Macrophages promote endothelial-to-mesenchymal transition via MT1-MMP/TGFβ1 after myocardial infarction
Figures
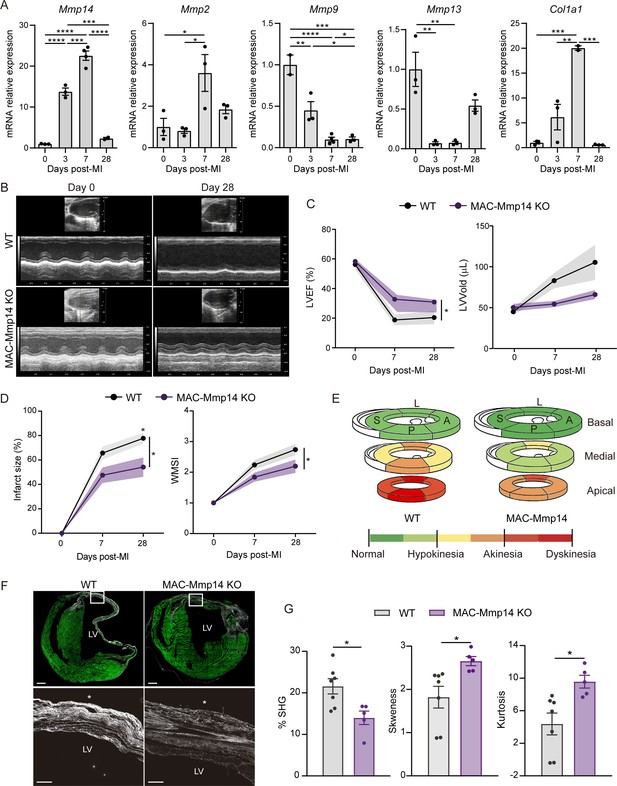
Mφ-restricted MT1-MMP deficiency attenuates LV dysfunction and dilation and reduces collagen deposition after MI.
(A) mRNA expression levels of genes related to ECM-remodeling assessed by qPCR in sorted Mφs at the indicated post-MI stages. Data are means ± SEM of three independent pools of 3–5 mice per time point. One-way ANOVA followed by Tukey's multiple comparisons test. (B) Representative LV M-mode long-axis echocardiography views at end-diastole on day 0 and day 28 post-MI in WT and MAC-Mmp14 KO mice. (C, D) Post-MI progression of LVEF and LVVold (C) and infarct size (percentage of LV with contractility alterations) and WMSI (D) assessed by echocardiography. Data are means ± SEM of 9–10 mice per genotype. Two-way ANOVA followed by Tukey's multiple comparisons test. (E) Quantitative assessment of LV contractility at 28 days post-MI, showing mean scores for every LV segment at the basal, medial, and apical levels throughout all samples. L, lateral; A, anterior; P, posterior; S, septal. Segment scores are colored-coded from green to red: green = normal, yellow = hypokinesia, orange = akinesia, and red = dyskinesia or aneurysm. (F) Representative SHG (white) and MPEF (green) microscopy images of transverse cardiac sections at 28 days post-MI. Scale bar, 500 µm. Magnified views of boxed areas within the infarct are shown in the lower panels. Scale bar, 100 µm. Asterisks mark the epicardium. (G) Percentage SHG, skewness, and kurtosis in infarcts at 28 days post-MI. Data are means ± SEM of 5–7 mice per genotype. Unpaired t-test.
-
Figure 1—source data 1
Mφ-restricted MT1-MMP deficiency attenuates LV dysfunction and dilation and reduces collagen deposition after MI.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig1-data1-v3.xlsx
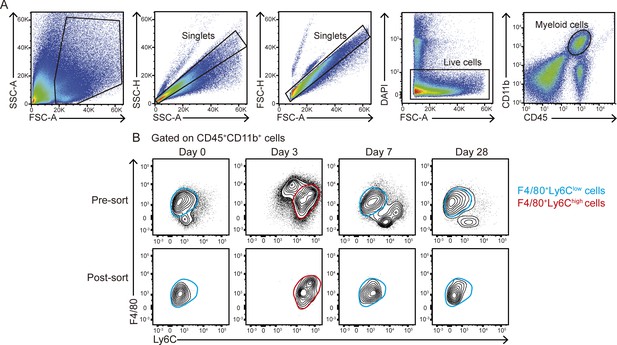
Isolation of cardiac Mφs after MI.
(A) FACS gating strategy to purify cardiac Mφs from C57BL/6 mice. (B) From the myeloid cell population (CD45+CD11b+ cells), F4/80+Ly6Clow cells (blue) were isolated at 0, 7, and 28 days post-MI, and F4/80+/Ly6Chigh cells (red) were purified at 3 days post-MI. Representative post-sort plots of Mφs are shown in the lower panel. Sorting was performed with a pool of 3–5 mice per time point.
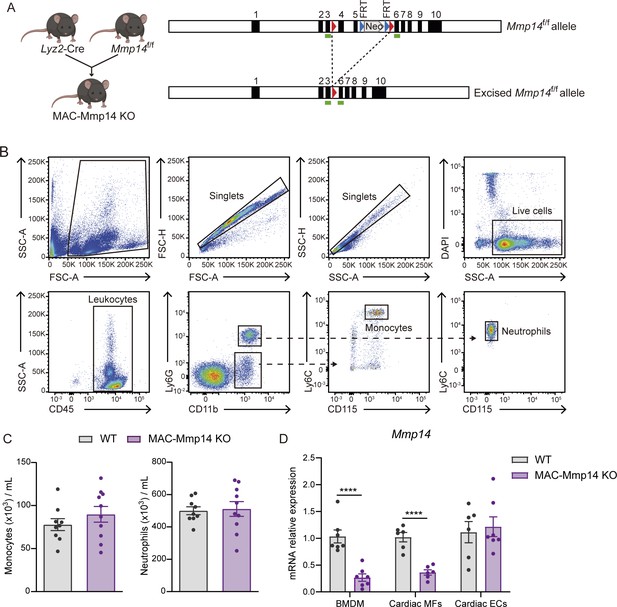
Mouse model of Mφ-specific MT1-MMP inactivation.
(A) Strategy for generating the transgenic mouse line used for Mφ inactivation of Mmp14. LoxP sites (red arrowheads) were introduced flanking exons 4 and 5, and an FRT-PGK-Neo-FRT-cassette was inserted between exons 5 and 6 to generate the Mmp14f/f construct (top, right). To obtain Mφ inactivation of Mmp14, Mmp14f/f mice were crossed with Lyz2-Cre mice, yielding the floxed Mmp14 allele (bottom, right) in MAC-Mmp14 KO mice. Green rectangles indicate alignment position for genotyping primers. (B) Representative plots showing the gating strategy for the identification of circulating monocytes (CD45+CD11b+Ly6G-CD115+Ly6Chigh cells) and neutrophils (CD45+CD11b+Ly6G+CD115-Ly6Clow cells) in peripheral blood from WT and MAC-Mmp14 KO mice. (C) Total numbers of baseline circulating monocytes and neutrophils as depicted in B. Data are means ± SEM of 9–10 mice per genotype. (D) qPCR analysis of Mmp14 inactivation efficiency in bone marrow-derived Mφs (BMDMs) and FACS-sorted 7-day-post-MI cardiac Mφs (CD45+CD11b+F4/80+Ly6Clow cells) and ECs (CD45-CD11b-CD31+ cells) from WT and MAC-Mmp14 KO mice. Data are means ± SEM of 7 mice per genotype (for BMDMs) or 6–7 independent pools of two mice per genotype (for cardiac Mφs and cardiac ECs). Unpaired t-test.
-
Figure 1—figure supplement 2—source data 1
Mouse model of Mφ-specific MT1-MMP inactivation.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig1-figsupp2-data1-v3.xlsx
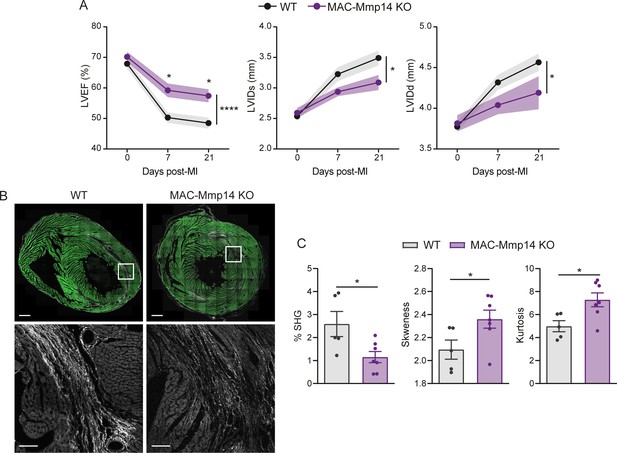
The inactivation of Mφ MT1-MMP ameliorates cardiac dysfunction and reduces collagen deposition in a model of transient ischemia.
(A) Progression of LVEF, LVIDs, and LVIDd in WT and MAC-Mmp14 KO mice after ischemia/reperfusion (I/R). Data are means ± SEM of 14 mice per genotype. Two-way ANOVA followed by Tukey's multiple comparisons test. (B) Representative SHG (white) and MPEF (green) microscopy images of transverse cardiac sections from WT and MAC-Mmp14 KO mice at 21 days after I/R. Scale bar, 1 mm. Magnified views of boxed areas in the infarct are shown in the lower panels. Scale bar, 100 µm. (C) Percentage SHG, skewness, and kurtosis in infarcts at 21 days post-I/R. Data are means ± SEM of 5–7 mice per genotype. Unpaired t-test.
-
Figure 1—figure supplement 3—source data 1
The inactivation of Mφ MT1-MMP ameliorates cardiac dysfunction and reduces collagen deposition in a model of transient ischemia.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig1-figsupp3-data1-v3.xlsx
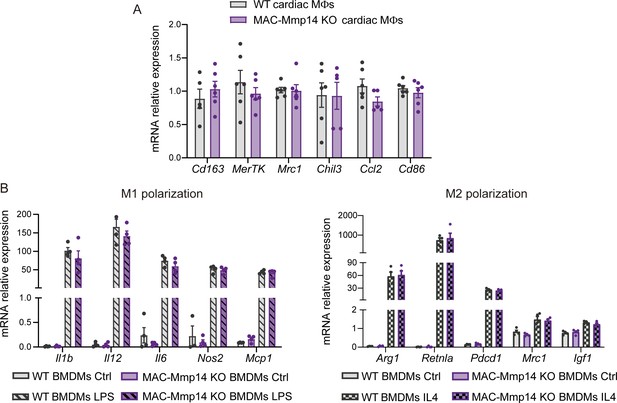
MT1-MMP inactivation does not alter the Mφ phenotype following MI.
(A) mRNA expression levels of anti- and pro-inflammatory genes assessed by qPCR in FACS-sorted 7-day-post-MI cardiac Mφs (CD45+CD11b+F4/80+Ly6Clow cells) from WT and MAC-Mmp14 KO mice. Data are means ± SEM of six independent pools of two mice per genotype. Unpaired t-test. (B) mRNA expression levels of M1 genes in WT and MAC-Mmp14 KO BMDMs activated with LPS (left) and M2 genes in BMDMs activated with IL4 (right). Data are means ± SEM of four mice per condition. Two-way ANOVA followed by Tukey's multiple comparisons test.
-
Figure 1—figure supplement 4—source data 1
MT1-MMP inactivation does not alter the Mφ phenotype following MI.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig1-figsupp4-data1-v3.xlsx
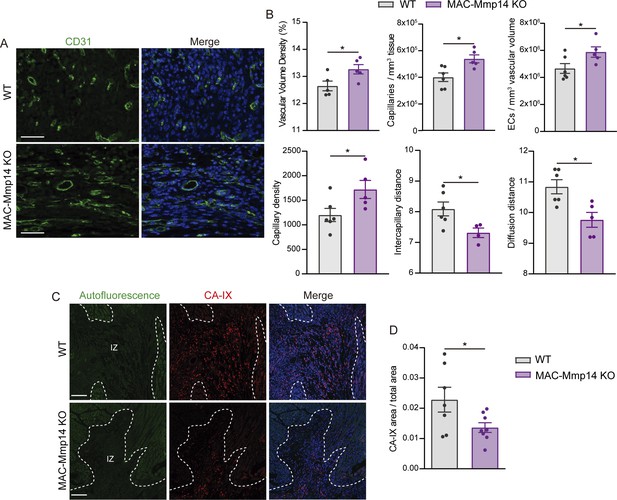
MAC-Mmp14 KO mice have a preserved microvasculature network and better myocardial oxygenation after ischemic injury.
(A) Representative confocal microscopy images showing immunostaining for CD31 (green) and nuclei (blue) within the infarction in WT and MAC-Mmp14 KO hearts at 7 days post-MI. Scale bar, 50 µm. (B) Vasculature-related parameters within the infarction at 7 days post-MI. Data are means ± SEM of 5–6 mice per genotype. Unpaired t-test. (C) Representative confocal immunofluorescence microscopy images of CA-IX (red) in the infarcted region of WT and MAC-Mmp14 KO hearts at 7 days post-MI. Nuclei are stained with DAPI (blue). Scale bar, 100 µm. (D) CA-IX+ area:total area ratio in the infarcted zone. Data are means ± SEM of 7–8 mice per genotype. Unpaired t-test.
-
Figure 2—source data 1
MAC-Mmp14 KO mice have a preserved microvasculature network and better myocardial oxygenation after ischemic injury.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig2-data1-v3.xlsx
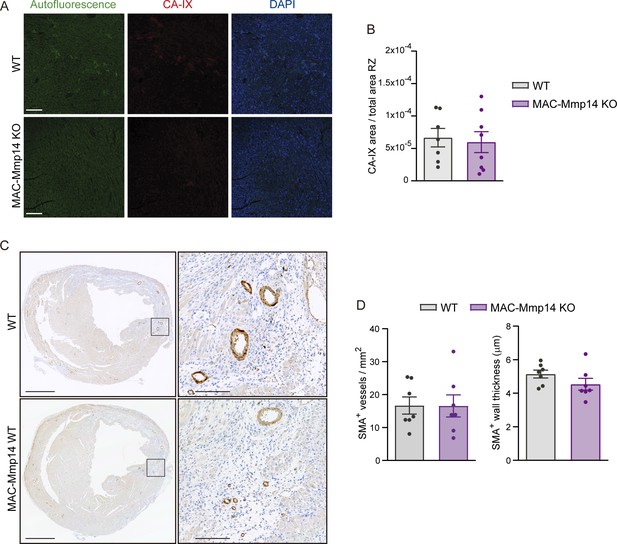
Remodeling of cardiac vasculature in MAC-Mmp14 KO mice after MI.
(A) Representative confocal immunofluorescence microscopy images of CA-IX (red) in WT and MAC-Mmp14 KO hearts at 7 days post-MI in the remote zone (RZ). Nuclei are stained with DAPI (blue). Scale bar, 100 µm. (B) CA-IX+ area:total area ratio in the RZ. Data are means ± SEM of 7–8 mice per genotype. Unpaired t-test. (C) Representative images of SMA-stained transverse sections of WT and MAC-Mmp14 KO hearts at 7 days post-MI. Scale bar, 1 mm. Magnified views of boxed areas in the infarct are shown on the right. Scale bar, 100 µm. (D) SMA+ vessel density and SMA+ vessel wall thickness in the IZ at 7 days post-MI. Data are means ± SEM of 5–7 per group. Unpaired t-test.
-
Figure 2—figure supplement 1—source data 1
Remodeling of cardiac vasculature in MAC-Mmp14 KO mice after MI.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig2-figsupp1-data1-v3.xlsx
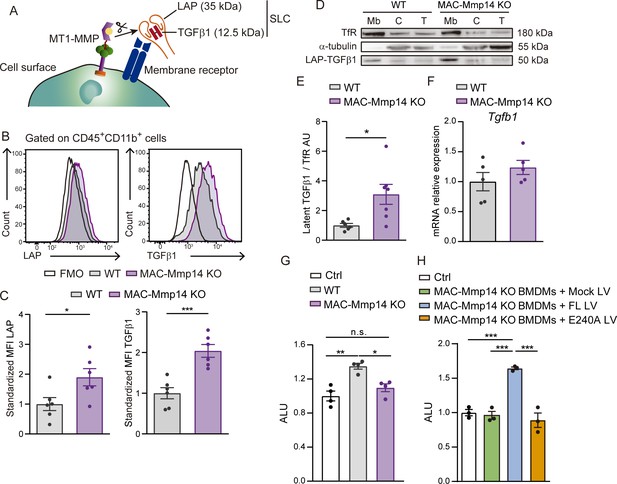
Mφ-inactivation of MT1-MMP impairs active TGFβ1 release from LAP-TGFβ1 complex.
(A) Scheme of LAP-TGFβ1 complex retention in the cell surface through LAP-binding to membrane receptors. (B) Representative flow cytometry histogram plots of LAP and TGFβ1 staining in LPS-activated WT and MAC-Mmp14 KO BMDMs. (C) Standardized mean fluorescence intensity (MFI) of LAP and TGFβ1 in experiments as in B. Data are means ± SEM of seven mice per group. Unpaired t-test. (D) Western blot of transferrin receptor (TfR), α-tubulin, and LAP-TGFβ1 complex in membrane fraction (Mb), cytosolic fraction (C), and total lysate (T) from LPS-activated WT and MAC-Mmp14 KO BMDMs. (E) Quantification of LAP-TGFβ1 complex in the membrane fraction. Data are means ± SEM of 6–7 mice per genotype. Unpaired t-test. (F) Tgfb1 mRNA expression in LPS-activated WT and MAC-Mmp14 KO BMDMs. Data are means ± SEM of five mice per genotype. Unpaired t-test. (G) Arbitrary luciferase units (ALU) in HEK293 cells co-cultured with or without LPS-activated WT or MAC-Mmp14 KO BMDMs. Control corresponds to transfected HEK293 cells cultured alone. Data are means ± SEM of a representative experiment of three performed with four technical replicates per condition. One-way ANOVA followed by Tukey's multiple comparisons test. (G) ALU in HEK293 cells co-cultured with or without conditioned media from LPS-activated MAC-Mmp14 KO BMDMs transduced with mock lentivirus (GFP), or lentivirus containing full-length MT1-MMP (FL) or catalytic MT1-MMP mutant (E240A). Control corresponds to transfected HEK293 cells cultured alone. Data are means ± SEM of a representative experiment of three performed with three technical replicates per condition. One-way ANOVA followed by Tukey's multiple comparisons test.
-
Figure 3—source data 1
Mφ-inactivation of MT1-MMP impairs active TGFβ1 release from the LAP-TGFβ1 complex.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig3-data1-v3.xlsx
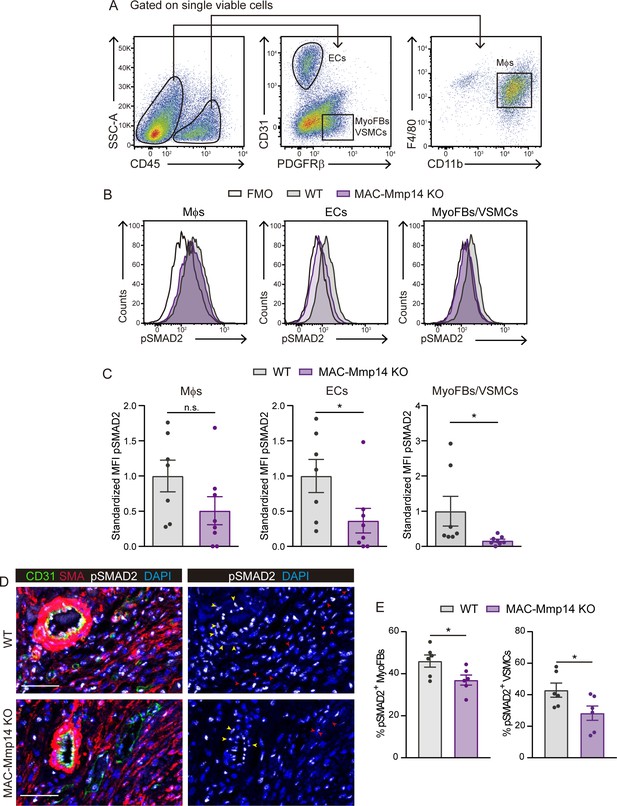
The inactivation of Mφ MT1-MMP reduces TGFβ1-pSMAD2 signaling in cardiac ECs, MyoFBs, and VSMCs after MI.
(A) Gating strategy used to assess pSMAD2 signaling in ECs, MyoFBs/VSMCs, and Mφs. (B) Representative flow cytometry histogram plots of pSMAD2 staining in the indicated cells from 7-day-post-MI hearts. (C) Standardized MFI of pSMAD2 in experiments as in B. Data are means ± SEM of 7–8 mice per genotype. Unpaired t-test. (D) Representative immunofluorescence staining of CD31 (green), SMA (red), and pSMAD2 (white) in infarcted cardiac tissue from WT mice (top) and MAC-Mmp14 KO mice (bottom) at 7 days post-MI. Nuclei are stained with DAPI (blue). Red and yellow arrowheads point to pSMAD2+ MyoFBs and pSMAD2+ VSMCs, respectively. Scale bar, 50 µm. (E) Percentages of pSMAD2+ MyoFBs and pSMAD2+ VSMCs within the total MyoFB or VSMC populations, respectively in the infarcted zone. Data are means ± SEM of six mice per genotype. Unpaired t-test.
-
Figure 4—source data 1
The inactivation of Mφ MT1-MMP reduces TGFβ1-pSMAD2 signaling in cardiac ECs, MyoFBs, and VSMCs after MI.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig4-data1-v3.xlsx
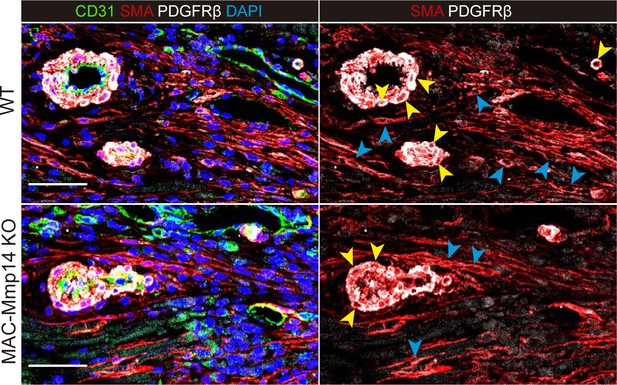
PDGFRβ is expressed by cardiac MyoFBs and VSMCs.
Representative immunofluorescence staining of CD31 (green), SMA (red), and PDGFRβ (white) in infarcted cardiac tissue from WT mice (top) and MAC-Mmp14 KO mice (bottom) at 7 days post-MI. Nuclei are stained with DAPI (blue). Blue and yellow arrowheads point to PDGFRβ+SMA+ MyoFBs and PDGFRβ+SMA+ pericytes, respectively. Scale bar, 50 µm.
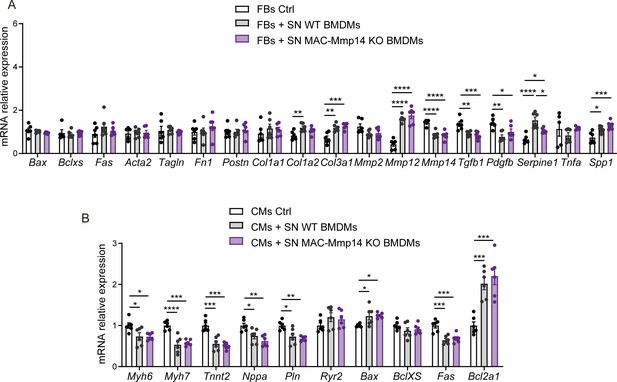
Effect of WT or MAC-Mmp14 KO Mφs on FBs and CMs.
(A) qPCR analysis of fibrosis-, apoptosis-, and FB activation-related genes in FBs treated with LPS-activated WT or MAC-Mmp14 KO BMDM supernatants (SN). (B) qPCR analysis of CM- and apoptosis-related genes in CMs treated with LPS-activated WT or MAC-Mmp14 KO BMDM SNs. Data are means ± SEM of a representative experiment of two performed. One-way ANOVA.
-
Figure 4—figure supplement 2—source data 1
Effect of WT or MAC-Mmp14 KO Mφs on FBs and CMs.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig4-figsupp2-data1-v3.xlsx
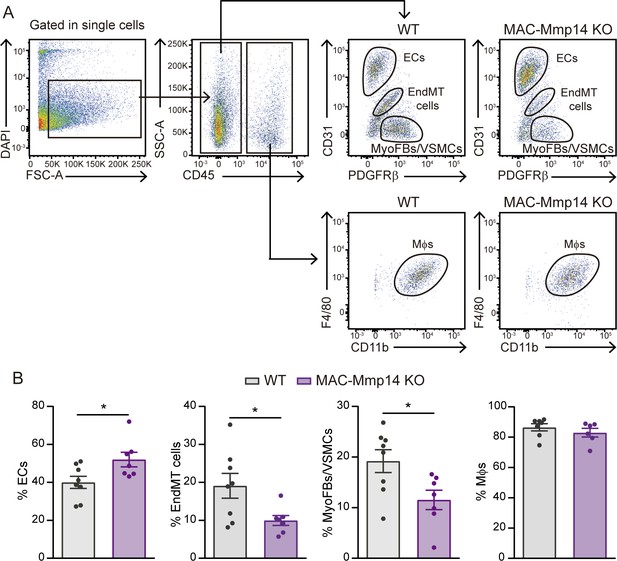
The inactivation of Mφ MT1-MMP alters myocardial cellular composition after MI.
(A) Flow cytometry gating strategy used to identify and quantify cardiac ECs, MyoFBs, cells undergoing EndMT, and Mφs in WT mice (left) and MAC-Mmp14 KO mice (right) on day 7 after MI. (B) Quantification of Mφs, ECs, MyoFBs, and cells undergoing EndMT in cardiac tissue 7 days after MI. Data are means ± SEM of at least 11 mice per genotype. Unpaired t-test.
-
Figure 5—source data 1
The inactivation of Mφ MT1-MMP alters myocardial cellular composition after MI.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig5-data1-v3.xlsx
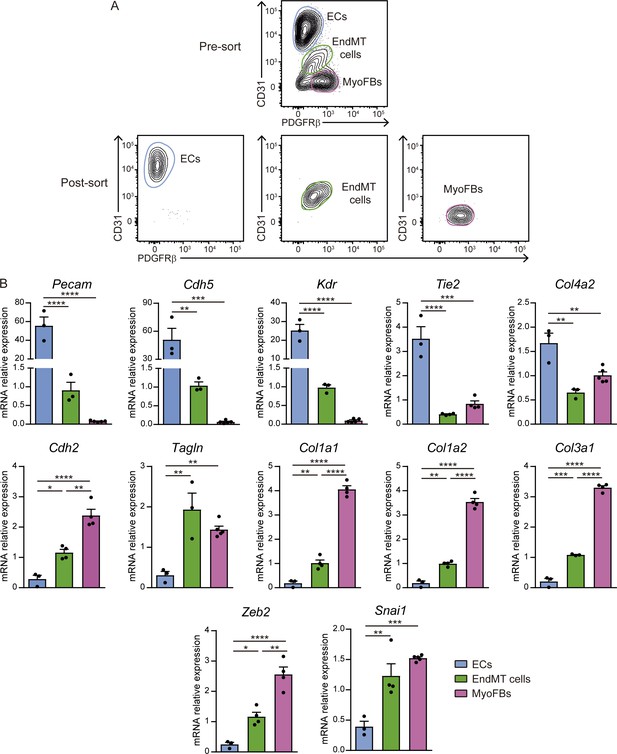
Endothelial-to-mesenchymal gene signature of CD31+PDGFRβ+ cells.
(A) FACS gating strategy to purify cardiac ECs, EndMT cells, and MyoFBs/VSMCs from 7 days post-MI WT hearts. Representative post-sort plots of Mφs are shown in the lower panel. Sorting was performed with a pool of two mice per time point. (B) qPCR analysis of ECs, EndMT cells, and MyoFBs/VSMCs for endothelial genes (Pecam, Cdh5, Kdr, Col4a1, Col4a2) and FB genes (Cdh2, Tagln, Col1a1, Col1a2, Col3a1), and EndMT-mediating transcription factors (Zeb2 and Snai1). Data are means ± SEM of at least three independent biological replicates per group. Two-way ANOVA followed by Tukey's multiple comparisons test.
-
Figure 5—figure supplement 1—source data 1
Endothelial-to-mesenchymal gene signature of CD31+PDGFRβ+ cells.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig5-figsupp1-data1-v3.xlsx
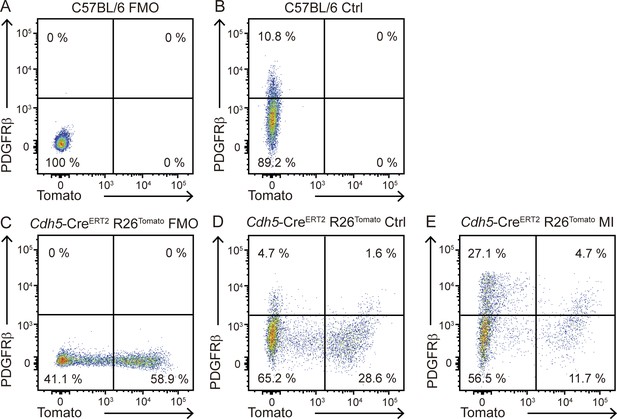
Lineage tracing of endothelial derived-mesenchymal cells.
(A–E) Representative flow cytometry plots showing expression of Tomato and PDGFRβ in CD45- cardiac cells in the indicated mice. (A) C57BL/6 FMO sample for PDGFRβ; (B) C57BL/6 sample stained with anti-PDGFRβ antibody; (C) Cdh5-CreERT2 R26Tomato FMO sample for PDGFRβ; (D, E) Cdh5-CreERT2 R26Tomato sample stained with anti-PDGFRβ antibody in control (D) or 7 day-post-MI hearts (E). Numbers indicate the percentage of cells within the CD45- cell population. Representative plots of n = 2 hearts per condition.
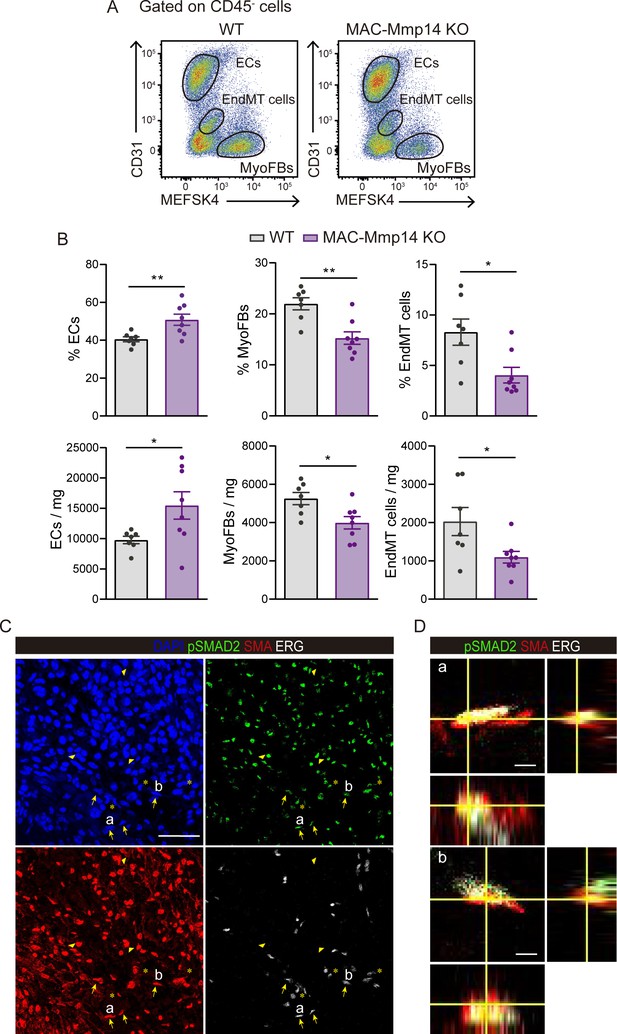
The inactivation of Mφ MT1-MMP attenuates post-MI EndMT.
(A) Complementary flow cytometry gating strategy used to identify and quantify cardiac ECs, MyoFBs/VSMCs, and cells undergoing EndMT in WT mice (left) and MAC-Mmp14 KO mice (right) on day 7 after MI. (B) Quantification of ECs, MyoFBs, and cells undergoing EndMT in cardiac tissue 7 days after MI. Data are means ± SEM of 7–8 mice per genotype. Unpaired t-test. (C) Representative confocal immunofluorescence microscopy images of pSMAD2 (green), SMA (red), ERG (white), and DAPI (blue) of the infarcted area of WT hearts at 7 days post-MI. Arrows indicate ERG+/SMA+/pSMAD2+ cells, asterisks indicate ERG+/SMA-/pSMAD2+ cells, and arrowheads indicate ERG-/SMA+/pSMAD2+ cells. Scale bar, 50 µm. (D) Triple positive cells named ‘a’ and ‘b’ are shown magnified with their orthogonal views in the boxes to the right. Scale bar, 5 µm.
-
Figure 5—figure supplement 3—source data 1
The inactivation of Mφ MT1-MMP attenuates post-MI EndMT.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig5-figsupp3-data1-v3.xlsx
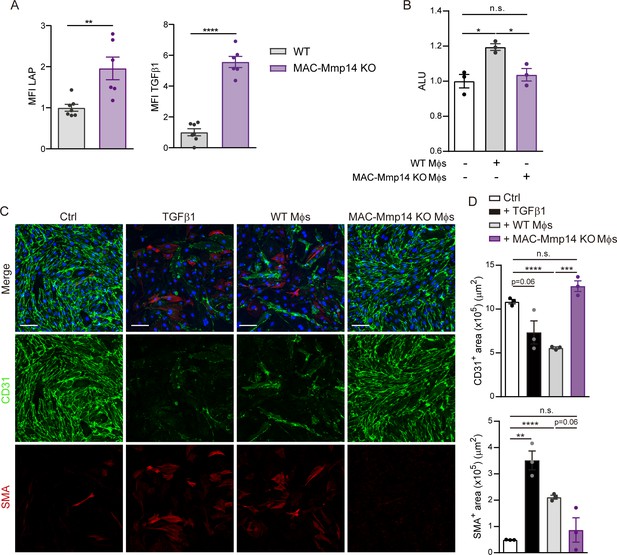
Cardiac Mφs induce post-MI EndMT through MT1-MMP-mediated TGFβ1 activation.
(A) Standardized MFI of LAP and TGFβ1 staining in WT and MAC-Mmp14 KO cardiac Mφs on day 7 after MI. Data are means ± SEM of 6–7 mice per group. Unpaired t-test. (B) Luciferase activity (ALU) in transfected HEK293 cells co-cultured with Mφs from 7-day-post-MI WT or MAC-Mmp14 KO hearts. Data are means ± SEM of three independent experiments performed with four technical replicates per condition. One-way ANOVA followed by Tukey's multiple comparisons test. (C) Representative immunofluorescence staining of CD31 (green) and SMA (red) in in vitro co-cultures of MAECs and cardiac Mφs from WT or MAC-Mmp14 KO 7-day-post-MI hearts. Nuclei are stained with DAPI (blue). Scale bar, 100 µm (D) CD31+ area (µm2) and SMA+ area (µm2) in the different conditions. Data are means ± SEM of a representative experiment of three performed with three technical replicates per condition. Unpaired t-test.
-
Figure 6—source data 1
Cardiac Mφs induce post-MI EndMT through MT1-MMP-mediated TGFβ1 activation.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig6-data1-v3.xlsx
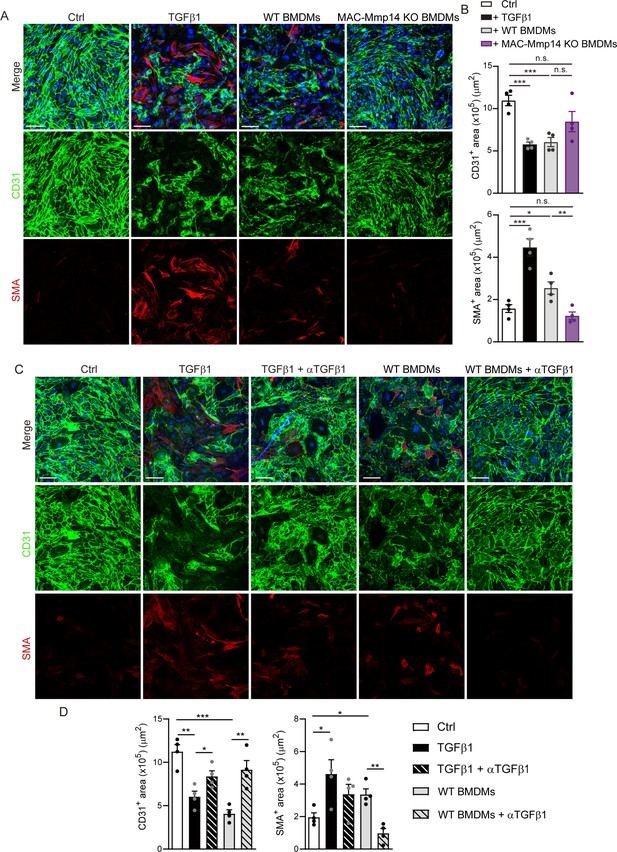
Mφs induce EndMT in vitro via MT1-MMP/TGFβ1.
(A) Representative immunofluorescence staining of CD31 (green) and SMA (red) in in vitro co-cultures of MAECs and LPS-activated WT or MAC-Mmp14 KO BMDMs. Nuclei are stained with DAPI (blue). MAECs were also treated with TGFβ1 (10 ng/mL) as an EndMT positive control. Scale bar, 100 µm (B) CD31+ area (µm2) and SMA+ area (µm2) in the different conditions. Data are means ± SEM of four independent experiments. Unpaired t-test. (C) Representative immunofluorescence staining of CD31 (green) and SMA (red) in in vitro co-cultures of MAECs and LPS-activated WT BMDMs. Nuclei are stained with DAPI (blue). MAECs were also treated with TGFβ1 (10 ng/mL) and/or neutralizing αTGFβ1 antibody (100 µg/mL). Scale bar, 100 µm (D) CD31+ area (µm2) and SMA+ area (µm2) in the different conditions. Data are means ± SEM of four independent experiments. Unpaired t-test.
-
Figure 6—figure supplement 1—source data 1
Mφs induce EndMT in vitro via MT1-MMP/TGFβ1.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig6-figsupp1-data1-v3.xlsx
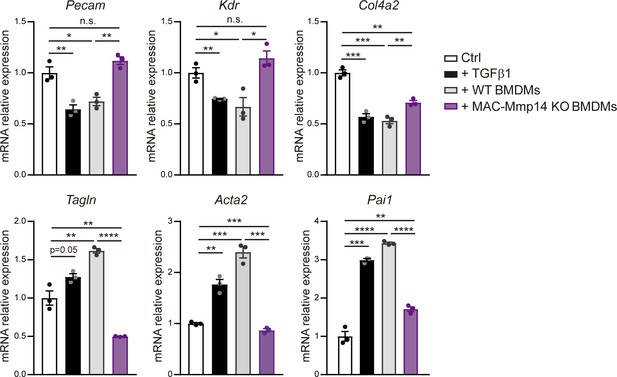
MT1-MMP is required for in vitro Mφ induction of EndMT.
qPCR analysis of endothelial and myoFB markers and TGFβ1 target genes in in vitro co-cultures of MAECs and LPS-activated WT or MAC-Mmp14 KO BMDMs. Data are means ± SEM of a representative experiment of two performed with three technical replicates per condition. Unpaired t-test.
-
Figure 6—figure supplement 2—source data 1
MT1-MMP is required for in vitro Mφ induction of EndMT.
- https://cdn.elifesciences.org/articles/57920/elife-57920-fig6-figsupp2-data1-v3.xlsx
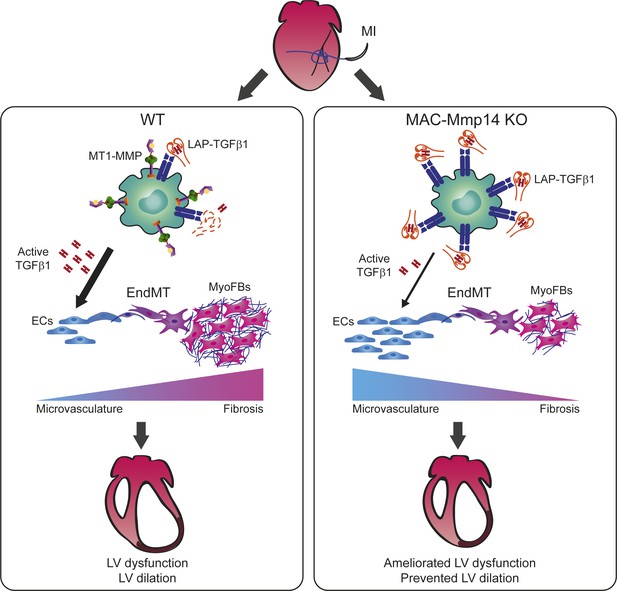
Mφ-inactivation of MT1-MMP preserves cardiac function after MI by impairing TGFβ1-mediated EndMT.
MI triggers MT1-MMP production by Mφs, contributing to the release of active TGFβ1 from SLC (LAP-TGFβ1) to the myocardium. Active TGFβ1 signals acting on ECs promote EndMT, contributing to adverse tissue remodeling. When Mφ MT1-MMP is absent, latent TGFβ1 accumulates, and the availability of active TGFβ1 in the myocardium decreases. In this scenario, the impairment of Mφ-mediated EndMT results in enhanced angiogenesis and reduced fibrosis, limiting LV remodeling, and preserving cardiac function.
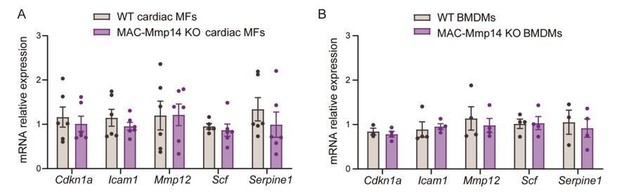
(A) mRNA expression levels of genes related to senescence-associated secretory phenotype (SASP) by qPCR in FACS-sorted 7-day-post-MI cardiac Mφs (CD45+CD11b+F4/80+Ly6Clow cells) from WT and MAC-Mmp14 KO mice.
Data are means ± SEM of 4 mice per genotype. Unpaired t-test. (B) mRNA expression levels of genes related to SASP by qPCR in BMDMs from WT and MAC-Mmp14 KO mice. Data are means ± SEM of 4 mice per genotype. Unpaired t-test.
Videos
Parasternal 2D long axis echocardiography view of WT (top) or MAC-Mmp14 KO (bottom) hearts at baseline (Day 0, left) and at 28 days post-MI (right) induced by LAD-ligation.
Tables
Mφ MT1-MMP inactivation does not affect homeostatic cardiac function.
Echocardiography and electrocardiography comparisons between 10-week-old WT and MAC-Mmp14 KO mice. Data are means ± SEM of 10 mice per group. Unpaired t-test. BW, body weight; FS, fraction shortening; HR, heart rate; HW, heart weight; LVEF, LV ejection fraction; LVIDd, LV end-diastolic internal diameter; LVIDs, LV end-systolic internal diameter; LVVold, LV end-diastolic volume; LVVols, LV end-systolic volume.
| WT | MAC-Mmp14 KO | |
|---|---|---|
| BW (g) | 18.62 ± 2.00 | 19.53 ± 2.25 |
| HW/BW (mg/g) | 4.91 ± 0.28 | 4.94 ± 0.19 |
| HR (beats/min) | 462 ± 14 | 463 ± 17 |
| PR (ms) | 39.82 ± 1.18 | 37.70 ± 1.21 |
| QRS (ms) | 25.41 ± 1.06 | 26.18 ± 0.88 |
| LVEF (%) | 53.52 ± 1.72 | 53.14 ± 2.31 |
| LVVols (µL) | 21.53 ± 1.36 | 19.98 ± 1.60 |
| LVVold (µL) | 25.26 ± 0.96 | 22.22 ± 0.90 |
| FS (%) | 26.58 ± 0.89 | 26.33 ± 1.51 |
| LVIDs (mm) | 3.67 ± 0.06 | 3.54 ± 0.08 |
| LVIDd (mm) | 2.7 ± 0.06 | 2.62 ± 0.10 |
Mφ MT1-MMP inactivation does not affect circulating bone marrow-derived populations.
Hematograms from 10-week-old WT and MAC-Mmp14 KO mice. Data are means ± SEM of eight mice per group.
| WT | MAC-Mmp14 KO | |||
|---|---|---|---|---|
| % | Cells (×103)/mL | % | Cells (×103)/mL | |
| Neutrophils | 9.50 ± 0.98 | 0.74 ± 0.11 | 11.01 ± 1.43 | 0.88 ± 0.13 |
| Lymphocytes | 86.75 ± 1.29 | 6.97 ± 1.00 | 84.79 ± 1.68 | 6.84 ± 0.54 |
| Monocytes | 0.89 ± 0.14 | 0.08 ± 0.02 | 1.20 ± 0.13 | 0.10 ± 0.01 |
| Eosinophils | 2.45 ± 0.44 | 0.21 ± 0.06 | 2.48 ± 0.51 | 0.20 ± 0.04 |
| Basophils | 0.41 ± 0.08 | 0.03 ± 0.01 | 0.53 ± 0.09 | 0.05 ± 0.01 |
Quantitative analysis of microvasculature parameters in infarcted cardiac tissue from WT and MAC-Mmp14 KO mice on day 7 after MI.
Capillaries correspond to CD31+SMA- vessels of diameter <3 µm. Data are means ± SEM of 5–6 mice per genotype. Unpaired t-test. Significant differences are indicated as *p<0.05.
| Minkowski-based metrics | WT | MAC-Mmp14 KO |
|---|---|---|
| Vascular volume density (%) | 12.65 ± 0.36 | 13.26 ± 0.34 * |
| Vascular surface area density (× 10−3) (µm2/µm3) | 21.24 ± 2.5 | 23.13 ± 1.92 |
| Graph-based metrics | ||
| Vascular segment length (µm) | 6.97 ± 0.74 | 6.2 ± 0.18 |
| Vascular segment surface (µm2) | 36.41 ± 5.52 | 29.5 ± 1.6 |
| Vascular segment volume (µm3) | 17 ± 3.8 | 12.54 ± 1.53 |
| Tortuosity (µm/µm) | 1.61 ± 0.02 | 1.61 ± 0.03 |
| Vascular segments (× 105)a | 4.12 ± 0.68 | 5.48 ± 0.53 |
| Vascular segmentsb | 144.17 ± 14.15 | 158.27 ± 2.31 |
| Vessels of diameter <= 3 (µm) (%) | 95.48 ± 2.55 | 96.97 ± 1.85 * |
| Vessels of diameter between 3 and 6 (µm) (%) | 4.51 ± 2.52 | 3.03 ± 1.85 |
| Vessels of diameter > 6 (µm) (%) | 0.012 ± 0.001 | 0.028 ± 0.001 |
| Vessels of diameter <= 3 (µm) (× 105)a | 3.97 ± 0.71 | 5.33 ± 0.59 * |
| Vessels of diameter between 3 and 6 (µm) (× 105)a | 0.16 ± 0.06 | 0.15 ± 0.09 |
| Vessels of diameter > 6 (µm)a | 32.81 ± 0.0001 | 73.36 ± 0.0001 |
| Branching nodes (× 104)a | 22.39 ± 4.28 | 30.43 ± 2.51 |
| Blind-ends/sprouts (× 104)a | 6.06 ± 0.92 | 7.93 ± 1.2 |
| Branching nodesb | 77.38 ± 9.59 | 87.31 ± 2.14 |
| Blind-ends/sproutsb | 24.09 ± 4.07 | 25.9 ± 1.66 |
| SMA-related metrics | ||
| Vessels covered with SMA (%) | 47.2 ± 17.67 | 53.54 ± 12 |
| SMA+ layer thikness (µm) | 2.98 ± 0.76 | 2.72 ± 0.43 |
| Damage index | 0.21 ± 0.11 | 0.19 ± 0.09 |
| Myofibroblasts (× 104)a | 2.51 ± 1.18 | 2.05 ± 0.64 |
| Myofibroblastsb | 8.9 ± 3.8 | 5.97 ± 1.47 |
| Myofibroblasts (× 105)d | 19.1 ± 10 | 15.3 ± 4.7 |
| SMA+ perivascular cells (× 104)a | 4.74 ± 2.36 | 5.25 ± 1.59 |
| SMA+ perivascular cellsb | 16.16 ± 7.31 | 15.48 ± 3.65 |
| SMA+ perivascular cells (× 105)d | 36.3 ± 19.3 | 38.9 ± 10.8 |
| Efficiency in oxygen diffusion | ||
| Maximal extravascular distance (µm) | 51.47 ± 10.83 | 45.47 ± 7.11 |
| Median extravascular distance (µm) | 14.28 ± 2.64 | 13 ± 1.38 |
| Capillary densityc | 1201 ± 298.12 | 1720 ± 368.61 * |
| Intercapillary distance | 8.09 ± 0.51 | 7.31 ± 0.27 * |
| Diffusion distance | 10.84 ± 0.51 | 9.76 ± 0.49 * |
| Additional cell-related metrics | ||
| Endothelial cells (× 104)a | 6.2 ± 1.05 | 7.91 ± 1.21 |
| Endothelial cellsb | 22.45 ± 3.91 | 23.16 ± 1.34 |
| Endothelial cells (× 105)d | 46.6 ± 8.1 | 58.8 ± 7.7 * |
-
a per mm3 of tissue,b per mm vessel length,c per mm2 of tissue,d per mm3 vessel volume.
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6 | Charles Rivers | ||
| Genetic reagent (Mus musculus) | Mmp14f/f | Gutiérrez-Fernández et al., 2015 | MGI:5694577 | Dr. Carlos López-Otín |
| Genetic reagent (Mus musculus) | Lyz2-Cre | Clausen et al., 1999 | MGI:1934631 | |
| Genetic reagent (Mus musculus) | Cdh5-CreERT2 | Sörensen et al., 2009 | MGI:3848982 | |
| Genetic reagent (Mus musculus) | R26TdTomato | Madisen et al., 2010 | MGI:3809524 | |
| Cell line (Homo sapiens) | HEK293 | Sigma | ||
| Antibody | Anti-CD16/CD32 (rat monoclonal) | BD Biosciences | 553141 | (1:100) |
| Antibody | Anti-CD45 (rat monoclonal) | Biolegend | 103132 | (1:100) |
| Antibody | Anti-CD45 (rat monoclonal) | Biolegend | 103116 | (1:100) |
| Antibody | Anti-CD45 (rat monoclonal) | eBioscience | 48–0451 | (1:100) |
| Antibody | Anti-CD11b (rat monoclonal) | BD Biosciences | 552850 | (1:100) |
| Antibody | Anti-CD11b (rat monoclonal) | BD Biosciences | 557395 | (1:100) |
| Antibody | Anti-CD11b (rat monoclonal) | Biolegend | 101206 | (1:100) |
| Antibody | Anti-Ly6C (rat monoclonal) | BD Biosciences | 560595 | (2:100) |
| Antibody | Anti-Ly6C (rat monoclonal) | BD Biosciences | 553104 | (1:100) |
| Antibody | Anti-CD31 (rat monoclonal) | BD Biosciences | 551262 | (1:100) |
| Antibody | Anti-CD31 (rat monoclonal) | BD Biosciences | 553372 | (1:100) |
| Antibody | Anti-PDGFR-β (rat monoclonal) | BioLegend | 136005 | (2:100) |
| Antibody | Anti-PDGFR-β (rat monoclonal) | BioLegend | 136007 | (2:100) |
| Antibody | Anti-Feeder Cells (MEFSK4) (rat monoclonal) | Miltenyi | 130-120-166 | (2:100) |
| Antibody | Anti-Feeder Cells (MEFSK4) (rat monoclonal) | Miltenyi | 130-120-802 | (2:100) |
| Antibody | Anti-F4/80 (rat monoclonal) | BioLegend | 123114 | (3:100) |
| Antibody | Anti-F4/80 (rat monoclonal) | BioLegend | 123110 | (3:100) |
| Antibody | Anti-Phospho-Smad2 (rabbit polyclonal) | Cell Signaling | 3104 | (1:100) |
| Antibody | Streptavidin-Alexa 488 conjugate | ThermoFisher Scientific | S11223 | (1:500) |
| Antibody | Anti-LAP (mouse monoclonal) | Biolegend | 141405 | (3:100) |
| Antibody | Anti-TGFβ1 (rabbit polyclonal) | Abcam | ab92486 | (1:100) |
| Antibody | anti-rabbit IgG (chicken polyclonal) | ThermoFisher | A-21441 | (1:500) |
| Antibody | anti- rabbit IgG (goat polyclonal) | ThermoFisher | A-21245 | (1:500) |
| Antibody | anti-CAI-IX (rabbit polyclonal) | Abcam | ab15086 | (1:100) |
| Antibody | anti-rabbit IgG (goat polyclonal) | ThermoFisher | A-11035 | (1:500) |
| Antibody | anti-CD31 (rat monoclonal) | Dianova | DIA-310 | (1:200) |
| Antibody | anti-Rat IgG (goat polyclonal) | ThermoFisher | A-11006 | (1:500) |
| Antibody | anti-pSMAD2 (rabbit monoclonal) | Cell Signaling | 3108 | (1:100) |
| Antibody | anti-SMA (mouse monoclonal) | Sigma-Aldrich | C6198 | (1:400) |
| Antibody | anti-ERG (rabbit monoclonal) | Abcam | ab196149 | (1:100) |
| Antibody | Anti-PDGFRβ (rat monoclonal) | Thermofisher | 14-1402-82 | (1:100) |
| Antibody | Anti-rat IgG (goat polyclonal) | Thermofisher | A-11006 | (1:500) |
| Antibody | Anti-TGFβ1 (rabbit polyclonal) | Santa Cruz | sc-146 | (1:500) |
| Antibody | Anti-TfR (rabbit polyclonal) | Abcam | ab84036 | (1:1000) |
| Antibody | Anti-α-tubulin (mouse monoclonal) | Sigma | T6074 | (1:1000) |
| Antibody | Anti-TGFβ1 (mouse monoclonal) | inVivoMab/Bio X Cell | BE0057 | 100 µg/mL |
| Antibody | anti-ICAM2 (rat monoclonal) | BD Biosciences | 553325 | (1:500) |
| Antibody | anti-rabbit IgG (goat polyclonal) | Jackson | 111-035-003 | (1:7500) |
| Antibody | anti-mouse IgG (goat polyclona) | Jackson | 115-035-003 | (1:7500) |
| Recombinant DNA reagent | p3TP-lux | Wrana et al., 1992 | RRID:Addgene_11767 | Dr. Carmelo Bernabeu |
| Sequence-based reagent | qPCR primers | This paper | Supplement file 1 | |
| Peptide, recombinant protein | Human TFGβ1 | Peprotech | 100–21 | |
| Peptide, recombinant protein | IL4 | Peprotech | 214–14 | |
| Peptide, recombinant protein | Murine M-CSF | Peprotech | 315–02 | |
| Chemical compound, drug | LPS | Sigma | L2654 | |
| Chemical compound, drug | Tamoxifen | Sigma | T5648 | |
| Chemical compound, drug | Collagenase type IV | Sigma | C5138 | |
| Chemical compound, drug | Collagenase type I | Worthington | LS004194 | |
| Chemical compound, drug | Collagenase type II | Worthington | LS004174 | |
| Chemical compound, drug | Porcine pancreatin | Sigma | P3292 | |
| Chemical compound, drug | RBC Lysis buffer solution | eBioscience | 00-4333-57 | |
| Chemical compound, drug | Passive Lysis 5× Buffer | Promega | E1941 | |
| Chemical compound, drug | TRIzol Reagent | Thermofisher | 15596026 | |
| Chemical compound, drug | Fluoromont-G | Southern Biotech | 0100–01 | |
| Commercial assay or kit | Dynabeads Sheep Anti-Rat IgG | Thermofisher | 11035 | |
| Commercial assay, kit | Foxp3/Transcription Factor Staining Buffer Set | eBioscience | 00-5523-00 | |
| Commercial assay, kit | High Capacity cDNA Reverse Transcription Kit | Applied Biosystems | 4368814 | |
| Commercial assay, kit | Bradford Assay | Bio-Rad | 5000001 | |
| Commercial assay, kit | Luciferase Assay System | Promega | PR-E1500 | |
| Software, algorithm | Vevo 2100 | Visual Sonics | RRID:SCR_015816 | |
| Software, algorithm | Fiji | fiji.sc | RRID:SCR_002285 | |
| Software, algorithm | GraphPad Prism | www.graphpad.com | RRID:SCR_002798 | |
| Software, algorithm | FlowJo | www.flowjo.com | RRID:SCR_008520 | |
| Software, algorithm | qBASE+ (Biogazelle) | www.qbaseplus.com | RRID:SCR_003370 | |
| Software, algorithm | NDP.view2 | Hamamatsu Photonics | ||
| Software, algorithm | Zen2 | Zeiss | RRID:SCR_013672 | |
| Software, algorithm | 3D fully automated image analysis | Gkontra et al., 2018 |
Additional files
-
Supplementary file 1
List of primer sequences used in the study.
- https://cdn.elifesciences.org/articles/57920/elife-57920-supp1-v3.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/57920/elife-57920-transrepform-v3.pdf





