IER5, a DNA damage response gene, is required for Notch-mediated induction of squamous cell differentiation
Figures
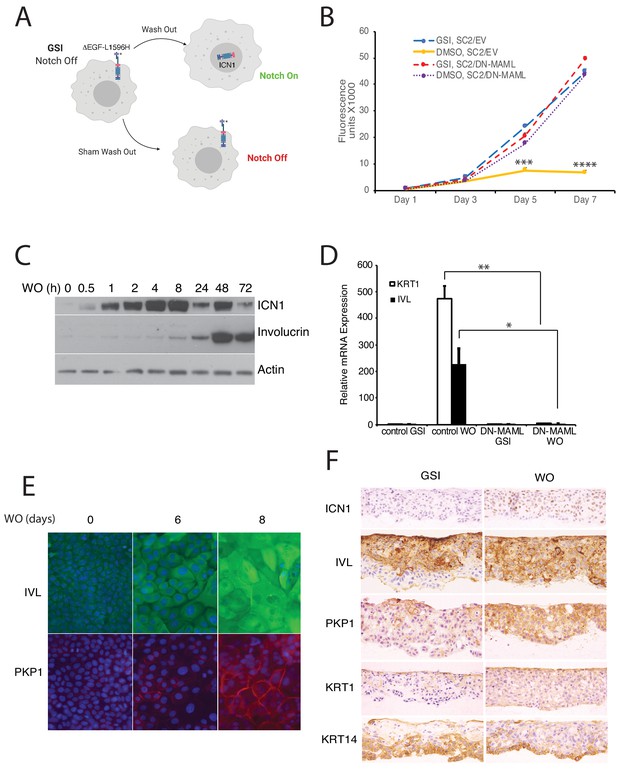
Notch activation induces growth arrest and differentiation of squamous carcinoma cells.
(A) Strategy used to activate Notch in a tightly regulated fashion. (B) Notch-induced suppression of SC2 cell growth in standard cultures is abrogated by DN-MAML, a specific inhibitor of canonical Notch signaling. SC2 cells were transduced with empty MigRI virus (EV) or with MigRI virus encoding DN-MAML. Cell numbers at various times post-GSI washout (DMSO vehicle alone) or sham GSI-washout (GSI) were assessed using Cell Titer-Blue on biological replicates performed in quadruplicate. Error bars represent standard deviations. Timepoints with significantly different cell growth between Notch-on cells (DMSO, empty vector) and Notch-off cells (GSI, empty vector; DMSO, DN-MAML1; and GSI, DN-MAML1) are denoted with *** (p<0.005) or **** (p<0.0005) (two-tailed student t test). (C) Western blot showing the kinetics of activated intracellular NOTCH1 (ICN1) generation and increases in involucrin (IVL) following GSI washout in SC2 cells in standard cultures. (D) Notch-induced differentiation of SC2 cells is abrogated by DN-MAML. Transcripts for involucrin (IVL) and keratin1 (KRT1) were measured in the presence of GSI and 3 days after GSI washout in SC2 cells transduced with empty virus or with DN-MAML. Transcript abundance in biological replicates performed in triplicate was measured by RT-PCR and normalized against GAPDH. Error bars represent standard deviations of the mean. **, p<0.005; *, p<0.05; two-tailed student t-test. (E) Indirect immunofluorescence microscopy showing staining for involucrin (IVL, green) and plakophilin-1 (PKP1, red) in SC2 cells at time 0 and 6 and 8 days after GSI washout. Nuclei in each image were counterstained with DAPI. (F) Immunohistochemical staining of SC2 cells grown in skin raft cultures for 14 days in the presence and absence of GSI.
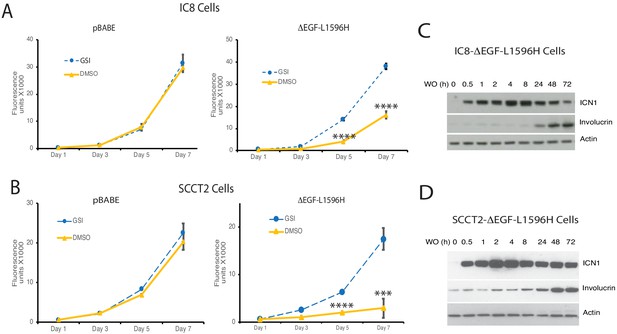
Notch activation induces differentiation and growth arrest of the squamous carcinoma cell lines IC8 and SCCT2.
(A, B) Effects of GSI and sham washout on the growth of IC8 and SCCT2 cells transduced with either empty vector (pBABE) or ΔEGF-L1596H. Cell numbers at various times post-GSI washout (DMSO) or sham GSI-washout (GSI) were assessed with Cell Titer-Blue in biological replicates prepared in quadruplicate. Timepoints with significantly different cell growth between Notch-on cells (DMSO, empty vector) and Notch-off cells (GSI) are denoted with *** (p<0.005) or **** (p<0.0005) (two-tailed student t test). (C, D) Western blot showing the kinetics of ICN1 generation and increases in involucrin (IVL) following GSI washout (WO) in IC8 and SCCT2 cells transduced with ΔEGF-L1596H.
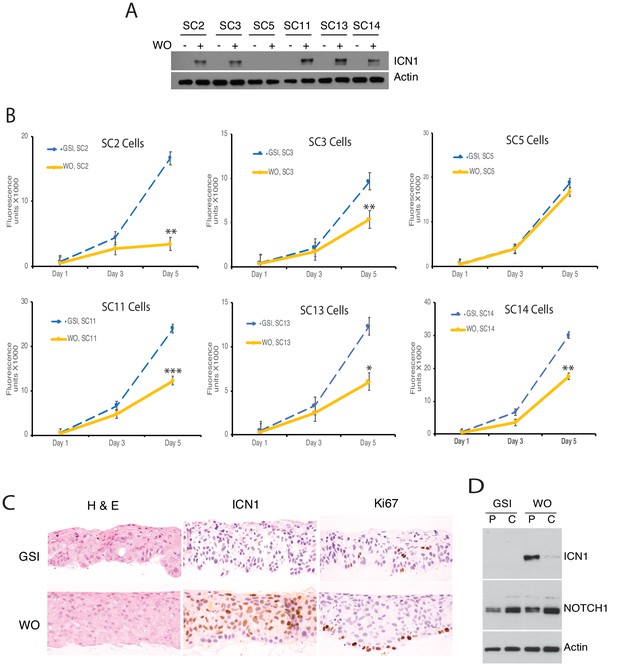
Characterization of clones derived from single IC8 cells transduced with ΔEGF-L1596H.
(A) Western blot showing ICN1 levels 4 hr post-GSI washout in IC8-ΔEGF-L1596H clones. (B) Effects of Notch activation on growth of IC8-ΔEGF-L1596H clones. Cell numbers at various times post-GSI washout (WO) or sham GSI-washout (GSI) were assessed with Cell Titer-Blue in biological replicates prepared in quadruplicate. Error bars represent standard deviations. Timepoints with significantly different cell growth between Notch-on cells (DMSO, empty vector) and Notch-off cells (GSI) are denoted with * (p<0.05), ** (p<0.005), or *** (p<0.0005) (two-tailed student t test). (C) Formation of a 3D epidermal layer by SC2 cells in skin raft culture in the presence of GSI and following GSI washout. Raft sections were stained with H and E (hematoxylin and eosin) or with antibodies specific for ICN1 (activated intracellular NOTCH1) or Ki67. (D) Effect of matrix on accumulation of ICN1 following GSI washout. SC2 cells were plated on plastic (P) or collagen (C) in the present of GSI and then subjected to sham (GSI) or true GSI washout (WO). Cell extracts were prepared 24 hr after washout and analyzed by western blot.
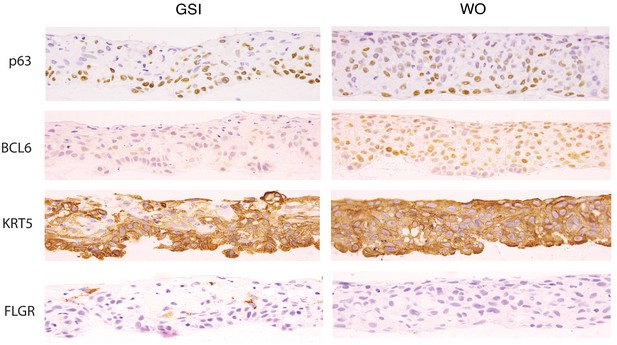
Immunohistochemical assessment of p63, BCL6, keratin5 (KRT5), and filaggrin (FLGR) protein levels in S2 cells grown in the Notch-off (GSI) or Notch-on (WO) states in 3D rafts.
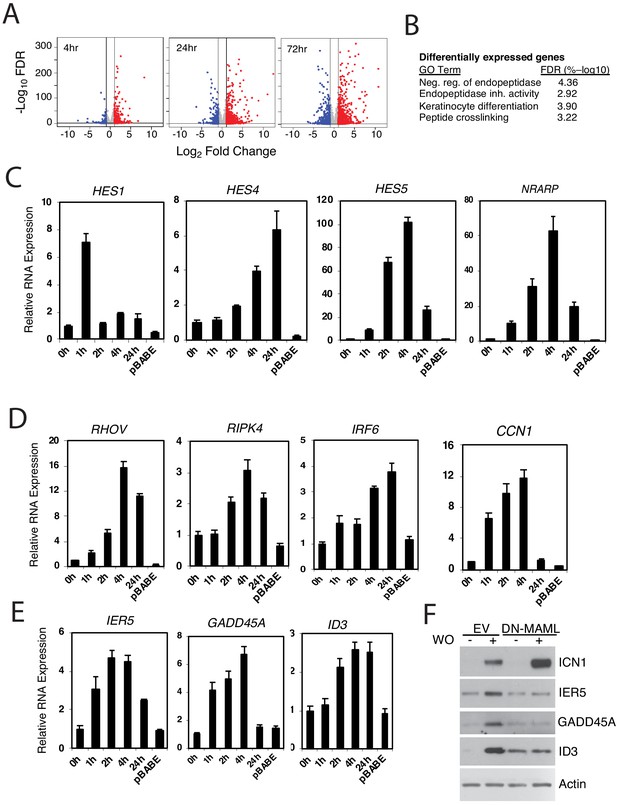
Identification of Notch-induced genes in squamous carcinoma cells.
(A) Volcano plots showing changes in RNA transcript read counts induced by Notch activation in SC2 cells for 4, 24, and 72 hr as compared to control cells treated with sham GSI washout. RNA-seq for each treatment group was performed in triplicate on biological replicates. Vertical lines denote a twofold change in read count, while the horizontal line denotes a false discovery rate (FDR) of 5%. (B) Gene ontogeny (GO) annotation of differentially expressed genes in ‘Notch-on’ SC2 cells. The most highly associated GO terms are shown; other significant associated annotated gene sets (FDR < 5%) are listed in Supplementary file 7. (C-E) Transcriptional responses of selected ‘canonical’ Notch target genes (C), genes linked to keratinocyte differentiation (D), and genes associated with DNA damage responses (E), to Notch activation in IC8-ΔEGF-L1596H cells. Transcript abundance in technical replicates prepared in triplicate was measured by RT-PCR and normalized against GAPDH. Error bars represent standard deviations of the mean. (F) Western blots of cell lysates prepared from IC8-ΔEGF-L1596H cells transduced with empty virus (EV) or DN-MAML following sham GSI washout (-) or 24 hr post-GSI washout (+).
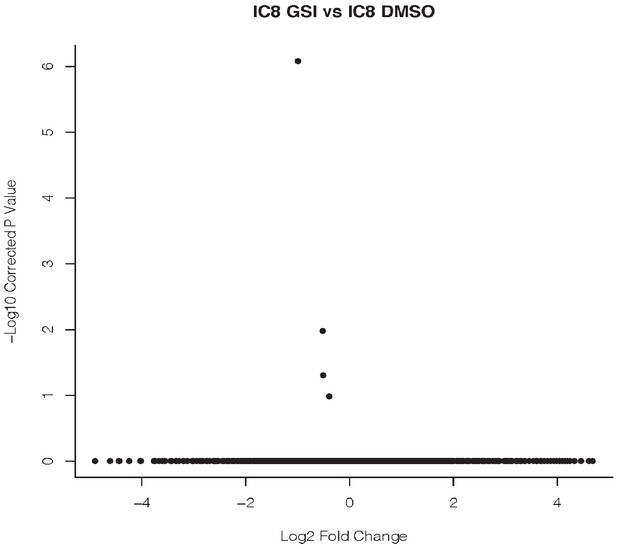
GSI treatment has little effect on gene expression in IC8 squamous carcinoma cells.
Duplicate cultures of IC8 cells were treated with GSI or vehicle (DMSO) for 24 hr and transcript abundance was assessed by RNA-seq in two biological replicates. A volcano plot shows no differentially expressed genes using cutoffs of adjusted p-value<0.0001 and log2 fold change >1.
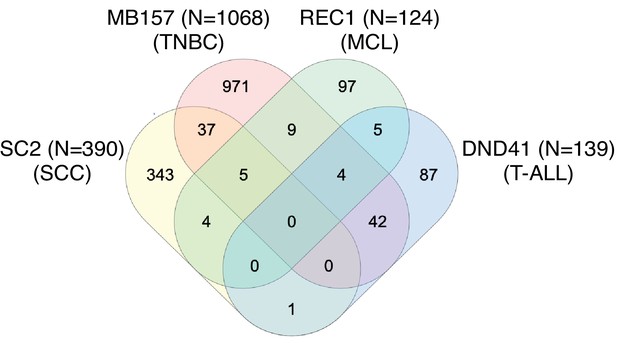
Venn diagram showing the overlap in Notch target genes (defined as log2 change >1 and FDR < 0.05 following GSI washout) in IC8 squamous carcinoma cells (S2 subclone), MB157 triple negative breast carcinoma cells (Petrovic et al., 2019), REC1 mantle cell lymphoma cells (Ryan et al., 2017), and DND-41 T-ALL cells (Petrovic et al., 2019).
The lists of overlapping genes are provided in Supplementary file 6.
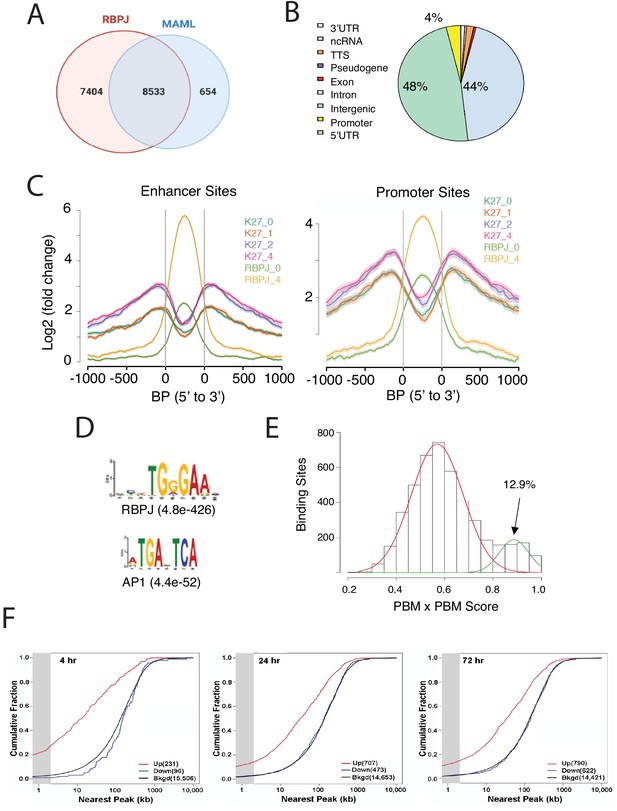
Characterization of Notch transcription complex (NTC) binding sites in IC8-ΔEGF-L1596H cells.
(A) Number and overlap of RBPJ and MAML1 binding sites determined by ChIP-Seq of chromatin prepared 4 hr after Notch activation. (B) Genomic distribution of RBPJ/MAML1 co-binding sites 4 hr after Notch activation. TTS, transcription termination sites; ncRNA, non-coding RNA. (C) Effect of NTC loading on histone3 lysine27 acetylation (H3K27ac), based on ChIP-Seq for H3K27ac in cells maintained in GSI and in cells 1, 2, and 4 hr after GSI washout. (D) Transcription factor motifs enriched within 300 bp of RBPJ/MAML1 ChIP-Seq signal peaks. (E) Protein-binding matrix (PBM) X PBM scores for NTC-binding sites. Sites with scores in the right-hand Gaussian distribution correspond to likely sequence paired sites. (F) Kolmogorov-Smirnov analysis showing spatial relationships between NTC-binding sites and transcriptional start sites (TSSs) of genes that increase, decrease, or are unchanged in expression following Notch activation. The gray zone denotes genes with TSSs within 2 kb of RBP/MAML1 peaks.
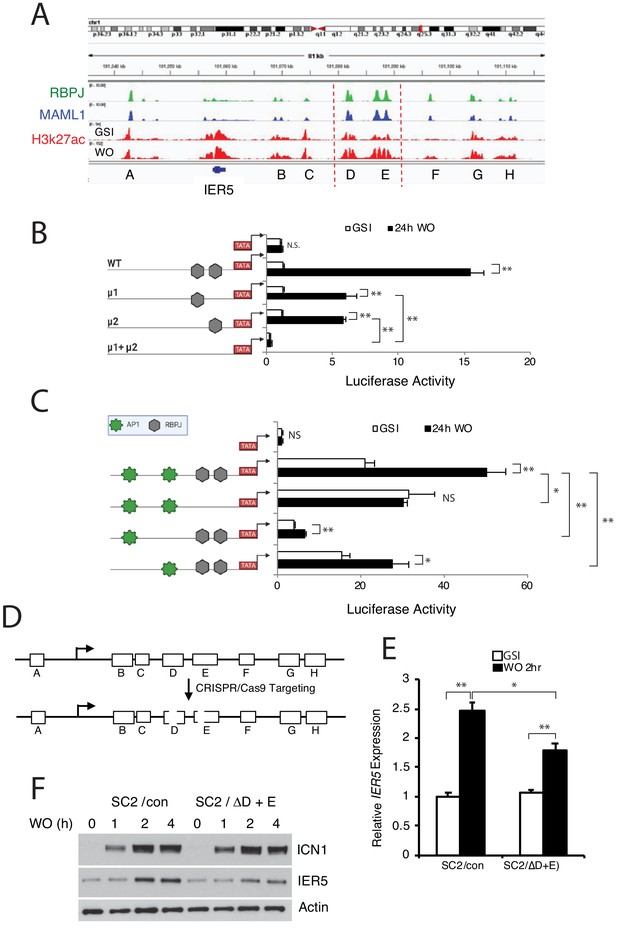
IER5 is a direct Notch target gene.
(A) Chromatin landscapes around IER5 in IC8-ΔEGF-L1596H cells. ChIP-Seq signals for RBPJ, MAML1, and H3K27ac for cells maintained in and 4 hr after GSI washout (WO) are shown. (B, C) Activities of a WT IER5 enhancer E luciferase reporter gene and derivatives bearing mutations (μ) in two RBPJ consensus motifs (B) and a WT IER5 enhancer D luciferase reporter gene and derivatives bearing mutations in two RBPJ consensus motifs or in flanking AP1 consensus motifs (C). Reporter gene assays were performed in SC2 cells maintained in GSI or 24 hr after GSI washout (WO). Luciferase reporter gene activity was determined in biological replicates prepared in triplicate and normalized to the activity of a Renilla luciferase internal control gene. Error bars represent standard deviations. (D) Cartoon showing the CRISPR/Cas9 targeting strategy for IER5 enhancers D and E. (E) Relative IER5 transcript levels in SC2 cells targeted with control AAVS1 CRISPR/Cas9 plasmids (SC2/con) or with CRISPR/Cas9 plasmids that remove the RPBJ sites in enhancers D and E (SC2/ΔD+E). Cells were either maintained in GSI or were harvested 2 hr following GSI washout (WO). Transcript abundance was measured in experimental triplicates by RT-PCR and normalized against GAPDH. Error bars represent standard errors of the mean. (F) Western blots showing IER5 protein levels in SC2/con cells and SC2/ΔD+E cells that were either maintained in GSI or harvested 1, 2, or 4 hr following GSI washout (WO). In B, C, and E, *, p<0.05; **, p<0.005; ***, p<0.0005 (all two-tailed student t test); NS, not significant.
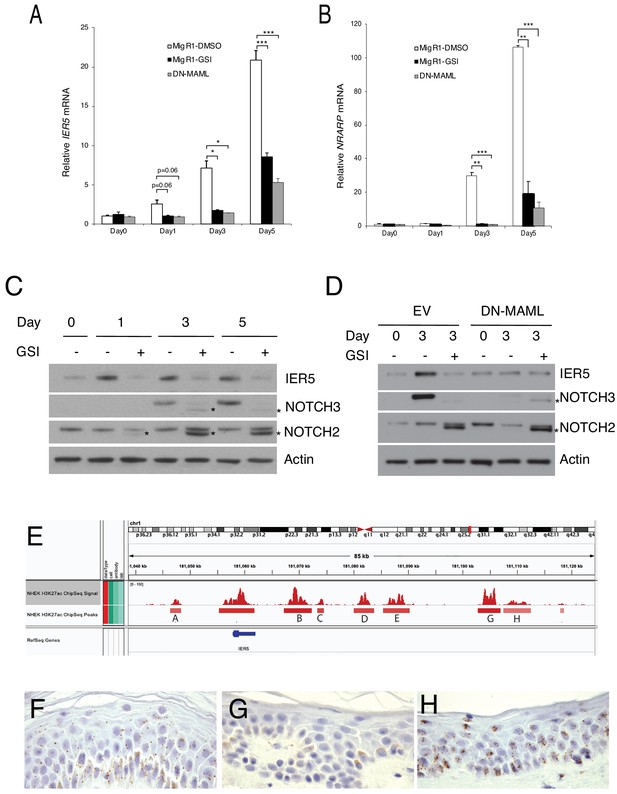
IER5 is regulated by Notch in non-transformed keratinocytes.
(A, B) TERT-immortalized NOK1 keratinocytes transduced with either empty MigRI retrovirus or MigRI encoding dominant negative MAML1 (DN-MAML) were grown in low Ca2+ medium (Day 0) or were shifted to high Ca2+ differentiation medium for 1–5 days. IER5 and NRARP transcript abundance in biological replicates prepared in triplicate was measured by RT-PCR and normalized against GAPDH. Error bars represent standard deviations of the mean. *, p<0.05; **, p<0.005; ***, p<0.0005 (all two-tailed student t test). (C) Western blot showing changes in IER5, NOTCH2, and NOTCH3 polypeptides following transfer of NOK1 cells to differentiation medium in the absence (-) and presence (+) of GSI. Increased levels of smaller NOTCH2 and NOTCH3 polypeptides consistent with ADAM-metalloprotease cleaved products seen in differentiation medium in the presence of GSI are denoted with asterisks. (D) Western blot showing changes in IER5, NOTCH2, and NOTCH3 polypeptides following transfer of NOK1 cells transduced with either empty MigRI retrovirus (EV) or MigRI retrovirus encoding DN-MAML1 to differentiation medium in the absence (-) and presence (+) of GSI. Increased levels of smaller NOTCH2 and NOTCH3 polypeptides consistent with ADAM-metalloprotease cleaved products seen in differentiation medium in the presence of GSI are denoted with asterisks. (E) H3K27ac landscapes near the IER5 gene body in normal human epidermal keratinocytes (NHEK cells). ChIP-Seq data are from ENCODE. (F-H) Detection of IER5 transcripts in normal human epidermis by in situ hybridization (ISH). (F) IER5-specific probe; (G) DapB-specific negative control probe; (H) PPIB-specific positive control probe. Positive signals correspond to brown spots in cells that are counterstained with hematoxylin.
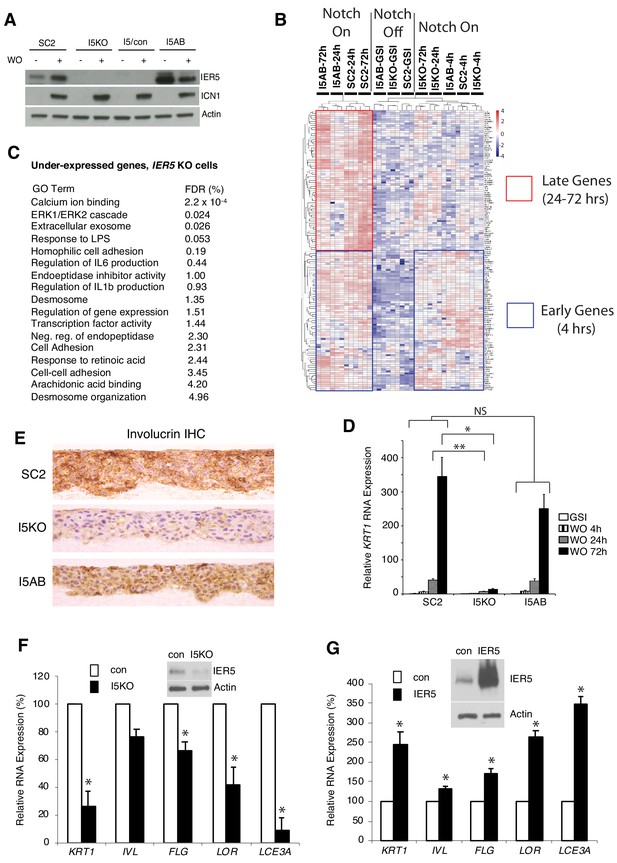
Effect of IER5 on Notch-dependent changes in gene expression in SC2 cells and NOK1 cells.
(A) Western blots showing IER5 and ICN1 protein levels in SC2 cells, a single-cell clone derived from SC2-IER5 knockout cells (I5KO), I5KO cells transduced with empty virus (I5/con), and pooled I5KO cells transduced with IER5 cDNA (I5AB) that were maintained in GSI (-) or harvested 48 hr post-GSI washout (+). (B) Heat map showing Notch-induced changes in gene expression in SC2 cells, I5KO cells, and I5AB cells. RNA-seq was performed in biological replicates in triplicate at time 0, 4 hr, 24 hr, and 72 hr after GSI washout. Samples were subjected to unsupervised clustering using a gene set containing all genes that were significantly upregulated at any time point after Notch activation in SC2 cells. The blue boxes highlight genes that are upregulated at 4, 24, and 72 hr after Notch activation, whereas the red box highlights genes that are under-expressed in IER5 knockout cells (I5KO) and rescued by re-expression of IER5 (I5AB) at later timepoints (24 and 72 hr). (C) Gene ontogeny (GO) terms associated with the set of under-expressed genes in I5KO cells following Notch activation. FDR = false discovery rate. (D) Diminished induction of KRT1 expression at 24 and 72 hr after GSI WO in I5KO cells is prevented by IER5 addback (I5AB cells). Transcript abundance in biological replicates prepared in triplicate was measured by RT-PCR and normalized against GAPDH. Error bars represent standard deviations of the mean. *, p<0.05; **, p<0.005; NS, not significant (two-tailed student t test). (E) Immunohistochemical staining for involucrin in SC2, I5KO, and I5AB cells in raft cultures grown in the absence of GSI. (F, G) Effect of CRISPR/Cas9 targeting of IER5 and enforced IER5 expression on differentiation-associated transcripts in NOK1 cells. In F, NOK1 cells were transduced with CRISPR/Cas9, GFP, and IER5 (I5KO) gRNA or AAVS1 control (con) gRNA and sorted for GFP positivity. In G, NOK1 cells were transduced with empty GFP-expressing retrovirus (con) or IER5 and GFP and sorted. In F and G, analyses were done on pooled GFP-positive transductants, which were moved to high Ca2+ medium for 3 days (F) or 5 days (G) prior to harvest. Inset western blots show the extent of IER5 loss (F) and IER5 overexpression (G) relative to control cells. In F and G, transcript abundance was measured in biological replicates prepared in triplicate by RT-PCR and normalized against GAPDH. Error bars represent standard deviations of the mean. *, p<0.05, student two-sided t test.
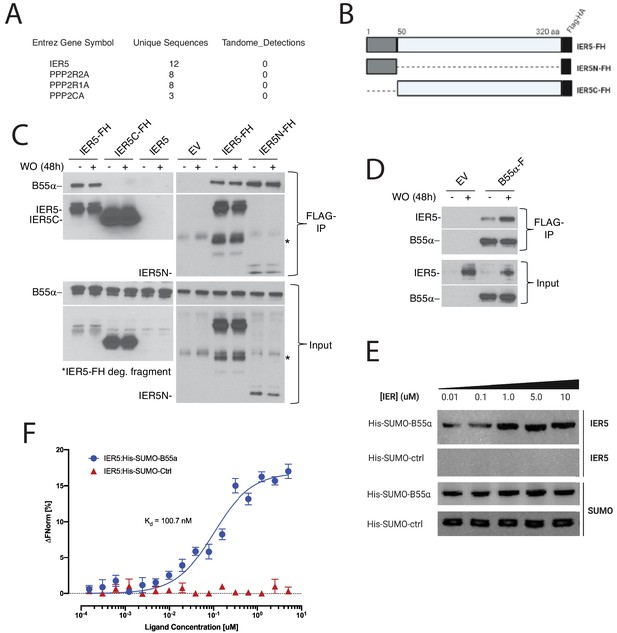
IER5 binds to B55α.
(A) Polypeptides identified by mass spectroscopy in immunoprecipitates prepared from I5 cells expressing tandem-tagged IER5. (B) Cartoon showing the structure of tandem-tagged IER5 polypeptides. FH, FLAG-HA tag. (C) Western blot analysis of immunoprecipitates prepared from I5 cells expressing the indicated forms of tagged IER5. WO, washout. (D) Western blot analysis of immunoprecipitates prepared from SC2 cells expressing FLAG-tagged B55α. (E) Western blot showing that IER5 binds His-Sumo-tagged B55α immobilized on beads. The upper two panels were stained for IER5, while the lower two panels were stained for SUMO. (F) Microscale thermophoresis showing saturable binding of IER5 to His-Sumo-tagged B55α.
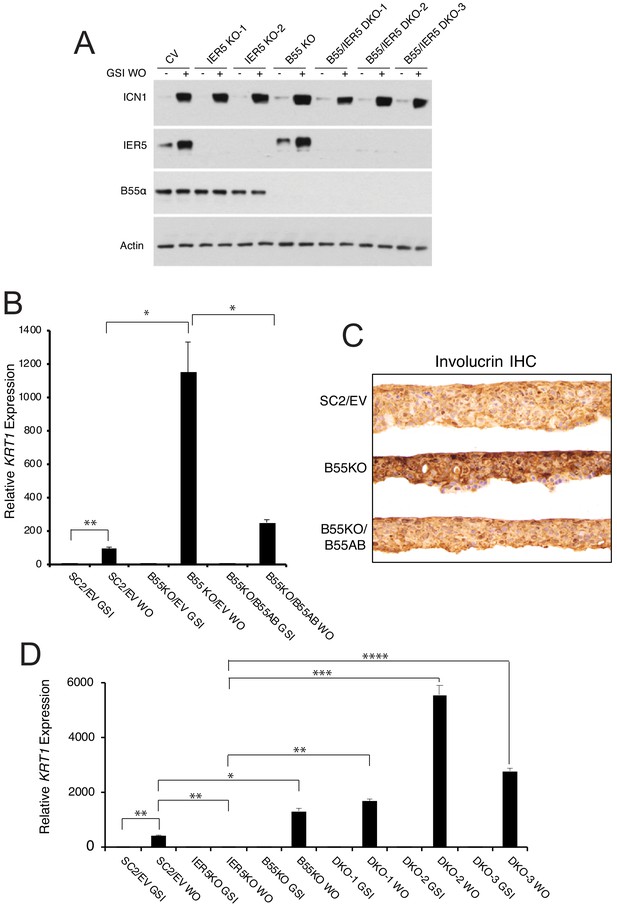
PPP2R2A is epistatic to IER5.
(A) Western blot showing IER5 and B55α protein levels in single (KO) and double (DKO) PPP2R2A and IER5 knockout clones in the presence of GSI (-) and 4 hr after GSI washout (+). (B) PPP2R2A knockout enhances Notch-dependent expression of KRT1. RT-PCR analysis of KRT1 expression in SC2 cells transduced with an empty retrovirus (SC2/EV); PPP2R2A knockout cells (B55 KO) transduced with empty retrovirus (B55KO/EV); and PPP2R2A knockout cells transduced with B55α-expressing retrovirus (B55KO/B55AB). WO = GSI washout. *, p<0.05; **, p<0.005 (two-tailed student t test). (C) Immunohistochemical (IHC) staining for involucrin of SC2 control, B55KO, and B55KO/B55AB cells in raft cultures in GSI-free medium. (D) B55α knockout negates the requirement for IER5 for Notch-dependent upregulation of KRT1. Results are shown for SC2 control cells (SC2/EV); an IER5 knockout clone; a PPP2R2A knockout clone (B55KO); and three IER5/PPP2R2A double knockout (DKO) clones. Cells were maintained in GSI or harvested 72 hr following GSI washout (WO). KRT1 transcript abundance was measured in biological replicates prepared in triplicate by RT-PCR and normalized against GAPDH. Error bars represent standard errors of the mean. *, p<0.05; **, p<0.005; ***, p<0.0005; ****, p<0.00005 (all two-tailed student t test).
Tables
Sequence variants, IC8* and SCCT2** squamous cell carcinoma cell lines.
| Gene | Variant | Variant allele frequency |
|---|---|---|
| IC8 cell | ||
| CASP8 | c.971T > C(p.M324T) | 66% of 411 reads |
| FBXW7 | c.1633T > C(p.Y545H) | 33% of 195 reads |
| KMT2D | c.7412G > A(p.R2471Q) | 42% of 255 reads |
| MGA | c.5599G > A(p.V1867I) | 39% of 710 reads |
| MTOR | c.4828G > A(p.E1610K) | 56% of 280 reads |
| NOTCH1 | c.5059C > T (p.Q1687*) | 100% of 412 reads |
| PAXIP1 | c.2023C > T(p.H675Y) | 17% of 384 reads |
| PMS1 | c.566_567delTCinsAT(p.V189D) | 36% of 108 reads |
| RIF1 | c.658G > A(p.E220K) | 62% of 251 reads |
| ROS1 | c.1144T > G(p.Y382D) | 87% of 169 reads |
| ROS1 | c.1164+2_1164+8delTTAGTCC () | 19% of 191 reads |
| SDHA | c.1627T > C(p.Y543H) | 56% of 668 reads |
| SF3B1 | c.2549T > C(p.I850T) | 31% of 246 reads |
| TERT | CC242-243TT promoter mutation | 50% of 26 reads |
| TP53 | c.451C > T(p.P151S) | 100% of 366 reads |
| WHSC1 | c.2185C > T(p.R729C) | 66% of 410 reads |
| WWTR1 | c.551T > G (p.V184G) | 64% of 256 reads |
| ZNF217 | c.2590C > T(p.L864F) | 39% of 835 reads |
| ZNF217 | c.1162delC(p.H388Tfs*77) | 55% of 822 reads |
| SCCT2 Cell | ||
| ALK | c.2854G > A (p.G952R) | 50% of 441 reads |
| ASXL1 | c.3959C > T (p.A1320V) | 31% of 930 reads |
| BRD3 | c.533C > T (p.S178F) | 49% of 281 reads |
| BRD4 | c.3915_3917dupTGC (p.A1306dup) | 45% of 170 reads |
| CDH4 | c.1801C > T (p.L601F) | 30% of 447 reads |
| CDKN2A | c.*151–1G > A () | 100% of 172 reads |
| CDKN2A | c.212A > T (p.N71I) | 100% of 184 reads |
| CREBBP | c.5842C > T (p.P1948S) | 74% of 77 reads |
| CREBBP | c.2116G > A (p.G706R) | 45% of 172 reads |
| DDB1 | c.327+6G > A () | 47% of 451 reads |
| DICER1 | c.775C > T (p.P259S) | 42% of 301 reads |
| DOCK8 | c.185T > A (p.V62E) | 100% of 597 reads |
| EGFR | c.1955G > A (p.G652E) | 48% of 518 reads |
| EGFR | c.298C > T (p.P100S) | 49% of 595 reads |
| ERCC2 | c.886A > T (p.S296C) | 48% of 165 reads |
| ERCC5 | c.264+1G > A () | 50% of 442 reads |
| ETV4 | c.1298C > G (p.P433R) | 45% of 302 reads |
| FANCF | c.494C > T (p.T165I) | 50% of 644 reads |
| FANCL | c.155+1G > A () | 51% of 220 reads |
| FAT1 | c.9076–1G > A () | 49% of 367 reads |
| FH | c.681G > T (p.Q227H) | 4% of 756 reads |
| FLT4 | c.2224G > A (p.D742N) | 49% of 346 reads |
| GALNT12 | c.1035+5G > A () | 52% of 523 reads |
| GLI2 | c.1859C > A (p.T620K) | 45% of 351 reads |
| HNF1A | c.1640C > T (p.T547I) | 54% of 392 reads |
| JAZF1 | c.477C > T (p.I159I) | 47% of 606 reads |
| JAZF1 | c.328C > T (p.P110S) | 44% of 211 reads |
| KMT2D | c.10355+1G > A () | 49% of 622 reads |
| LIG4 | c.1271_1275delAAAGA (p.K424Rfs*20) | 40% of 659 reads |
| MAP2K1 | c.568+1G > A () | 53% of 239 reads |
| MED12 | c.2080G > A (p.E694K) | 100% of 269 reads |
| MYB | c.1461+5G > A () | 41% of 430 reads |
| NF1 | c.2608G > A (p.V870I) | 50% of 615 reads |
| NF2 | c.813T > G (p.F271L) | 48% of 168 reads |
| NOTCH1 | c.1226G > T (p.C409F) | 44% of 519 reads |
| NOTCH1 | c.1406A > G (p.D469G) | 50% of 912 reads |
| NOTCH1 | c.1245G > T (p.E415D) | 42% of 495 reads |
| NOTCH2 | c.5252G > A (p.G1751D) | 44% of 459 reads |
| NOTCH2 | c.1298G > A (p.C433Y) | 50% of 484 reads |
| NOTCH2 | c.1108+1G > A () | 53% of 305 reads |
| NSD1 | c.7669G > A (p.G2557R) | 49% of 743 reads |
| PDGFRB | c.2586+2T > A () | 43% of 380 reads |
| PHOX2B | c.181A > T (p.T61S) | 52% of 222 reads |
| POLQ | c.6565G > A (p.A2189T) | 27% of 462 reads |
| POLQ | c.1634G > A (p.S545N) | 33% of 667 reads |
| PPARG | c.819+6T > C () | 100% of 134 reads |
| PRKDC | c.6436G > A (p.A2146T) | 42% of 471 reads |
| RAD51C | c.996G > A (p.Q332Q) | 45% of 302 reads |
| RHEB | c.443C > T (p.S148F) | 46% of 120 reads |
| ROS1 | c.6871C > T (p.P2291S) | 45% of 605 reads |
| ROS1 | c.3342A > T (p.Q1114H) | 48% of 274 reads |
| ROS1 | c.137A > T (p.D46V) | 42% of 215 reads |
| RPTOR | c.2992G > A (p.V998I) | 48% of 352 reads |
| RUNX1T1 | c.1039G > A (p.D347N) | 45% of 715 reads |
| SDHA | c.1151C > T (p.S384L) | 53% of 446 reads |
| SLC34A2 | c.1700T > A (p.I567N) | 50% of 460 reads |
| SMARCA4 | c.3947T > G (p.F1316C) | 55% of 431 reads |
| SMARCE1 | c.395C > T (p.A132V) | 51% of 587 reads |
| STAT3 | c.1852G > A (p.G618S) | 47% of 527 reads |
| TDG | c.166+4G > A () | 48% of 329 reads |
| TP53 | c.375+1G > T | 47% of 173 reads |
| TP53 | c.832_833delCCinsTT (p.P278F) | 46% of 418 reads |
| UIMC1 | c.971T > C (p.V324A) | 48% of 745 reads |
| XPC | c.571C > T (p.R191W) | 100% of 219 reads |
-
*Based on analysis of 16,131,317 unique, high-quality sequencing reads (mean, 406 reads per targeted exon, with 98% of exons having more than 30 reads).
†Based on analysis of 20,972,158 unique, high-quality sequencing reads (mean, 413 reads per targeted exon, with 99% of exons having more than 30 reads).
Copy number variants, squamous cell carcinoma cell lines.
| Chromosome | Type | Genes affected |
|---|---|---|
| IC8 cell line | ||
| 1q | Gain | MCL1, GBA, RIT1, NTRK1, DDR2, PVRL4, SDHC, CDC73, MDM4, PIK3C2B, UBE2T, PTPN14, H3F3A, EGLN1, AKT3, EXO1, FH |
| 2 | Loss | XPO1, FANCL, REL, MSH6, EPCAM, MSH2, SOS1, ALK, BRE, DNMT3A, GEN1, MYCN, TMEM127, GLI2, ERCC3, CXCR4, RIF1, ACVR1, ABCB11, NFE2L2, PMS1, CASP8, SF3B1, CTLA4, ERBB4, IDH1, BARD1, XRCC5, DIS3L2 |
| 3p | Loss | MITF, BAP1, PBRM1, COL7A1, RHOA, SETD2, CTNNB1, MLH1, MYD88, XPC, PPARG, RAF1, FANCD2, OGG1, VHL |
| 3q | Gain | NFKBIZ, CBLB, POLQ, GATA2, MBD4, TOPBP1, FOXL2, ATR, MECOM, PRKCI, TERC, PIK3CA, SOX2, ETV5, BCL6 |
| 4 | Loss | PHOX2B, RHOH, SLC34A2, FGFR3, WHSC1, KDR, KIT, PDGFRA, FAM175A, HELQ, TET2, FBXW7, NEIL3, FAT1 |
| 5 | Gain | RICTOR, IL7R, SDHA, TERT, MAP3K1, PIK3R1, XRCC4, RASA1, APC, RAD50, CTNNA1, PDGFRB, ITK, NPM1, TLX3, FGFR4, NSD1, UIMC1, FLT4 |
| 6 | Gain | CCND3, NFKBIE, POLH, VEGFA, CDKN1A, PIM1, RNF8, FANCE, DAXX, HFE, HIST1H3B, HIST1H3C, ID4, PRDM1, ROS1, RSPO3, MYB, TNFAIP3, ESR1, ARID1B, PARK2, QKI |
| 7 | Gain | EGFR, IKZF1, JAZF1, ETV1, PMS2, RAC1, CARD11, SBDS, CDK6, SLC25A13, CUX1, RINT1, MET, POT1, SMO, BRAF, PRSS1, EZH2, RHEB, XRCC2, PAXIP1 |
| 8p | Loss | KAT6A, POLB, FGFR1, WHSC1L1, NRG1, WRN, NKX3-1, PTK2B, GATA4, NEIL2 |
| 8q11.21-q21.11 | Loss | PRKDC, MYBL1, TCEB1 |
| 8q21.3-q24.3 | Gain | NBN, RUNX1T1, RAD54B, RSPO2, EXT1, RAD21, MYC, RECQL4 |
| 9p13.2-p21.3 | Loss | PAX5, FANCG, RMRP, CDKN2A, CDKN2B, MTAP |
| 9p24.1-p24.3 | Gain | CD274, JAK2, PDCD1LG2, DOCK8 |
| 11p11.2-p13 | Gain | EXT2, LMO2 |
| 13q33.1 | Loss | ERCC5 |
| 15q | Gain | FAN1, GREM1, BUB1B, MGA, RAD51, TP53BP1, B2M, USP8, MAP2K1, PML, NEIL1, FAH, NTRK3, BLM, FANCI, IDH2, IGF1R |
| 16p13.3 | Loss | CREBBP, SLX4 |
| 19 | Loss | BABAM1, CRTC1, JAK3, KLF2, MEF2B, BRD4, NOTCH3, CALR, KEAP1, SMARCA4, ELANE, GNA11, MAP2K2, STK11, TCF3, CCNE1, C19orf40, CEBPA, AKT2, AXL, CIC, XRCC1, ARHGAP35, ERCC1, ERCC2, BCL2L12, PNKP, POLD1, PPP2R1A |
| 20 | Gain | MCM8, ASXL1, BCL2L1, MAFB, AURKA, ZNF217, GNAS, CDH4 |
| SCCT2 Cell Line | ||
| 1q32.1 | Loss | UBE2T |
| 1q42.12-q42.2 | Gain | H3F3A, EGLN1 |
| 1q43 | Loss | AKT3, EXO1 |
| 1q43 | Gain | FH |
| 3 p Arm level | Loss | MITF, BAP1, PBRM1, COL7A1, RHOA, SETD2, CTNNB1, MLH1, MYD88, XPC, PPARG, RAF1, FANCD2, OGG1, VHL |
| 3q Arm level | Gain | NFKBIZ, CBLB, POLQ, GATA2, MBD4, TOPBP1, FOXL2, ATR, MECOM, PRKCI, TERC, PIK3CA, SOX2, ETV5, BCL6 |
| 8q Arm level | Gain | PRKDC, MYBL1, TCEB1, NBN, RUNX1T1, RAD54B, RSPO2, EXT1, RAD21, MYC, RECQL4 |
| 9q Arm level | Gain | GNAQ, NTRK2, FANCC, PTCH1, GALNT12, XPA, KLF4, TAL2, ENG, ABL1, TSC1, BRD3, NOTCH1 |
| 18q11.2 | Gain | GATA6, RBBP8 |
| 18q11.2-q21.33 | Gain | SS18, SETBP1, SMAD2, SMAD4, BCL2 |
| 20 | Gain | MCM8, ASXL1, BCL2L1, MAFB, AURKA, ZNF217, GNAS, CDH4 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Rabbit monoclonal anti-MAML1 | Cell Signaling Technology | Cat. #: 12166 | ChIP, 1 ml per1 × 106 cells |
| Antibody | Rabbit monoclonal anti-RBPJ | Cell Signaling Technology | Cat. #: 5313 | ChIP, 2.5 ml per1 × 106 cells |
| Antibody | Rabbit polyclonal anti-histone H3 acetyl K27 | Abcam | Cat. #: ab4729 | ChIP, 9 ml per1 × 106 cells |
| Antibody | Mouse monoclonal anti-involucrin | Sigma | Cat. #: I9018 | IF, 1:500; IHC, 1:10,000 |
| Antibody | Rabbit polyclonal anti-plakophilin-1 | Sigma | Cat. #: HPA027221 | IF, 1:300; IHC, 1:500 |
| Antibody | Rabbit monoclonal anti-activated NOTCH1 (ICN1) | Cell Signaling Technology | Cat. #: 4147 | IHC, 1:50; WB, 1:1000 |
| Antibody | Rabbit monoclonal anti-keratin-1 | Abcam | Cat. #: ab185628 | IHC, 1:1000 |
| Antibody | Rabbit monoclonal anti-Ki-67 | Biocare | Cat. #: CRM325 | IHC, 1:100 |
| Antibody | Mouse monoclonal anti-B55a | Cell Signaling Technology | Cat. #: 5689 | WB, 1:1000 |
| Antibody | Rabbit monoclonal anti-NOTCH2 | Cell Signaling Technology | Cat. #: 5732 | WB, 1:1000 |
| Antibody | Rabbit monoclonal anti-NOTCH3 | Cell Signaling Technology | Cat. #: 5276 | WB, 1:1000 |
| Antibody | Rabbit monoclonal anti-GADD45A | Cell Signaling Technology | Cat. #: 4632 | WB, 1:1000 |
| Antibody | Rabbit monoclonal anti-ID3 | Cell Signaling Technology | Cat. #: 9837 | WB, 1:1000 |
| Antibody | Horse polyclonal anti-mouse Ig linked to HRP | Cell Signaling Technology | Cat. #: 7076 | WB, 1:1,000-1:20,000 |
| Antibody | Goat polyclonal anti-rabbit Ig linked to HRP | Cell Signaling Technology | Cat. #: 7074 | WB, 1:1000 |
| Antibody | Mouse monoclonal anti-actin | Sigma | Cat. #: A1978 | WB, 1:10,000 |
| Antibody | Mouse monoclonal anti-FLAG | Sigma | Cat. #: F3165 | WB, 1:1000 |
| Antibody | Rabbit polyclonal anti-IER5 | Sigma | Cat. #: HPA029894 | WB, 1:1000 |
| Antibody | Mouse monoclonal anti-filaggrin | Santa Cruz Biotechnology | Cat. #: sc-66192 | IHC, 1:100 |
| Antibody | Mouse monoclonal anti-p63 | Biocare Medical | Cat. #: CM163A | IHC, 1:250 |
| Antibody | Rabbit polyclonal anti-loricrin | BioLegend | Cat. #: 905103 | IHC, 1:800 |
| Antibody | Mouse monoclonal anti-BCL6 | Cell Marque Tissue Diagnostics | Cat. #: 227 M-95 | IHC, 1:500 |
| Antibody | Rabbit monoclonal anti-keratin5 | Cell Signaling Technology | Cat. #: 71536 | IHC, 1:2000 |
| Antibody | Chicken polyclonal anti-keratin14 | BioLegend | Cat. #: 906004 | IHC, 1:800 |
| Antibody | Chicken polyclonal anti-SUMO | Lifesensors | Cat. #: AB7002 | WB, 1:2000 |
| Antibody | Sheep polyclonal anti-rabbit Ig linked to Dynabeads | ThermoFisher Scientific | Cat. #: 11203D | ChIP, 100 μl beads per 20 × 106 cells |
| Antibody | Mouse monoclonal anti-FLAG epitope linked to magnetic beads | Sigma | Cat #: M8823 | Tandem purification,40 μl to 1 ml beads |
| Cell line (Homo sapiens) | IC8 | 10.1038/s41467-018-06027-1 | Dr. Andrew South (Thomas Jefferson University) | |
| Cell line (H. sapiens) | SCCT2 | 10.1038/s41467-018-06027-1 | Dr. Andrew South (Thomas Jefferson University) | |
| Cell line (H. sapiens) | NOK1 | Piboonniyom et al., 2003; 63:476–83 | Dr. Karl Munger (Tufts University) | |
| Commercial assay or kit | CellTiter Blue | Promega | Cat. #: G8080 | |
| Commercial assay or kit | ChIP Assay Kit | Millipore | Cat. #: 17–295 | |
| Commercial assay or kit | Next Ultra II DNA Library Prep Kit | New England BioLabs | Cat. #: E7645 | |
| Commercial assay or kit | Next Ultra II RNA Library Prep Kit | New England BioLabs | Cat. #: E7775 | |
| Commercial assay or kit | QuickChange II Kit | Agilent Technologies | Cat. #: 200523 | |
| Commercial assay or kit | Dual Luciferase Kit | Promega | Cat. #: E1910 | |
| Chemical compound, drug | Compound E | Tocris | Cat. #: CAS 209986-17-4 | |
| Recombinant DNA reagent | pL-CRISPR. SFFV.GFP | Addgene | Cat. #: #57827 | |
| Recombinant DNA reagent | pL-CRISPR.SFFV.tRFP | Addgene | Cat. #: #57826 | |
| Recombinant DNA reagent | lentiCRISPRv2 neo | Addgene | Cat. #: 98292 | |
| Recombinant DNA reagent | lentiCRISPRv2 hygro | Addgene | Cat. #: 98291 | |
| Recombinant DNA reagent | pVL1392 | Expression Systems | Cat. #: 91–012 |
Additional files
-
Supplementary file 1
Differentially expressed genes after 4 hr of Notch activation, SC2 cells.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp1-v2.xls
-
Supplementary file 2
Differentially expressed genes after 24 hr of Notch activation, SC2 cells.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp2-v2.xls
-
Supplementary file 3
Differentially expressed genes after 72 hr of Notch activation, SC2 cells.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp3-v2.xls
-
Supplementary file 4
Unclustered GO annotations of Notch-sensitive genes, SC2 cells, at 72 hr of Notch activation.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp4-v2.xls
-
Supplementary file 5
Clustered GO annotations of Notch-sensitive genes, SC2 cells, at 72 hr of Notch activation.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp5-v2.xls
-
Supplementary file 6
Comparison of Notch-responsive genes in SC2 cells, MB157 triple-negative breast cancer cells, REC1 mantle cell lymphoma cells, and DND41 T-cell acute lymphoblastic leukemia cells.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp6-v2.xlsx
-
Supplementary file 7
Unclustered GO annotations of IER5-dependent Notch-sensitive genes, SC2 cells.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp7-v2.xls
-
Supplementary file 8
Clustered GO annotations of IER5-dependent Notch-sensitive genes, SC2 cells.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp8-v2.xls
-
Supplementary file 9
IER5-interacting proteins identified by tandem affinity purification.
- https://cdn.elifesciences.org/articles/58081/elife-58081-supp9-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58081/elife-58081-transrepform-v2.dotx





