ neurons exhibit an inhibitory effect with an unknown mechanism (to be discussed later), the inactivation of nAChRs in the absence of acetylcholine results in the activation of dsx+ Tdc2+ neurons in mated females. As a consequence, OA is released from dsx+ Tdc2+ neurons, received by Oamb, induces [Ca2+]i in the escort cells, and finally activates the Mmp2 enzymatic activity. The activity of Mmp2 positively regulates the Dpp-mediated niche signaling, thereby leading to mating-induced GSC increase (Figure 7).
The octopamine- and its receptor-dependent GSC control
Our proposed model is that the OA from dsx+ Tdc2+ neurons is directly received by the escort and follicle cells in the germarium. Our model is supported by two of our observations. First, mating-induced GSC increase is impaired by Oamb RNAi using a GAL4 driver that is active specifically in the germarium cells but not mature follicle cells. Second, OA treatment evokes [Ca2+]i elevation in these germarium cells in an Oamb-dependent manner. However, in this study, we did not address whether the escort cells and/or the follicle cells in the germarium express Oamb, as we failed to observe any clear GAL4 expression in two independent Oamb-T2A-GAL4 drivers (Figure 1—figure supplement 2A,B,C and D). We surmise that this may be due to lower amounts of Oamb transcript in the germarium.
We have shown that the activation of the ovary-projecting dsx+ Tdc2+ neurons is necessary and sufficient to induce GSC increase. However, from an anatomical point of view, the dsx+ Tdc2+ neurons project to the distal half of the ovary but not to the germarium (Figure 5—figure supplement 1E). Considering our model described above, this disagreement can be attributed to the characteristic volume transmission of monoamine neurotransmitters. In other words, neurotransmitters act at a distance well beyond their release sites from cells or synapses (Fuxe et al., 2010). Therefore, the OA secreted from the terminals of dsx+ Tdc2+ neurons could reach the germarium located at the most proximal part of the ovary.
Several previous studies have revealed that OA signaling has a pivotal role in reproductive tissues other than germarium, such as mature follicle cells, oviduct, and ovarian muscle, to promote ovulation, oviduct remodeling, and ovarian-muscle contraction, respectively (Deady and Sun, 2015; Heifetz et al., 2014; Lee et al., 2009; Middleton et al., 2006; Rezával et al., 2014). Therefore, it is likely that the dsx+ Tdc2+ neurons orchestrate multiple different events during oogenesis in response to mating stimulus. Because a mated female needs to activate oogenesis to continuously produce eggs in concert with sperm availability, it is reasonable that the ovary-projecting neurons switch on the activity of the entire process of reproduction.
The role of Mmp2 in mating-induced GSC increase
Based on our present study and several previous studies (Deady et al., 2015; Deady and Sun, 2015; Knapp and Sun, 2017), the OA-Oamb-Ca2+-Mmp2 axis is required for GSC increase and follicle rupture, both of which are induced by mating stimuli in D. melanogaster. In both cases, Mmp2 enzymatic activity is likely to be essential, as the overexpression of Timp encoding a protein inhibitor of Mmp2 suppresses GSC increase, as well as follicle rupture. Mmp2 in mature follicle cells cleaves and downregulates Viking/collagen VI (Deady et al., 2017; Wang et al., 2008). In fact, several previous studies have revealed that Viking/collagen VI is required for GSC maintenance in female D. melanogaster (Van De Bor et al., 2015; Wang et al., 2008). However, we observed no significant change in Viking/Collagen VI levels in the germarium between the control and Mmp2 RNAi flies (Figure 4—figure supplement 1E). Therefore, we concluded that Viking/collagen VI is not a substrate of Mmp2 in the regulation of mating-induced GSC increase. Besides Viking/Collagen VI, Dally-like (Dlp) is another basement membrane protein associated with extracellular matrix and known as the Mmp2 substrate (Wang and Page-McCaw, 2014). Interestingly, dlp is expressed in the escort cells (Wang and Page-McCaw, 2014). Moreover, Dlp controls the distribution of Dpp and Wnts, both of which significantly affect GSC self-renewal and differentiation (Wang et al., 2015; Xie and Spradling, 1998). Future research should decipher the exact substrate by which Mmp2 controls Dpp and/or Wnts to modulate GSC behavior in response to mating stimulus.
Another remaining question to be addressed is how Mmp2 function is regulated in GSC increase. Ecdysteroid biosynthesis and signaling in the ovary are necessary but not sufficient for the OA-Oamb-Ca2+–mediated GSC increase and follicle rupture (Ameku and Niwa, 2016; Knapp and Sun, 2017). We found that in the regulation of mating-induced GSC increase, ecdysteroid signaling acts downstream of Ca2+ signaling (Figure 4F). On the other hand, in the follicle rupture process, ecdysteroid signaling either acts downstream, upstream, or both, of Ca2+ signaling. Further, the precise action of ecdysteroid has yet to be elucidated (Knapp and Sun, 2017). The Mmp2-GFP fusion protein level in the follicle cells is not changed in the loss-of-Ecdysone receptor-function flies, implying that ecdysteroid signaling might regulate Mmp2 enzymatic activity by an unknown mechanism (Knapp and Sun, 2017). Considering the involvement of both the OA-Oamb-Ca2+-Mmp2 axis and ecdysteroid biosynthesis, it is very likely that the Mmp2 enzymatic activity is also regulated by the same, unknown mechanism not only in the mature follicle cells to control follicle rupture, but also in the germarium to control mating-induced GSC increase.
SPSN-mediated suppression of dsx+/Tdc2+ neurons
In many animals, reproduction involves significant behavioral and physiological shifts in response to mating. In female D. melanogaster, several post-mating responses are coordinated by SPSNs and their downstream afferent neuronal circuit (Wang et al., 2020), including Stato-Acoustic Ganglion neurons, the ventral abdominal lateral Myoinhibitory peptide neurons, and the efferent dsx+ Tdc2+ neurons (Feng et al., 2014; Häsemeyer et al., 2009; Jang et al., 2017; Rezával et al., 2014). Our GRASP analysis indicates a direct synaptic connection between cholinergic SPSNs and OAergic neurons. Moreover, we demonstrated that nAChRs in dsx+ Tdc2+ neurons are responsible for the suppression of their neuronal activity in virgin females. However, nAChRs are the cation channels leading to depolarization upon acetylcholine binding, and therefore usually activate neurons (Corringer et al., 2000; Lee and O’Dowd, 1999; Perry et al., 2012). How is the opposite role of nAChRs in dsx+ Tdc2+ neuronal activity achieved? One possibility is that acetylcholine-nAChR signaling does not evoke a simple depolarization but rather generates a virgin-specific temporal spike pattern in dsx+ Tdc2+ neurons. Interestingly, recent studies demonstrated that the pattern, instead of the frequency, of neuronal firing is significant in adjusting the neuronal activity of clock neurons in D. melanogaster (Tabuchi et al., 2018). The firing pattern relies on control of ionic flux by the modulation of Ca2+-activated potassium channel and Na+/K+ ATPase activity. Because whether mating changes the firing pattern of dsx+ Tdc2+ neurons remains to be examined, the neuronal activity in SPSNs and the dsx+ Tdc2+ neuronal circuit between virgin and mated females are future research areas.
Interorgan communication among multiple organs to regulate the increase and maintenance of female GSCs
In the last decades, there is growing evidence that GSCs and their niche are influenced by multiple humoral factors (Drummond-Barbosa, 2019; Yoshinari et al., 2019). Based on the data from our current study and previous studies, there are at least four crucial humoral factors for regulating the increase and/or maintenance of D. melanogaster female GSCs, including DILPs (Hsu et al., 2008; Hsu and Drummond-Barbosa, 2009; LaFever, 2005), ecdysteroids (Ables and Drummond-Barbosa, 2010; Ameku et al., 2017; Ameku and Niwa, 2016; König et al., 2011), Neuropeptide F (NPF) (Ameku et al., 2018), and OA (this study). Notably, all of these come from different sources: DILPs are from the insulin-producing cells located in the pars intercerebralis of the central brain; ecdysteroids from the ovary; NPF from the midgut; and OA from the neurons located in the abdominal ganglion. In addition to these identified humoral factors, recent studies also imply that adiponectin and unknown adipocyte-derived factor(s) are essential for GSC maintenance (Armstrong and Drummond-Barbosa, 2018; Laws et al., 2015; Matsuoka et al., 2017). These data clearly indicate that D. melanogaster female GSCs are systemically regulated by interorgan communication involving multiple organs. The additional interorgan communication mechanisms that ensure the faithful coupling of the increase and maintenance of GSC to the organism’s external and physiological environments are essential to be investigated in future studies.
To modulate the increase and maintenance of GSC, ecdysteroids are received by both GSCs and niche cells (Ables and Drummond-Barbosa, 2010; König et al., 2011), whereas DILPs, NPF, and OA are received by niche cells. A major signal transduction mechanism of each of these humoral factors have been well characterized, namely phosphoinositide 3-kinase pathway for DILPs-InR signaling, EcR/Ultraspiracle-mediated pathway for ecdysteroid signaling, cAMP pathway for NPF-NPFR signaling (Garczynski et al., 2002), and Ca2+ pathway for OA-Oamb signaling. However, it remains unclear whether and how each of these signaling pathways control the production and secretion of the niche signal, as well as its distribution and transduction. In addition, it is important to understand whether and how the multiple system signals are integrated to control the mating-induced increase and maintenance of GSCs.
Evolutionarily conservation of monoamine-steroid hormone axis to control female reproduction
In recent years, many studies have revealed that not only local niche signals but also systemic and neuronal factors play indispensable roles in regulating GSC behavior (Ables and Drummond-Barbosa, 2017; Drummond-Barbosa, 2019; Yoshinari et al., 2019). In D. melanogaster, ecdysteroid signaling is essential for the proliferation and maintenance of GSCs and neural stem cells (Ables and Drummond-Barbosa, 2010; Homem et al., 2014; König et al., 2011). In this study, we have identified the ovary-projecting OAergic neurons as new regulators of stem cell homeostasis. Both steroid hormones and OA-like monoamines, such as noradrenaline, are also involved in stem cell regulation in mammals. For example, the mammalian steroid hormone, estrogen, is important in regulating cell division and/or maintenance of hematopoietic stem cells, mammary stem cell, neural stem cells, and hematopoietic stem cells (Asselin-Labat et al., 2010; Bramble et al., 2019; Kim et al., 2016; Nakada et al., 2014). Moreover, noradrenergic neurons, which directly project to the bone marrow, regulate the remodeling of hematopoietic stem cells niche (Ho et al., 2019; Méndez-Ferrer et al., 2010; Méndez-Ferrer et al., 2008). Therefore, the steroid hormone- and noradrenergic nerve-dependent control of stem cell homeostasis are likely conserved across animal species. In this regard, the D. melanogaster reproductive system will further serve as a powerful model to unravel the conserved systemic and neuronal regulatory mechanisms for stem cell homeostasis in animals.
Materials and methods
Drosophila strains
Request a detailed protocolFlies were raised on cornmeal-yeast-agar medium at 25°C. EcRA483T, temperature-sensitive mutants, were cultured at 31°C for 1 d prior to the assays. w1118 was used as the control strain.
The genetic mutant stocks used were EcRA483T (Bloomington Drosophila Stock Center [BDSC] #5799) and EcRM554fs (BDSC #4894). The protein-trap GFP line of Vkg (Vkg::GFP) was obtained from Kyoto Stock Center (DGRC #110692). Dad-LacZ (Tsuneizumi et al., 1997) (a gift from Yoshiki Hayashi, University of Tsukuba, Japan).
The following GAL4 and LexA strains were used: c587-GAL4 (Manseau et al., 1997) (gift from Hiroko Sano, Kurume University, Japan), R44E10-GAL4 (Deady and Sun, 2015) (a gift from Jianjun Sun, University of Connecticut, USA), RS-GAL4 (Lee et al., 2009) (a gift from Kyung-An Han, Pennsylvania State University, USA), nSyb-GAL4 (BDSC #51941), nSyb-GAL80 (Harris et al., 2015) (a gift from James W. Truman, Janelia Research Campus, USA), tj-GAL4 (DGRC #104055), R13C06-GAL4 (BDSC #47860), 109–30 GAL4 (BDSC #7023), c355-GAL4 (BDSC #3750), c306-GAL4 (BDSC #3743), slbo-GAL4 (BDSC #6458), bab1-GAL4 (Bolívar et al., 2006) (a gift from Satoru Kobayashi, University of Tsukuba, Japan), nos-GAL4 (DGRC #107748), tub >FRT >GAL80>FRT (BDSC #38879), OambKI-RD-GAL4 (BDSC#84677) (Deng et al., 2019), Oamb-KI-T2A-GAL4, nAChRα1-T2A-GAL4, nAChRα2-T2A-GAL4, nAChRα3-T2A-GAL4, nAChRβ1-T2A-GAL4, nAChRβ2-T2A-GAL4 (Kondo et al., 2020; Ihara et al., 2020), ChaT-GAL4 (BDSC #6793), ppk-GAL4 (Grueber et al., 2007) (a gift from Hiroko Sano, Kurume University, Japan), and SPSNs-LexA (Feng et al., 2014) (a gift from Young-Joon Kim, Gwangju Institute of Science and Technology, South Korea).
The following UAS and LexAop strains were used: 20xUAS-6xGFP (BDSC #52261), UAS-GFP;UAS-mCD8::GFP (Ito et al., 1997; Lee and Luo, 1999) (a gift from Kei Ito, University of Cologne, Germany), UAS-Stingar (BDSC #84277), UAS-mCD8::RFP (BDSC #32219), UAS-CsChrimson (BDSC #55134), UAS-Insp3R (BDSC #30742), UAS-OambAS (Lee et al., 2009) (a gift from Kyung-An Han, Pennsylvania State University, USA), UAS-Timp (BDSC #58708) (a gift from Andrea Page-McCaw, Vanderbilt University, USA), UAS > stop >dTrpA1mcherry, UAS > stop >TNT, UAS > stop >TNTin (von Philipsborn et al., 2011; Yu et al., 2010), dsx-FLP (Rezával et al., 2014) (a gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan) TRiC; UAS-mCD8::RFP, LexAop2-mCD8::GFP;nSyb-MKII::nlsLexADBDo;UAS-p65AD::CaM (BDSC:61679), ppk-eGFP (Grueber et al., 2003) (a gift from Tadashi Uemura, Kyoto University, Japan), and LexAop-Kir2.1 (Feng et al., 2014) (a gift from Young-Joon Kim, Gwangju Institute of Science and Technology, South Korea).
The RNAi transgenic lines used were as follows: UAS-LacZRNAi (a gift from Masayuki Miura, The University of Tokyo, Japan), UAS-OambRNAi1(BDSC #31171), UAS-OambRNAi2(BDSC #31233), UAS-OambRNAi3 (Vienna Drosophila Resource Center [VDRC] #106511), UAS-Octβ1RRNAi(VDRC #110537), UAS-Octβ2RRNAi(VDRC #104524), UAS-Octβ3RRNAi (VDRC #101189), UAS-Insp3RRNAi (BDSC #25937), UAS-EcRRNAi (VDRC #37059), UAS-Mmp2RNAi1 (BDSC #31371), UAS-Mmp2RNAi2 (VDRC #330303), UAS-TimpRNAi1 (BDSC #61294), UAS-TimpRNAi2 (VDRC #109427), UAS-Tdc2RNAi1 (VDRC #330541), UAS-Tdc2RNAi2 (BDSC #25871), UAS-TβhRNAi1 (VDRC #107070), UAS-TβhRNAi2 (BDSC #67968), UAS-ChATRNAi1 (VDRC #330291), UAS-ChATRNAi2 (BDSC #25856), UAS-nAChRα1RNAi (VDRC #48159), UAS-nAChRα2RNAi (VDRC #101760), UAS-nAChRα3RNAi (VDRC #101806), UAS-nAChRβ1RNAi (VDRC #106570), UAS-nAChRβ2RNAi (VDRC #109450), UAS-nvdRNAi1, and UAS-nvdRNAi2 (Yoshiyama et al., 2006).
Generation of Oamb and nAChRα1 genetic loss-of-function mutant strains
Request a detailed protocolThe mutant alleles OambΔ (Figure 1—figure supplement 1F), nAChRα1228, and nAChRα1326 (Figure 6—figure supplement 2A) were created in a white (w) background using CRISPR/Cas9 as previously described (Kondo and Ueda, 2013). The following guide RNA (gRNA) sequences were used: Oamb, 5ʹ-GATGAACTCGAGTACGGCCA-3ʹ, and 5ʹ-GCGATCTCTGGTGCCGCATT-3ʹ; nAChRα1228, 5ʹ-GGACATCATGCGTGTGCCGG-3ʹ; nAChRα1326, 5ʹ-GGGCAGGTAGAAGACCAGAA-3ʹ. The breakpoint detail of OambΔ is described in Figure 1—figure supplement 1F, whereas those of nAChRα1228 and nAChRα1326 are described in Figure 6—figure supplement 2A.
Generation of UAS-nAChRα1 transgenic line
Request a detailed protocolThe pcDNA3.1 plasmid containing the wild-type D. melanogaster nAChRα1 coding sequences (nAChRα1-pcDNA3.1) was synthesized previously described (Ihara et al., 2018). Briefly, nAChRα1-pcDNA3.1 was digested with EcoRI and NotI, and then the digested nAChRα1 fragment was ligated with a EcoRI-NotI–digested pWALIUM10-moe plasmid (Perkins et al., 2015). Transformants were generated using the phiC31 integrase system in the P{CaryP}attP40 strain (Groth et al., 2004). The w+ transformants of pWALIUM10-moe were established using standard protocols.
Behavioral assays
Request a detailed protocolFlies were reared at 25°C and aged for 5–6 d. Virgin female flies were mated overnight to w1118 male flies at 25°C (10 males and 5–8 females per vial). For the thermal activation assays, flies were first reared at 17°C for 6 d and transferred to 29°C. In the case of EcR mutant assays, flies were transferred to 31°C for 24 hr before mating or ex vivo culture.
For OA feeding, newly eclosed virgin females were aged for 4 d in vials with standard food containing 7.5 mg/mL of OA (Monastirioti et al., 1996; Rubinstein and Wolfner, 2013).
Immunohistochemistry
Request a detailed protocolTissues were dissected in phosphor buffer serine (PBS) and fixed in 4% paraformaldehyde in PBS for 30 to 60 min at room temperature (RT). The fixed samples were washed three times in PBS supplemented with 0.2% Triton X-100, blocked in blocking solution (PBS with 0.3% Triton X-100% and 0.2% bovine serum albumin [BSA]) for 1 hr at RT, and incubated with a primary antibody in the blocking solution at 4°C overnight. The primary antibodies used were chicken anti-GFP (Abcam #ab13970; 1:4,000), rabbit anti-RFP (Medical and Biological Laboratories PM005; 1:2,000), mouse anti-Hts 1B1 (Developmental Studies Hybridoma Bank [DSHB]; 1:50), rat anti-DE-cadherin DCAD2 (DSHB; 1:50), rabbit anti-pH3 (Merck Millipore #06–570; 1:1000), rabbit monoclonal anti-pMad (Abcam #ab52903; 1:1000), mouse anti-Lamin C LC28.26 (DSHB; 1:10), rabbit cleaved Dcp-1 (Cell Signaling Technology #9578; 1:100), rat anti-Vasa (DSHB; 1:50), mouse anti-LacZ (β-galactosidase) (DSHB#40-1a; 1:50), rabbit anti-Tdc2 (Abcam #ab128225; 1:2000), Alexa Fluor 546 phalloidin (Thermo Fisher Scientific #A22283; 1:200), and Alexa Fluor 633 phalloidin (Thermo Fisher Scientific #A22284; 1:200). After washing, fluorophore (Alexa Fluor 488, 546 or 633)-conjugated secondary antibodies (Thermo Fisher Scientific) were used at a 1:200 dilution, and the samples were incubated for 2 hr at RT in the blocking solution. After another washing step, all samples were mounted in FluorSave reagent (Merck Millipore #345789). GSC numbers were determined based on the morphology and position of their anteriorly anchored spherical spectrosome (Ables and Drummond-Barbosa, 2010; Ameku et al., 2018; Ameku and Niwa, 2016). Cap cells were identified by immunostaining with anti-Lamin C antibody as previously described (Ables and Drummond-Barbosa, 2010).
Ex vivo ovary culture
Request a detailed protocolWe used 5–6-day-old females. The ovaries were dissected in Schneider’s Drosophila medium (Thermo Fisher Scientific #21720024) and isolated from oviduct using forceps. Approximately 5–6 ovaries were immediately transferred to a dish containing 3 mL of Schneider’s Drosophila medium supplemented with 15% fetal calf serum and 0.6% penicillin-streptomycin with/without the addition of OA (Sigma, final concentration of OA is 0–1000 μM) and 20E (Enzo Life Sciences; final concentration of 20 nM). The cultures were incubated at RT (except for EcR mutant flies, Figure 3D) for 16 hr, and the samples were immunostained to determine the GSC number.
Ex vivo calcium imaging
Request a detailed protocolWe employed the previously described imaging methods to visualize GSC behavior (Morris and Spradling, 2011; Reilein et al., 2018). For the live imaging, the ovaries dissected from adult virgin female flies were placed on a glass bottom dish (IWAKI #4970–041) with 3 mL of Schneider’s Drosophila medium and 100 µL of the test reagent (Schneider’s Drosophila medium containing 300 mM OA) placed directly at the center of each dish. The images were obtained with a × 40 objective lens (water-immersion) using a Zeiss LSM 700 confocal microscope and were recorded every 4 s. The GCaMP6s fluorescence intensity in the escort cell was then calculated for each time point. The ratio of fluorescence (ΔF) at each time point was calculated by normalizing the fluorescence with the initial fluorescence (F0). The initial fluorescence (F0) is the average GCaMP6s fluorescence intensity before adding the test reagent.
Optogenetic activation of the escort cells
Request a detailed protocolRed-shifted channelrhodopsin CsChrimson (Klapoetke et al., 2014) was used to increase the [Ca2+]i in the escort cells by light. UAS-CsChrimson was expressed using c587-GAL4 with nSyb-GAL80. All crosses and the early development of flies were performed under dark conditions. The experiment was done at 25°C. Adult flies were raised with standard food for 3 d after eclosion and then with standard food with 1 mM all-trans-retinal (ATR) for 3 d. Subsequently, they were kept in the presence of orange–red light from LED for 24 hr. LED light was shone from the outside of the plastic chamber covered by aluminum foil to enhance light intensity.
Statistical analysis
Request a detailed protocolAll experiments were performed independently at least twice. Fluorescence intensity in confocal sections was measured via ImageJ. For pMad quantification, signal intensity was calculated by measuring the fluorescence intensity in GSCs and CBs, which were co-stained with anti-Vasa antibody to visualize their cell boundaries. Sample sizes were chosen based on the number of independent experiments required for statistical significance and technical feasibility. The experiments were not randomized, and the investigators were not blinded. All statistical analyses were carried out using the ‘R’ software environment. The P value is provided in comparison with the control and indicated as * for p≤0.05, ** for p≤0.01, *** for p≤0.001, and ‘NS’ for non-significant (p>0.05).
Appendix 1
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | c587-GAL4 | Manseau et al., 1997 | FBal0150629 RRID:BDSC_67747 | A gift from Hiroko Sano, Kurume University, Japan |
| Genetic reagent (D. melanogaster) | OambΔ | This paper | Detail described in Figure 1—figure supplement 1F | |
| Genetic reagent (D. melanogaster) | nAChRa1228 | This paper | Detail described in Figure 6—figure supplement 2A | |
| Genetic reagent (D. melanogaster) | nAChRa1326 | This paper | Detail described in Figure 6—figure supplement 2A | |
| Genetic reagent (D. melanogaster) | EcRA483T | Bloomington Drosophila Stock Center | BDSC: #5799 RRID:BDSC_5799 FBal0083501 | |
| Genetic reagent (D. melanogaster) | EcRM554fs | Bloomington Drosophila Stock Center | BDSC: #4894 RRID:BDSC_4894 FBal0083490 | |
| Genetic reagent (D. melanogaster) | Vkg::GFP | KYOTO stock center | DGRC #110692 RRID:DGGR_110692 FBal0286156 | |
| Genetic reagent (D. melanogaster) | Dad-LacZ | Tsuneizumi et al., 1997 | FBal0065787 RRID:DGGR_118114 | A gift from Yoshiki Hayashi, University of Tsukuba, Japan |
| Genetic reagent (D. melanogaster) | R44E10-GAL4 | Deady and Sun, 2015 | FBal0252601 PMID:26473732 | A gift from Jianjun Sun, University of Connecticut, USA |
| Genetic reagent (D. melanogaster) | RS-GAL4 | Lee et al., 2009 | FBal0263794 PMID:19262750 | A gift from Kyung-An Han, Pennsylvania State University, USA |
| Genetic reagent (D. melanogaster) | nSyb-GAL4 | Bloomington Drosophila Stock Center | BDSC: #51941 RRID:BDSC_51941 FBti0154973 | FACS (5 ul per test) |
| Genetic reagent (D. melanogaster) | nSyb-GAL80 | Harris et al., 2015 | PMID:26193122 | A gift from James W. Truman, Janelia Research Campus, USA |
| Genetic reagent (D. melanogaster) | tj-GAL4 | KYOTO stock center | DGRC: #104055 RRID:DGGR_104055 FBti0034540 | |
| Genetic reagent (D. melanogaster) | R13C06-GAL4 | Bloomington Drosophila Stock Center | BDSC: #47860 RRID:BDSC_47860 FBal0249828 | |
| Genetic reagent (D. melanogaster) | 109–30 GAL4 | Bloomington Drosophila Stock Center | BDSC: #7023 RRID:BDSC_7023 FBti0027548 | |
| Genetic reagent (D. melanogaster) | c355-GAL4 | Bloomington Drosophila Stock Center | BDSC: #3750 RRID:BDSC_3750 FBti0002591 | |
| Genetic reagent (D. melanogaster) | c306-GAL4 | Bloomington Drosophila Stock Center | BDSC: #3743 RRID:BDSC_3743 FBal0048787 | |
| Genetic reagent (D. melanogaster) | slbo-GAL4 | Bloomington Drosophila Stock Center | BDSC: #6458 RRID:BDSC_6458 FBst0006458 | |
| Genetic reagent (D. melanogaster) | bab1-GAL4 | Bolívar et al., 2006 | FBal0242654 PMID:17013875 | A gift from Satoru Kobayashi, University of Tsukuba, Japan |
| Genetic reagent (D. melanogaster) | nos-GAL4 | KYOTO stock center | DGRC: #107748 RRID:DGGR_107748 FBst0306396 | |
| Genetic reagent (D. melanogaster) | tub > FRT > GAL80>FRT | Bloomington Drosophila Stock Center | BDSC: #38879 RRID:BDSC_38879 FBti0147580 | |
| Genetic reagent (D. melanogaster) | OambKI-RD-GAL4 | Deng et al., 2019 Bloomington Drosophila Stock Center | BDSC: #84677 RRID:BDSC_84677 FBti0209942 | |
| Genetic reagent (D. melanogaster) | Oamb-KI-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRα1-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRα2-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRα3-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRβ1-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | nAChRβ2-T2A-GAL4 | Kondo et al., 2020 | PMID:31914394 | |
| Genetic reagent (D. melanogaster) | ChaT-GAL4 | Bloomington Drosophila Stock Center | BDSC: #6793 RRID:BDSC_6793 FBst0006793 | |
| Genetic reagent (D. melanogaster) | ppk-GAL4 | Grueber et al., 2007 | FBtp0039691 PMID:17164414 | A gift from Hiroko Sano, Kurume University, Japan |
| Genetic reagent (D. melanogaster) | SPSNs-LexA | Feng et al., 2014 | FBtp0110869 PMID:24991958 | A gift from Young-Joon Kim, Gwangju Institute of Science and Technology, South Korea |
| Genetic reagent (D. melanogaster) | 20xUAS-6xGFP | Bloomington Drosophila Stock Center | BDSC: #52261 RRID:BDSC_52261 FBst0052261 | |
| Genetic reagent (D. melanogaster) | UAS-GFP;UAS-mCD8::GFP | Ito et al., 1997; Lee and Luo, 1999 | FBtp0002652 PMID:9043058 PMID:10457015 | A gift from Kei Ito, University of Cologne, Germany |
| Genetic reagent (D. melanogaster) | UAS-Stinger | Bloomington Drosophila Stock Center | BDSC: #84277 RRID:BDSC_84277 | |
| Genetic reagent (D. melanogaster) | UAS-mCD8::RFP | Bloomington Drosophila Stock Center | BDSC: #32219 RRID:BDSC_32219 FBti0131967 | |
| Genetic reagent (D. melanogaster) | UAS-CsChrimson | Bloomington Drosophila Stock Center | BDSC: #55134 RRID:BDSC_55134 FBti0160571 | |
| Genetic reagent (D. melanogaster) | UAS-Insp3R | Bloomington Drosophila Stock Center | BDSC: #30742 RRID:BDSC_30742 FBti0129829 | |
| Genetic reagent (D. melanogaster) | UAS-OambK3 | Lee et al., 2009 | FBtp0069415 PMID:19262750 | A gift from Kyung-An Han, Pennsylvania State University, USA |
| Genetic reagent (D. melanogaster) | UAS-Timp | Bloomington Drosophila Stock Center | BDSC: #58708 RRID:BDSC_58708 FBti0164930 | A gift from Andrea Page-McCaw, Vanderbilt University, USA |
| Genetic reagent (D. melanogaster) | UAS-nAChRα1 | This paper | Detail described in Material and method | |
| Genetic reagent (D. melanogaster) | UAS > stop > dTrpA1mcherry | von Philipsborn et al., 2011 | FBtp0064577 PMID:21315261 | A gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan |
| Genetic reagent (D. melanogaster) | UAS > stop > TNT | von Philipsborn et al., 2011 | FBtp0020863 PMID:21315261 | A gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan |
| Genetic reagent (D. melanogaster) | UAS > stop > TNTin | von Philipsborn et al., 2011 | FBtp0020863 PMID:21315261 | A gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan |
| Genetic reagent (D. melanogaster) | dsx-FLP | Rezával et al., 2014 | FBal0296301 PMID:24631243 | A gift from Daisuke Yamamoto, Advanced ICT Research Institute, National Institute of Information and Communications Technology, Japan |
| Genetic reagent (D. melanogaster) | TRiC; UAS-mCD8::RFP, LexAop2-mCD8::GFP;nSyb-MKII::nlsLexADBDo;UAS-p65AD::CaM | Bloomington Drosophila Stock Center | BDSC: #61679 RRID:BDSC_61679 FBst0061679 | |
| Genetic reagent (D. melanogaster) | ppk-eGFP | Grueber et al., 2003 | FBtp0041053 PMID:12699617 | A gift from Tadashi Uemura, Kyoto University, Japan |
| Genetic reagent (D. melanogaster) | LexAop-Kir2.1 | Feng et al., 2014 | FBtp0110870 PMID:24991958 | A gift from Young-Joon Kim, Gwangju Institute of Science and Technology, South Korea |
| Genetic reagent (D. melanogaster) | UAS-LacZRNAi | Kennerdell and Carthew, 2000 | FBtp0016505 PMID:10932163 | A gift from Masayuki Miura, The University of Tokyo, Japan |
| Genetic reagent (D. melanogaster) | UAS-OambRNAi1 | Bloomington Drosophila Stock Center | BDSC: #31171 RRID:BDSC_31171 FBst0031171 | |
| Genetic reagent (D. melanogaster) | UAS-OambRNAi2 | Bloomington Drosophila Stock Center | BDSC: #31233 RRID:BDSC_31233 FBst0031233 | |
| Genetic reagent (D. melanogaster) | UAS-OambRNAi3 | Vienna Drosophila Resource Center | VDRC: #106511 FBst0478335 | |
| Genetic reagent (D. melanogaster) | UAS-Octβ1RRNAi | Vienna Drosophila Resource Center | VDRC: #110537 FBst0482104 | |
| Genetic reagent (D. melanogaster) | UAS-Octβ2RRNAi | Vienna Drosophila Resource Center | VDRC: #104524 FBst0476382 | |
| Genetic reagent (D. melanogaster) | UAS-Octβ3RRNAi | Vienna Drosophila Resource Center | VDRC: #101189 FBst0473062 | |
| Genetic reagent (D. melanogaster) | UAS-Insp3RRNAi | Bloomington Drosophila Stock Center | BDSC: #25937 FBst0025937 | |
| Genetic reagent (D. melanogaster) | UAS-EcRRNAi | Vienna Drosophila Resource Center | VDRC: #37059 FBst0461818 | |
| Genetic reagent (D. melanogaster) | UAS-Mmp2RNAi1 | Bloomington Drosophila Stock Center | BDSC: #31371 RRID:BDSC_31371 FBst0031371 | |
| Genetic reagent (D. melanogaster) | UAS-Mmp2RNAi2 | Vienna Drosophila Resource Center | VDRC: #330203 FBst0490996 | |
| Genetic reagent (D. melanogaster) | UAS-TimpRNAi1 | Bloomington Drosophila Stock Center | BDSC: #61294 RRID:BDSC_61294 FBst0061294 | |
| Genetic reagent (D. melanogaster) | UAS-TimpRNAi2 | Vienna Drosophila Resource Center | VDRC: #109427 FBst0481116 | |
| Genetic reagent (D. melanogaster) | UAS-Tdc2RNAi1 | Vienna Drosophila Resource Center | VDRC: #330541 FBst0492256 | |
| Genetic reagent (D. melanogaster) | UAS-Tdc2RNAi2 | Bloomington Drosophila Stock Center | BDSC: #25871 RRID:BDSC_25871 FBst0025871 | |
| Genetic reagent (D. melanogaster) | UAS-TβhRNAi1 | Vienna Drosophila Resource Center | VDRC: #107070 FBst0478893 | |
| Genetic reagent (D. melanogaster) | UAS-TβhRNAi2 | Bloomington Drosophila Stock Center | BDSC: #67968 RRID:BDSC_67968 FBst0067968 | |
| Genetic reagent (D. melanogaster) | UAS-ChATRNAi1 | Vienna Drosophila Resource Center | VDRC: #330291 FBst0490951 | |
| Genetic reagent (D. melanogaster) | UAS-ChATRNAi2 | Bloomington Drosophila Stock Center | BDSC: #25856 RRID:BDSC_25856 FBst0025856 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRα1RNAi | Vienna Drosophila Resource Center | VDRC #48159 FBst0467755 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRα2RNAi | Vienna Drosophila Resource Center | VDRC: #101760 FBst0473633 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRα3RNAi | Vienna Drosophila Resource Center | VDRC: #101806 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRβ1RNAi | Vienna Drosophila Resource Center | VDRC: #106570 FBst0478394 | |
| Genetic reagent (D. melanogaster) | UAS-nAChRβ2RNAi | Vienna Drosophila Resource Center | VDRC: #109450 FBst0481138 | |
| Genetic reagent (D. melanogaster) | UAS-nvdRNAi1 | Yoshiyama et al., 2006 | FBal0193613 PMID:16763204 | |
| Genetic reagent (D. melanogaster) | UAS-nvdRNAi2 | Yoshiyama et al., 2006 | FBal0193614 PMID:16763204 | |
| Chemical, compound, drug | Octopamine | Sigma-Aldrich | #O0250 | |
| Chemical, compound, drug | Schneider’s Drosophila medium | Thermo Fisher Scientific | #21720024 | |
| Chemical, compound, drug | 20-hydroxyecdysone | Enzo Life Sciences | ALX-370–012 | |
| Antibody | anti-GFP (chicken polyclonal) | Abcam | #ab13970 | 1:4000 dilution |
| Antibody | anti-RFP (rabbit polyclonal) | Medical and Biological Laboratories | #PM005 | 1:2000 dilution |
| Antibody | anti-Hts 1B1 (mouse monoclonal) | Developmental Studies Hybridoma Bank | 1:50 dilution | |
| Antibody | anti-DE-cadherin DCAD2 (rat monoclonal) | Developmental Studies Hybridoma Bank | 1:50 dilution | |
| Antibody | anti-pH3 (rabbit polyclonal) | Merck Millipore | #06–570 | 1:2000 dilution |
| Antibody | anti-pMad (rabbit polyclonal) | Abcam | #ab52903 | 1:2000 dilution |
| Antibody | anti-Lamin C LC28.26 (mouse monoclonal) | Developmental Studies Hybridoma Bank | 1:10 dilution | |
| Antibody | anti-cleaved Dcp-1 (rabbit polyclonal) | Cell Signaling Technology | #9578 | 1:1000 dilution |
| Antibody | anti-Vasa (rat monoclonal) | Developmental Studies Hybridoma Bank | 1:50 dilution | |
| Antibody | anti-LacZ 40-1a (mouse monoclonal) | Developmental Studies Hybridoma Bank | 1:50 dilution | |
| Antibody | anti-Tdc2 (rabbit polyclonal) | Abcam | #ab128225 | 1:2000 dilution |
| Antibody | Alexa Fluor 546 phalloidin | Thermo Fisher Scientific | #A22283 | 1:200 dilution |
| Antibody | Alexa Fluor 633 phalloidin | Thermo Fisher Scientific | #A22284 | 1:200 dilution |
| Chemical, compound, drug | FluorSave reagent | Merck Millipore | #345789 | |
| Chemical, compound, drug | all trans-Retinal | Sigma-Aldrich | #R2500 | |
| Software, algorithm | ImageJ | https://imagej.nih.gov/ij; RRID:SCR_003070 PMID:22930834 | ||
| Software, algorithm | R | RRID:SCR_001905 |
Data availability
All data generated or analysed during this study are included in the manuscript and supporting files. Source data files have been provided for figures representing germline stem cell number, pMad signal intensity, GCaMP6 signal intensity, and TRIC signal intensity. Data has been deposited in Dryad under https://doi.org/10.5061/dryad.zkh189375.
-
Dryad Digital RepositoryData from: Neuronal octopamine signaling regulates mating-induced germline stem cell proliferation in female Drosophila.https://doi.org/10.5061/dryad.zkh189375
References
-
Genetic dissection of a stem cell niche: the case of the Drosophila ovaryDevelopmental Dynamics 235:2969–2979.https://doi.org/10.1002/dvdy.20967
-
Sex steroid hormone modulation of neural stem cells: a critical reviewBiology of Sex Differences 10:0242.https://doi.org/10.1186/s13293-019-0242-x
-
Molecular properties and functions of insect acetylcholine receptorsJournal of Insect Physiology 33:771–790.https://doi.org/10.1016/0022-1910(87)90025-4
-
A map of octopaminergic neurons in the Drosophila brainThe Journal of Comparative Neurology 513:643–667.https://doi.org/10.1002/cne.21966
-
Two functional but noncomplementing Drosophila tyrosine decarboxylase genes: distinct roles for neural tyramine and octopamine in female fertilityThe Journal of Biological Chemistry 280:14948–14955.https://doi.org/10.1074/jbc.M414197200
-
Nicotinic receptors at the amino acid levelAnnual Review of Pharmacology and Toxicology 40:431–458.https://doi.org/10.1146/annurev.pharmtox.40.1.431
-
The discovery of central monoamine neurons gave volume transmission to the wired brainProgress in Neurobiology 90:82–100.https://doi.org/10.1016/j.pneurobio.2009.10.012
-
A transcriptional reporter of intracellular Ca2+ in DrosophilaNature Neuroscience 18:917–925.https://doi.org/10.1038/nn.4016
-
Mutations of choline acetyltransferase and associated neural defectsJournal of Comparative Physiology ? A 137:83–92.https://doi.org/10.1007/BF00656920
-
Nicotinic acetylcholine receptors of the central nervous system of DrosophilaBiochimica Et Biophysica Acta (BBA) - Molecular Cell Research 1137:299–308.https://doi.org/10.1016/0167-4889(92)90150-A
-
A novel octopamine receptor with preferential expression in Drosophila mushroom bodiesThe Journal of Neuroscience 18:3650–3658.https://doi.org/10.1523/JNEUROSCI.18-10-03650.1998
-
The regulation of Drosophila ovarian stem cell niches by signaling crosstalkCurrent Opinion in Insect Science 37:23–29.https://doi.org/10.1016/j.cois.2019.10.006
-
Loops D, E and G in the Drosophila dα1 subunit contribute to high neonicotinoid sensitivity of Dα1-chicken β2 nicotinic acetylcholine receptorBritish Journal of Pharmacology 175:1999–2012.https://doi.org/10.1111/bph.13914
-
The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cellsDevelopment 124:761–771.
-
Heritable gene silencing in Drosophila using double-stranded RNANature Biotechnology 18:896–898.https://doi.org/10.1038/78531
-
Sex hormones establish a reserve pool of adult muscle stem cellsNature Cell Biology 18:930–940.https://doi.org/10.1038/ncb3401
-
The Drosophila ovary: an active stem cell communityCell Research 17:15–25.https://doi.org/10.1038/sj.cr.7310123
-
Independent optical excitation of distinct neural populationsNature Methods 11:338–346.https://doi.org/10.1038/nmeth.2836
-
Sex-peptides: seminal peptides of the Drosophila maleCellular and Molecular Life Sciences 60:1689–1704.https://doi.org/10.1007/s00018-003-3052
-
Octopamine receptor OAMB is required for ovulation in Drosophila melanogasterDevelopmental Biology 264:179–190.https://doi.org/10.1016/j.ydbio.2003.07.018
-
Fast Excitatory Synaptic Transmission Mediated by Nicotinic Acetylcholine Receptors in Drosophila NeuronsThe Journal of Neuroscience 19:5311–5321.https://doi.org/10.1523/JNEUROSCI.19-13-05311.1999
-
Regulation of adult female germline stem cells by nutrient-responsive signalingCurrent Opinion in Insect Science 37:16–22.https://doi.org/10.1016/j.cois.2019.10.005
-
Cooperation of β2- and β3-adrenergic receptors in hematopoietic progenitor cell mobilizationAnnals of the New York Academy of Sciences 1192:139–144.https://doi.org/10.1111/j.1749-6632.2010.05390.x
-
Characterization of Drosophila tyramine beta-hydroxylase gene and isolation of mutant flies Lacking octopamineThe Journal of Neuroscience 16:3900–3911.
-
A high signal-to-noise Ca2+ probe composed of a single green fluorescent proteinNature Biotechnology 19:137–141.https://doi.org/10.1038/84397
-
TGF-beta family signal transduction in Drosophila development: from mad to smadsDevelopmental Biology 210:251–268.https://doi.org/10.1006/dbio.1999.9282
-
Tyramine and octopamine: ruling behavior and metabolismAnnual Review of Entomology 50:447–477.https://doi.org/10.1146/annurev.ento.50.071803.130404
-
Dopamine and octopamine differentiate between aversive and appetitive olfactory memories in DrosophilaThe Journal of Neuroscience 23:10495–10502.https://doi.org/10.1523/JNEUROSCI.23-33-10495.2003
-
Germline stem cellsCold Spring Harbor Perspectives in Biology 3:a002642.https://doi.org/10.1101/cshperspect.a002642
-
A matrix metalloproteinase mediates long-distance attenuation of stem cell proliferationJournal of Cell Biology 206:923–936.https://doi.org/10.1083/jcb.201403084
-
Adrenaline stimulates the proliferation and migration of mesenchymal stem cells towards the LPS-induced lung injuryJournal of Cellular and Molecular Medicine 18:1612–1622.https://doi.org/10.1111/jcmm.12283
-
Endocrine regulation of female germline stem cells in the fruit fly Drosophila melanogasterCurrent Opinion in Insect Science 31:14–19.https://doi.org/10.1016/j.cois.2018.07.001
-
The Conserved Rieske Oxygenase DAF-36/Neverland Is a Novel Cholesterol-metabolizing EnzymeJournal of Biological Chemistry 286:25756–25762.https://doi.org/10.1074/jbc.M111.244384
-
Cellular organization of the neural circuit that drives Drosophila courtship behaviorCurrent Biology : CB 20:1602–1614.https://doi.org/10.1016/j.cub.2010.08.025
-
Signal transduction pathways regulating Drosophila ovarian germline stem cellsCurrent Opinion in Insect Science 37:1–7.https://doi.org/10.1016/j.cois.2019.10.002
-
A subset of octopaminergic neurons are important for Drosophila aggressionNature Neuroscience 11:1059–1067.https://doi.org/10.1038/nn.2164
Article and author information
Author details
Funding
Japan Agency for Medical Research and Development (19gm1110001h0003)
- Ryusuke Niwa
Takeda Science Foundation
- Ryusuke Niwa
Japan Society for the Promotion of Science (KAKENHI 18J20572)
- Yuto Yoshinari
Japan Society for the Promotion of Science (KAKENHI 15J00652)
- Tomotsune Ameku
Japan Society for the Promotion of Science (KAKENHI 26250001)
- Hiromu Tanimoto
Japan Society for the Promotion of Science (KAKENHI 17H01378)
- Hiromu Tanimoto
Japan Agency for Medical Research and Development (17gm6010011h0001)
- Takayuki Kuraishi
Japan Society for the Promotion of Science (KAKENHI 19H05240)
- Ryusuke Niwa
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We thank Toshiro Aigaki, Kendal S Broadie, Aki Ejima, Kyung-An Han, Yukako Hattori, Yoshiki Hayashi, Makoto Ihara, Young-Joon Kim, Satoru Kobayashi, Kazuhiko Matsuda, Masayuki Miura, Akira Nakamura, Takashi Nishimura, Andrea Page-McCaw, Hiroko Sano, Jianjun Sun, Nobuaki Tanaka, James W Truman, Tadashi Uemura, Daisuke Yamamoto, the Bloomington Stock Center, the Kyoto Stock Center (DGRC), the National Institute of Genetics, the Vienna Drosophila Resource Center, and the Developmental Studies Hybridoma Bank for providing stocks and reagents; Aki Hori and Reiko Kise for their technical assistance; and Satoru Kobayashi and Shosei Yoshida for their helpful discussion. YY and TA were recipients of the fellowship from the Japan Society for the Promotion of Science. This work was supported by grants from KAKENHI (19H05240 to RN, 26250001 and 17H01378 to HT, 18J20572 to YY, and 15J00652 to TA), AMED-PRIME, AMED (17gm6010011h0001 to TK), AMED-CREST, AMED (19gm1110001h0003 to RN), and the Takeda Science Foundation to RN.
Copyright
© 2020, Yoshinari et al.
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 4,456
- views
-
- 487
- downloads
-
- 43
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Citations by DOI
-
- 43
- citations for umbrella DOI https://doi.org/10.7554/eLife.57101






