Regulation of canonical Wnt signalling by the ciliopathy protein MKS1 and the E2 ubiquitin-conjugating enzyme UBE2E1
Figures
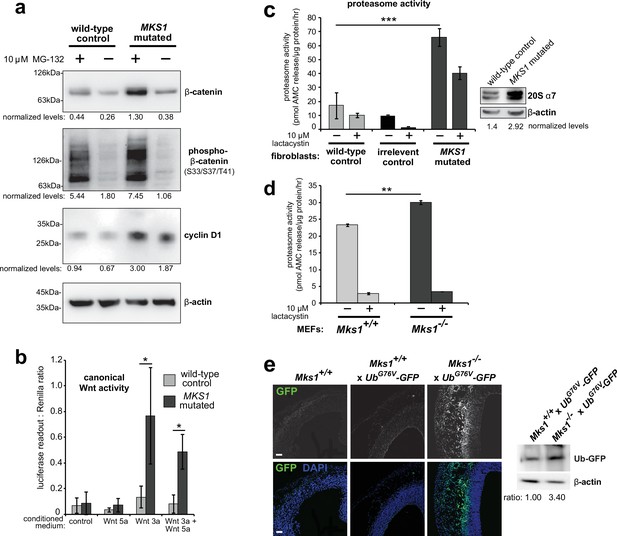
Deregulation of canonical Wnt signalling and proteasome activity following loss or mutation of MKS1.
(a) Immunoblots for total soluble β-catenin, phospho-β-catenin, cyclin D1 and β-actin (loading control) in either wild-type normal or MKS1-mutated immortalised human fibroblasts from an MKS patient (MKS-562) following treatment with MG-132 proteasome inhibitor (+) or vehicle control (-). (b) SUPER-TOPFlash assays of canonical Wnt signalling activity in human MKS1-mutated fibroblasts compared to wild-type control fibroblasts following treatment with control conditioned medium, Wnt5a, Wnt3a, or a mixture of Wnt3a and Wnt5a media, as indicated. Statistical significance of pairwise comparisons is shown (* indicates p < 0.05, paired two-tailed Student t-test). Error bars indicate s.e.m. with results shown for four independent biological replicates. (c) Proteasome activity assays for wild-type or MKS1-mutated human fibroblasts or an irrelevant control (ASPM-mutant fibroblasts), following treatment with c-lactacystin-β-lactone (+) or vehicle control (-). Statistical significance of pairwise comparison as for (b); *** indicates p < 0.001 for three independent biological replicates. Immunoblots show levels of the 20 S proteasome α7 subunit compared to β-actin loading control. (d) Protease activity assays of crude proteasome preparations from Mks1+/+ or Mks1-/- mouse embryonic fibroblasts (MEFs), expressed as pmol AMC released per µg proteasome per hr. Treatment with lactacystin is the assay control. Statistical analysis as for (b); ** indicates p < 0.01 for three independent biological replicates. (e) Accumulation of GFP-tagged ubiquitin (GFP; green) in Mks1-/- x UbG76V-GFP E12.5 embryonic cerebral neocortex treated with MG-262 proteasome inhibitor. Immunoblot for GFP in Mks1-/- x UbG76V-GFP and wild-type littermate E12.5 embryo protein lysates, with immunoblotting for β-actin as a loading control, showing accumulation of GFP-tagged ubiquitin (Ub-GFP) in Mks1-/-.
-
Figure 1—source data 1
Characterisation of MKS1-mutated human patient fibroblasts: full western blots.
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig1-data1-v2.pdf
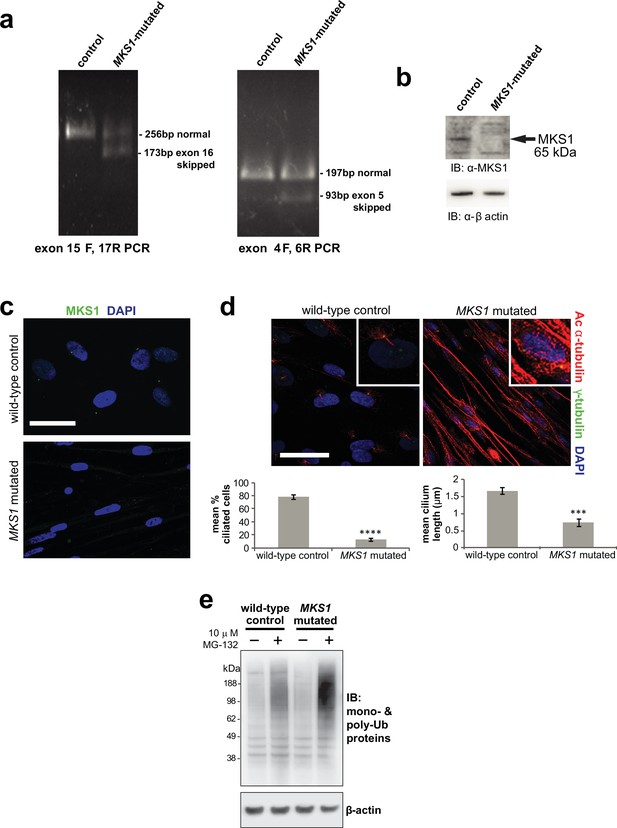
Characterisation of MKS1-mutated human patient fibroblasts.
(a) RT-PCR amplicons of exons 4–6 and 15–17 from cDNA of healthy control and MKS1 patient fibroblasts, compound heterozygote for the MKS1 mutations [c.472C > T]+[IVS15-7_35del29] causing the predicted nonsense and splice-site mutations [p.R158*]+[p.P470fs*562]. Additional smaller PCR products in MKS1 patient corresponds to skipping of exon 5 and exon 16, confirmed by Sanger sequencing, due to the frameshift mutation affecting splicing. (b) Immunoblot showing loss of MKS1 protein in MKS1-mutated patient fibroblasts compared to healthy controls; loading control is β-actin. (c) IF microscopy images of wild-type control and MKS1-mutated fibroblasts showing loss of cilia and disorganisation of cytoskeleton in patient cells. Bar graphs quantify reductions in incidence and length of cilia in patient cells. Statistical significance of pairwise comparisons with control for three independent biological replicates are shown (*** p < 0.001, **** p < 0.0001; paired two-tailed Student t-test; error bars indicate s.e.m.) (d) IF microscopy images showing loss of MKS1 ciliary localisation in MKS1-mutated patient fibroblasts compared to wild-type control fibroblasts (indicated by arrowheads). (e) Western blot showing increased levels of polyubiquitinated protein in MKS1-mutated fibroblasts after MG-132 proteasome inhibition. and shUbe2e1 after MG-132 treatment.
-
Figure 1—figure supplement 1—source data 1
Characterisation of MKS1-mutated human patient fibroblasts: full western blots & gels.
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig1-figsupp1-data1-v2.pdf
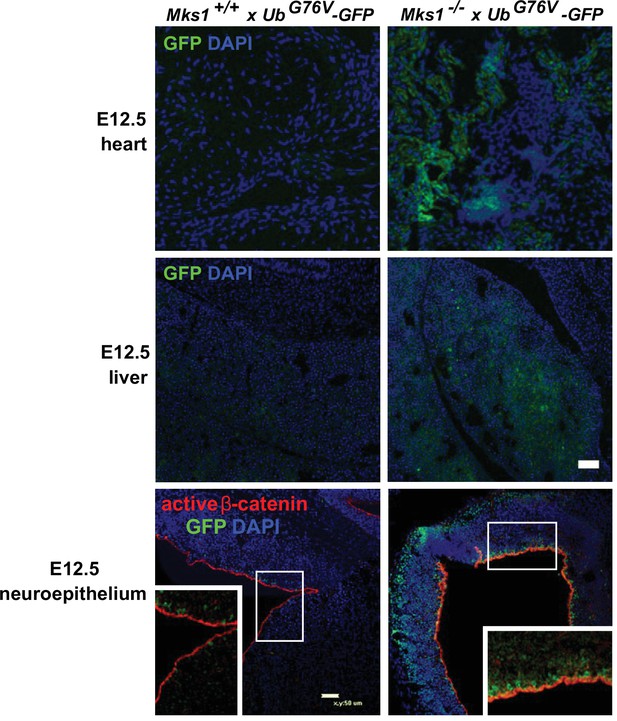
In vivo loss of MKS1 causes deregulated ubiquitin-proteasome processing.
Proteasome defects (GFP; green) in Mks1-/- x UbG76V-GFP E12.5 embryonic heart (top panels) and liver (middle panels) as indicated, with accumulation of active β-catenin (red) and GFP-tagged ubiquitin (green) in the neuroepithelium of E12.5 Mks1-/-xUbG76VGFP embryonic neocortex. Scale bars = 50 μm. White frames indicate magnified regions displayed in insets.
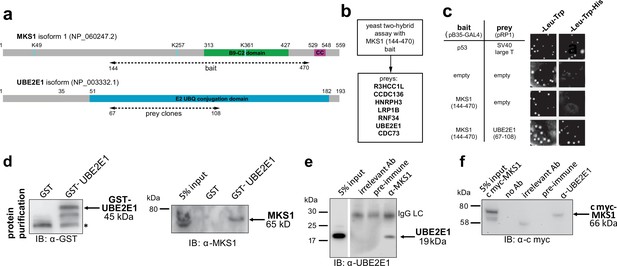
The E2 ubiquitin conjugation enzyme UBE2E1 interacts with MKS1.
(a) Domain structure of MKS1 and UBE2E1 proteins for the indicated isoform showing the locations of the B9/C2 domain, putative ubiquitinated lysines in blue (predicted by UbPred), a predicted coiled-coil (CC) motif, and the E2 ubiquitin (UBQ) conjugation domain in UBE2E1. Numbering indicates the amino acid residue. Dashed lines indicate the region used as ‘bait’ in MKS1 for the yeast two-hybrid assay and the ‘prey’ clones in the UBE2E1 interactant. (b) List of preys identified in the MKS1 Y2H screen (c) Left panel: yeast ‘one-to-one’ assays for the indicated bait, prey and control constructs. Right panel: only colonies for the positive control (p53+ SV40 large T) and MKS1 bait+ UBE2E1 prey grew on triple dropout (-Leu -Trp -His) medium. (d) GST-UBE2E1 purified from bacterial extracts (left panel) pulled down endogenous MKS1 from ARPE19 whole cell extract. (e) Co-immunoprecipitation (co-IP) of endogenous UBE2E1 by rabbit polyclonal anti-MKS1, but not pre-immune serum or an irrelevant antibody (Ab; anti cmyc); IgG light chain (LC) is indicated. (f) Co-IP of exogenously expressed cmyc-MKS1 by anti-UBE2E1 but not pre-immune serum or an irrelevant antibody.
-
Figure 2—source data 1
The E2 ubiquitin conjugation enzyme UBE2E1 interacts with MKS1: full western blots.
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig2-data1-v2.pdf
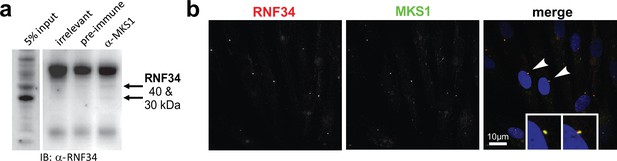
The E3 ubiquitin ligase RNF34 interacts with MKS1 and co-localises at the basal body.
(a) Co-immunoprecipitation of 30 and 40 kDa isoforms of endogenous RNF34 by rabbit polyclonal anti-MKS1, but not by an irrelevant antibody or the pre-immune. (b) Co-localisation of RNF34 (red) with MKS1 (green) at the basal body (arrowheads) in mIMCD3 cells. Scale bar = 10 μm.
-
Figure 2—figure supplement 1—source data 1
The E3 ubiquitin ligase RNF34 interacts with MKS1 and co-localizses at the basal body: full western blot.
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig2-figsupp1-data1-v2.pdf
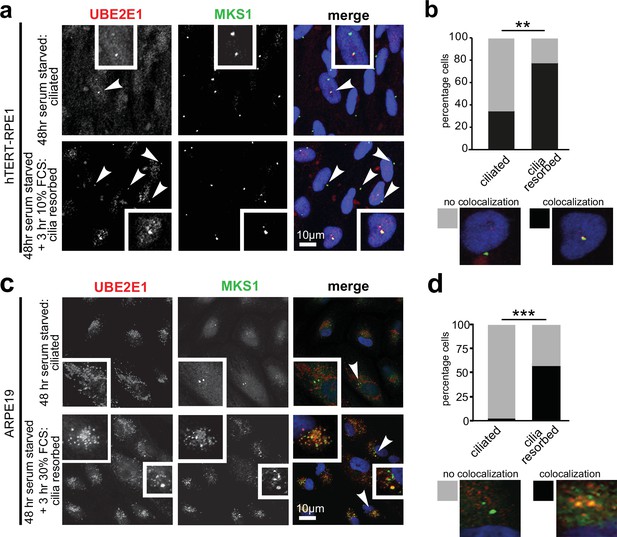
Co-localisation of MKS1 and UBE2E1 under conditions of ciliary resorption.
(a) MKS1 (green) and UBE2E1 (red) partially colocalise at the basal body/centrosome in human wild-type hTERT-RPE1 cells, particularly when induced to resorb cilia by treatment with 10% FCS after 72 hr of serum starvation. White arrowheads indicate cells magnified in insets. Scale bar = 10 µm. (b) Bar graph indicates the percentage of cells in which MKS1 and UBE2E1 co-localise at the basal body (black), and the percentage without co-localisation (grey) for three independent biological replicates, with examples shown of representative cells. (c) Figure details as for (a) showing partial co-localisation of MKS1 and UBE2E1 in human ARPE19 cells. (d) Bar graph details as for (b). Data in (b) and (d) were analysed by two-way ANOVA followed by Tukey’s multiple comparison test (statistical significance of comparisons indicated by ** p < 0.01, *** p < 0.001).
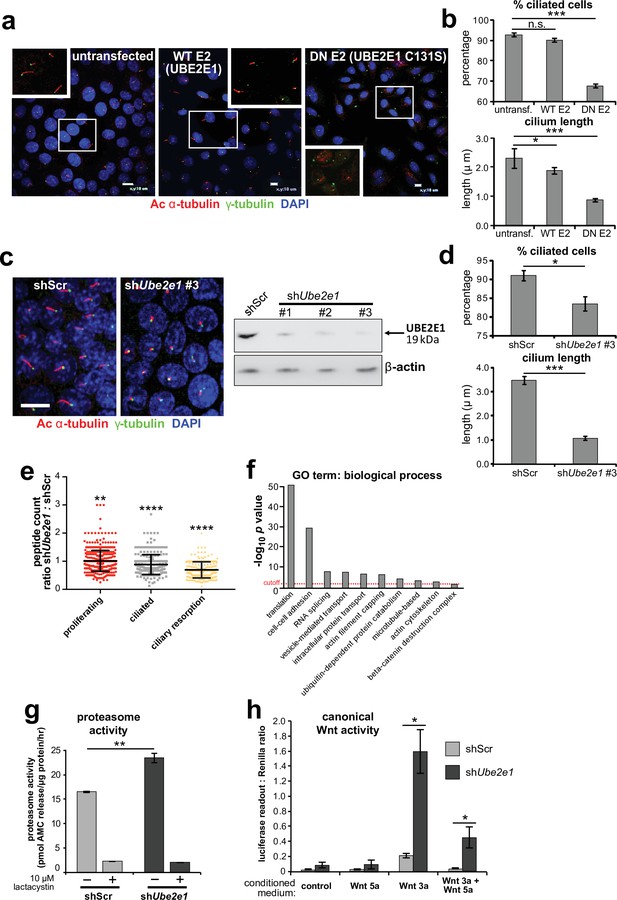
UBE2E1 is required for regulation of ciliogenesis, proteasome activity, and canonical Wnt signalling.
(a) Primary cilia in mIMCD3 cells following transfection with either wild-type (WT) UBE2E1 (E2) or dominant negative (DN) UBE2E1 carrying the active site mutation C131S, compared to mock-transfected negative control. Scale bars = 10 μm. (b) For experiments shown in (a), statistical significance of pairwise comparisons with control (untransf.) for three independent biological replicates are shown (n.s. not significant, * p < 0.05, ** p < 0.01, *** p < 0.001; unpaired two-tailed Student t-test; error bars indicate s.e.m.). (c) shRNA-mediated knockdown of Ube2e1 in stably-transfected mIMCD3 cell-line #3 causes decreased ciliary incidence and length. Scale bar = 10 μm. Immunoblot shows loss of UBE2E1 protein expression compared to β-actin loading control following shRNA knockdown. (d) Bar graphs quantifying decreased ciliary incidence and length with statistical analysis as for (b). (e) Scatter plot of relative differences in the proteins pulled-down by anti-MKS1 immunoprecipitations, under different conditions of ciliogenesis (proliferating cells, ciliated cells, cells undergoing ciliary resorption), expressed as the ratios of peptide counts for shScr: shUbe2e1 knockdowns. Statistical significance of pairwise comparisons for each set of ratios was calculated as for (b) (paired two-tailed Student t-tests). Error bars indicate s.d. Full data-sets are available in Figure 4—source data 1. (f) Bar graph of -log10 p values for significantly enriched GO terms (biological processes) for proteins included in (e), with cut-off for p < 0.05 indicated by the red dashed line. Enrichment for GO terms was analyzed by using DAVID (https://david.ncifcrf.gov/). (g) Protease activity assays of crude proteasome preparations from shScr and shUbe2e1 mIMCD3 knockdown cells, showing increased proteasomal activity in shUbe2e1 as assayed by pmol AMC released per μg proteasome per hour. Treatment with lactacystin is the assay control. Statistical significance of pairwise comparisons as for (b). (h) SUPER-TOPFlash assays of canonical Wnt signalling activity in shUbe2e1 cells compared to shScr following treatment with control conditioned medium, Wnt5a, Wnt3a, or a mixture of Wnt3a and Wnt5a media, as indicated. Statistical significance of pairwise comparisons of at least four independently replicated experiments as for (b).
-
Figure 4—source data 1
Mass spectrometry results for MKS1 pull-downs from mIMCD3 cells across different conditions of ciliogenesis.
The data-set lists proteins (identified by ≥3 unique peptide counts) pulled-down by anti-MKS1 immunoprecipitations, under different conditions of ciliogenesis (proliferating cells, ciliated cells, cells undergoing ciliary resorption). Columns A and B: protein and gene name. Column C: protein accession number. Columns G to L: peptide counts identified by LC-MS/MS mass spectrometry (columns F and M indicating non-specific peptide counts following BSA washes), with counts heat-mapped red (high) to green (low). Column N: χ2 tests of peptide counts for shScr compared to shUbe2e1 knockdown cells, across different conditions of ciliogenesis (red highlighted cells indicate χ2 test p < 0.05). Columns Q to S: shScr:shUbe2e1 peptide count ratios (derived from columns G to L), with values < 1 indicating decreased peptide counts following shUbe2e1 knockdown. Columns U to Z: indicate if a particular protein was identified in a significantly enriched biological process under the indicated GO terms (row 2).
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig4-data1-v2.pdf
-
Figure 4—source data 2
UBE2E1 is required for regulation of ciliogenesis, proteasome activity, and canonical Wnt signalling: full western blot.
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig4-data2-v2.xlsx
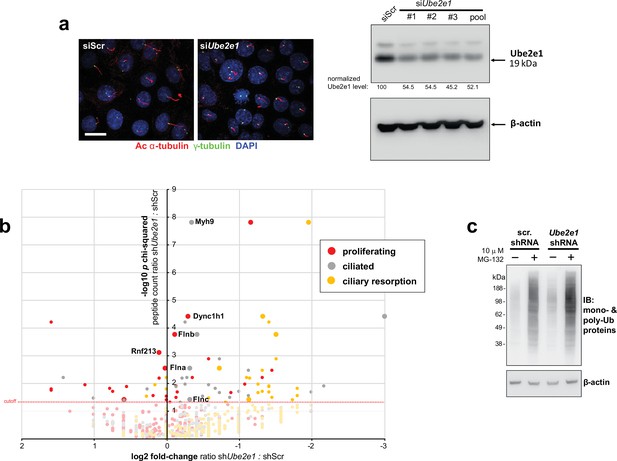
Validation of Ube2e1 siRNA knockdown in mIMCD3 cells and effect of shRNA Ube2e1 knockdowns on MKS1 interacting proteins under different conditions of ciliogenesis.
(a) Left panels: mouse Ube2e1 pooled siRNA duplexes (siUbe2e1) prevent ciliogenesis compared to scrambled (siScr) control. Primary cilia and basal bodies are visualised by staining for acetylated α-tubulin (red) and γ-tubulin (green). Scale bar = 10 μm. Right panel: immunoblot to confirm reduction of Ube2e1 protein levels following knockdown in mIMCD3 cells using individual siRNA duplexes (#1 to #3) and pooled duplexes, with knockdown efficiency indicated normalised to β-actin loading control levels. (b) Scatter plot representing log2 fold-difference values for peptide ratios shScr: shUbe2e1 in mIMCD3 knockdown cells versus -log10 p χ2 tests for shUbe2e1: shScr rations across different conditions of ciliogenesis. These conditions comprise: proliferating cells grown in media containing serum (red); ciliated cells grown in the absence of serum (grey); and cells undergoing ciliary resorption grown in the absence of serum followed by 2 hr incubation in media with serum (gold). The proteins indicated in the plot are members of the enriched biological process GO term ‘actin filament capping’ term which includes known interactants of ciliopathy proteins such as filamin A and B (Flna and Flnb). Full data-sets for this plot are available in Figure 4e–f, Figure 4—source data 1. (c) Western blot showing increased levels of polyubiquitinated protein in shUbe2e1 knock-down cells after MG-132 proteasome inhibition, compared to scrambled (scr.) controls.
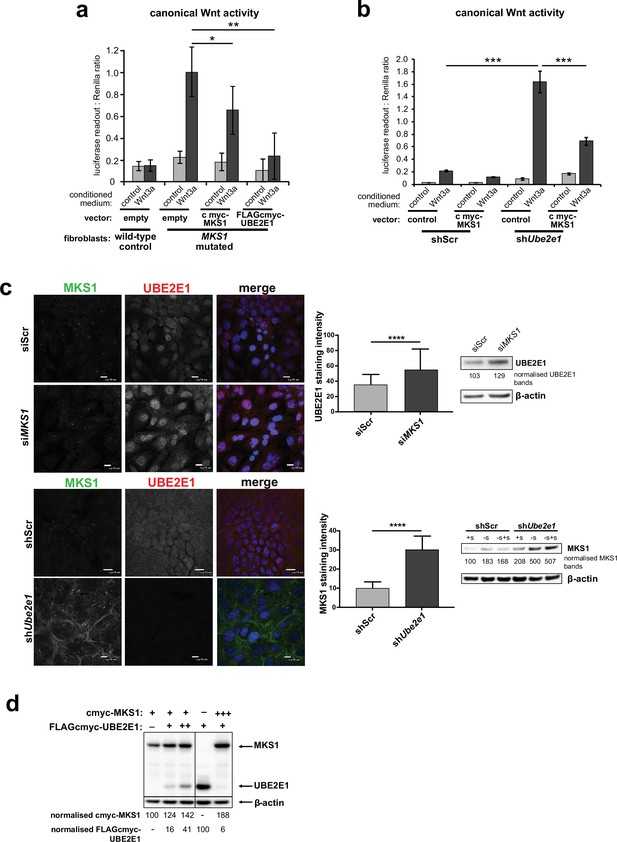
Co-dependant regulation of MKS1 and UBE2E1.
(a) SUPER-TOPFlash assays in wild-type or MKS1-mutated fibroblasts, following transient co-transfection with either exogenous control, MKS1-cmyc or UBE2E1-FLAG-cmyc, and treatment with either Wnt3a or control conditioned medium. Statistical significance of the indicated pairwise comparisons with control for three independent biological replicates are shown (* p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001; unpaired two-tailed Student t-test; error bars indicate s.e.m.) (b) SUPER-TOPFlash assays in shScr and shUbe2e1 cell-lines, following transient co-transfection with either exogenous cmyc-MKS1 or empty plasmid construct (control) and treatment with either Wnt3a or control conditioned medium, as indicated. Statistical comparisons as for (a). (c) Top panel: increased per cell staining intensity for UBE2E1 following MKS1 siRNA knockdown Bottom panel: increased per cell staining intensity for MKS1 in Ube2e1 mIMCD3 knockdown cells Scale bars = 10 µm. Bar graphs quantitate staining intensities for three independent biological replicates. Statistical significance of pairwise comparisons as for (a), error bars indicate s.e.m. Western blots (panels on right) show increased UBE2E1 protein levels for siMKS1 knockdown cells, and increased MKS1 protein levels for shUbe2e1 cells. Quantitation of band intensities were normalised to β-actin loading control. (d) HEK293 cells were transiently transfected with control vector (-), constant (+) or high (+++) levels of cmyc-MKS1 and/or FLAG-cmyc-UBE2E1. Levels were normalised to β-actin loading control. MKS1 levels moderately decreased with increasing levels of UBE2E1, whereas high levels of MKS1 caused loss of UBE2E1.
-
Figure 5—source data 1
Co-dependant regulation of MKS1 and UBE2E1: full western blots.
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig5-data1-v2.pdf
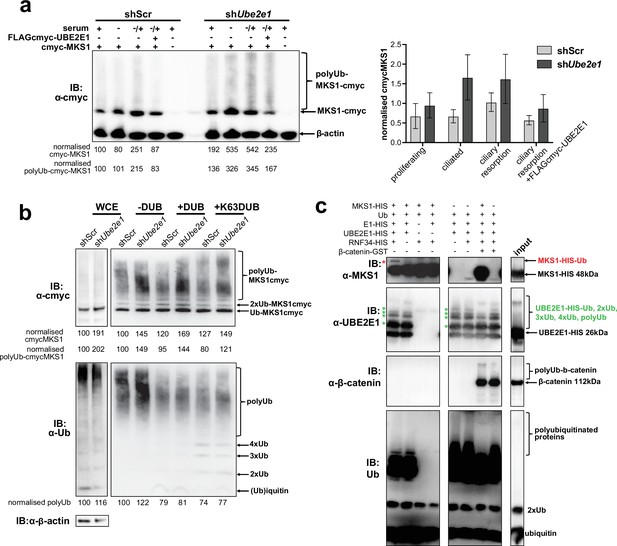
MKS1 is ubiquitinated and its ubiquitynation depends on UBE2E1.
(a) shScr and shUbe2e1 mIMCD3 knockdown cells transiently transfected with cmyc-MKS1 and/or FLAG-cmyc-UBE2E1 under different conditions of ciliogenesis: proliferating cells grown in media containing serum (+); ciliated cells grown in the absence of serum (-); and cells undergoing ciliary resorption grown in the absence of serum followed by 2 hr incubation in media with serum (-/+). Increased levels of cmyc-MKS1 and smears representing poly-ubiquitinated (polyUb) cmyc-MKS1 in shUbe2e1 cells are indicated. Addition of exogenous FLAG-cmyc-UBE2E1 partially rescued correct MKS1 levels and ubiquitination. Normalised band intensities for the whole cmyc-MKS1 staining and only polyUb-cmyc-MKS1 are shown below the blots. Bar graph quantitates cmyc-MKS1 levels normalised to β-actin levels for three independent biological replicates. Data was analysed by two-way ANOVA followed by Tukey’s multiple comparison test (statistical significance of comparison between shScr and shUbe2e1 is p < 0.05, error bars represent s.d.). (b) TUBE experiment confirming ubiquitination of cmyc-MKS1. Consistently increased levels of polyubiquitinated cmyc-MKS1 were observed in shUbe2e1 knockdown cells. Broad-range deubiquitinating enzymes (+ DUB) and K63-specific (+ K63 DUB) deubiquitinating enzyme were used to assess the type of MKS1 ubiquitination. Normalised band intensities are shown below the blots. (c) In vitro ubiquitination assay for MKS1-HIS, UBE2E1-HIS, RNF34-HIS, E1-HIS, Ub, and β-catenin-GST fusion proteins. The MKS1 blot shows possible mono-ubiquitination of MKS1 (red asterisk) in the presence of UBE2E1 and RNF34. Auto-ubiquitination of UBE2E1 (green asterisks indicate the addition of one, two, three, four and poly-ubiquitin chains) was inhibited by MKS1. This was further inhibited by addition of β-catenin, but β-catenin addition by itself did not affect UBE2E1 polyubiquitination. although β-catenin was polyubiquitinated by UBE2E1.
-
Figure 6—source data 1
MKS1 is ubiquitinated and its ubiquitynation depends on UBE2E1: full western blots.
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig6-data1-v2.pdf
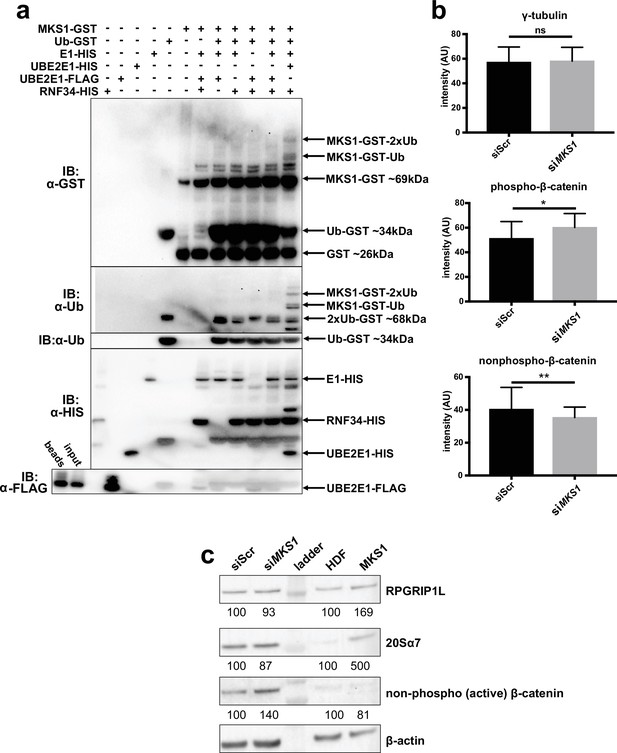
MKS1 is mono-ubiquitinated in presence of UBE2E1 and RNF34.
(a) In vitro ubiquitination assay for MKS1-GST fusion protein, for the indicated controls and reaction conditions, showing the addition of one (Ub) and two (2xUb) ubiquitins in the presence of both UBE2E1 and RNF34. (b) Immunofluorescence staining intensity measurements (arbitrary units; AU) for the indicated proteins in control compared to siMKS1 knock-down cells. There was significant increase in phosphorylated-β-catenin staining in these cells (* = 0.0013, Student unpaired t-test with Welch’s correction) at the basal body, and significantly decreased nonphosphorylated (active) β-catenin staining in cell nuclei in siMKS1 cells (** = 0.004 Student unpaired t-test with Welch’s correction). (c) Western blot analysis of siMKS1 hTERT-RPE1 cells and MKS1 patient fibroblast showing no overall changes in RPGRIP1L following either knock-down (in siMKS1 cells) or MKS1-mutated patient fibroblast. Levels of the proteasome marker 20Sα7 were increased in MKS1-mutated fibroblasts compared to normal control (HDF). There were no consistent changes in nonphosphorylated (active) β-catenin in siMKS1 cells and MKS1-mutated fibroblasts. Protein levels were normalised to the loading control, β-actin.
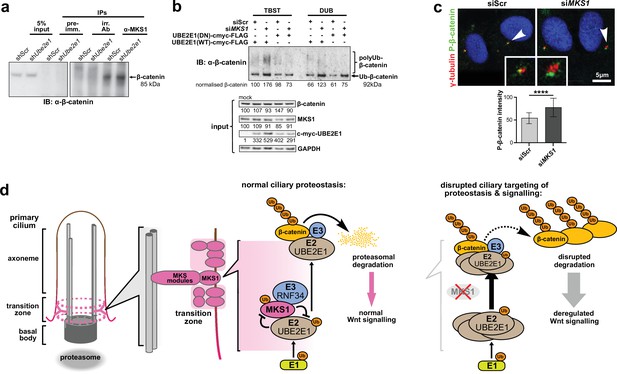
MKS1 and UBE2E1 interact to regulate β-catenin ubiquitination.
(a) Immunoblot showing increased co-immunoprecipitation of β-catenin by anti-MKS1 in shUbe2e1 knockdown cells compared to shScr control cells. (b) TUBE pulldown followed by β-catenin immunoblotting, showing polyubiquitination of β-catenin was increased following MKS1 knockdown and in the presence of the wild-type (WT) UBE2E1. Inactive form of UBE2E1 (DN) had not had effect on polyubiquitination of β-catenin, highlighting the importance of this UBE2E1 in β-catenin degradation. (c) Immunofluorescence staining of hTERT-RPE1 cells showing co-localisation of phosphorylated (P)-β-catenin (green) with γ-tubulin (red) at the base of cilia (arrowheads). P-β-catenin localisation significantly increased following siMKS1 knockdown (paired two-tailed Student t-test, **** p < 0.0001 for three independent biological replicates; > 40 cells quantified per replicate). Scale bar = 5 μm. (d) Schematic representation of UPS regulation of MKS1 and β-catenin protein levels at the ciliary apparatus. Protein levels of MKS1 (pink) and UBE2E1 (light brown) are co-dependant through regulation at the base of the cilium. MKS1 localises to the TZ (dashed pink lines) and is mono-/bi-ubiquitinated by a complex that includes UBE2E1 and RNF34 (blue). MKS1 and UBE2E1 regulate each other, what has an effect on downstream UBE2E1 role in regulation of polyubiquitination of β-catenin (yellow). The correct regulation between these proteins facilitates normal proteasomal function and canonical Wnt signalling (small pink arrow). Both processes are de-regulated following MKS1 mutation of loss (red cross), causing aberrant accumulation of UBE2E1 and polyubiquitinated β-catenin and disrupted tethering to the ciliary apparatus.
-
Figure 7—source data 1
MKS1 and UBE2E1 interact to regulate β-catenin ubiquitination: full western blots.
- https://cdn.elifesciences.org/articles/57593/elife-57593-fig7-data1-v2.pdf
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | B6;129P2-Mks1tm1a(EUCOMM)Wtsi | Wellcome Trust Sanger Institute | EM:05429 | RRID:IMSR_EM:05429 |
| Strain, strain background (Mus musculus) | B6.Cg-Tg(ACTB-Ub*G76V/GFP)1Dant/J | Jackson Laboratory, Maine, USA | 008111 | RRID:IMSR_JAX:008111 |
| Cell line (Mus musculus) | mIMCD3 | ATCC | CRL-2123 | RRID:CVCL_0429 |
| Cell line (Homo sapiens) | hTERT-RPE1 | ATCC | CRL-4000 | RRID:CVCL_4388 |
| Cell line (Homo sapiens) | ARPE-19 | ATCC | CRL-2302 | RRID:CVCL_0145 |
| Cell line (Homo sapiens) | HEK293 | ATCC | ACS-4500 | RRID:CVCL_4V93 |
| Cell line (Homo sapiens) | MKS-562 fibroblasts | Khaddour et al., 2007 | MKS1 compound heterozygote mutations | |
| Transfected construct (Homo sapiens) | pCMV-cmyc-MKS1 | Dawe et al., 2009 | full-length cDNA (NM_017777); see Cloning, plasmid constructs and transfection | |
| Transfected construct (Homo sapiens) | pGEX5X-1-UBE2E1 | Hong et al., 2008 | construct used to generate UBE2E1 protein | |
| Transfected construct (Homo sapiens) | pCMV-UBE2E1-FLAG-cmyc | Hong et al., 2008 | ||
| Transfected construct (Homo sapiens) | pCMV-UBE2E1 (DN) -FLAG-cmyc | this paper | c.341T > A, p.C131S UBE2E1 active site dominant negative (DN) mutation; see Cloning, plasmid constructs and transfection | |
| Transfected construct (Mus musculus) | Ube2e1 shRNA | Origene | TR502364 | Cells selected using 0.5 μg/ml puromycin for five passages |
| Transfected construct (Mus musculus) | Ube2e1 siRNA | Dharmacon ON-TARGET PLUS siRNA | L-062416-01-0005 | |
| Transfected construct (Mus musculus) | Mks1 siRNA | Dharmacon ON-TARGET PLUS siRNA | L-063962-01-0005 | |
| Antibody | Anti-cmyc, clone 9E10 (mouse monoclonal) | Sigma-Aldrich Co. Ltd. | M4439 | RRID:AB_439694WB: 1:1,000 |
| Antibody | Anti-acetylated-α-tubulin, clone 6-11B-1 (mouse monoclonal) | Sigma-Aldrich Co. Ltd. | MABT868 | RRID:AB_2819178IF: 1:1,000 |
| Antibody | Anti-HA, clone HA-7 (mouse monoclonal) | Sigma-Aldrich Co. Ltd. | H9658 | RRID:AB_260092WB: 1:100 |
| Antibody | Anti-GFP (rabbit polyclonal) | Living Colors A.v. Peptide Antibody | 632,377 | RRID:AB_2313653IF: 1:100 |
| Antibody | Anti-UBE2E1, clone 42/UbcH6 (mouse monoclonal) | BD Biosciences Inc | 611,218 | RRID:AB_398750IF: 1:100WB: 1:500 |
| Antibody | Anti-UBE2E1 (rabbit polyclonal) | Aviva Systems Biology | ARP43012_P050 | RRID:AB_2048646IF: 1:100WB: 1:500 |
| Antibody | Anti-γ-tubulin (rabbit polyclonal) | Sigma-Aldrich Co. Ltd. | T5192 | RRID:AB_261690IF: 1:500 |
| Antibody | Anti-β-actin, clone AC-15 (mouse monoclonal) | Abcam Ltd. | ab6276 | RRID:AB_2223210WB: 1:5,000 |
| Antibody | Anti-cyclin D1, clone A-12 (mouse monoclonal) | Santa Cruz Biotechnology Inc | sc-8396 | RRID:AB_627344WB: 1:1,000 |
| Antibody | Anti-phospho-β-catenin (rabbit polyclonal) | Cell Signalling Technology Inc | 9,561 | RRID:AB_331729WB: 1:1,000IF: 1:100 |
| Antibody | Anti-β-catenin, clone D10A8 (rabbit monoclonal) | Cell Signalling Technology Inc | 8,480 | RRID:AB_2798305WB: 1:1,000IF: 1:100 |
| Antibody | Anti-mono- and polyubiquitinylated conjugates, clone FK2 (mouse monoclonal) | Enzo Life Sciences, Inc | ENZ-ABS840 | RRID:AB_10541840WB: 1:1,000 |
| Antibody | Anti-20S proteasome α7 subunit, clone MCP72 (rabbit monoclonal) | Enzo Life Sciences Inc | BML-PW8110 | RRID:AB_10538395WB: 1:1,000IF: 1:100 |
| Antibody | Anti-MKS1 (rabbit polyclonal) | Dawe et al., 2007; Näthke et al., 1996 | WB: 1:500IF: 1:100 | |
| Antibody | Anti MKS1 (rabbit polyclonal) | Proteintech | 16206–1-AP | RRID:AB_10637856WB: 1:500IF: 1:100 |
| Antibody | Anti-ubiquitin, clone P4D1 (mouse monoclonal) | Santa Cruz Biotechnology, Inc | sc-8017 | RRID:AB_2762364 |
| Peptide, recombinant protein | MKS1-HIS | Proteintech Group, Inc | Ag9504 | |
| Peptide, recombinant protein | UBE2E1-HIS | Enzo Life Sciences, Inc | UW8710 | |
| Peptide, recombinant protein | RNF34-HIS | Novus Biologicals | NBP2-23440 | |
| Peptide, recombinant protein | β-catenin-GST | Abcam | Ab63175 | |
| Commercial assay or kit | 20 S fluorophore substrate Suc-LLVY-AMC | Enzo Life Sciences Inc | BML-P802-0005 | |
| Commercial assay or kit | Dual-Luciferase Reporter Assay system | Promega Corp. | E1910 | |
| Commercial assay or kit | Ubiquitination kit | Enzo Life Sciences, Inc | BML-UW0400 | |
| Commercial assay or kit | TUBE assays | LifeSensors, Malvern, PA, USA | UM-402 | |
| Chemical compound, drug | MG-132 | Sigma-Aldrich Co. Ltd. | C2211 | treatment at 10 μM for 3 hr |
| Software, algorithm | Prism7 | GraphPad Software Inc |
Additional files
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57593/elife-57593-transrepform1-v2.docx
-
Source code 1
Source data files for gels and blots displayed in Figures 1-7 & figure supplements.
- https://cdn.elifesciences.org/articles/57593/elife-57593-code1-v2.zip





