Structural insight into toxin secretion by contact-dependent growth inhibition transporters
Figures
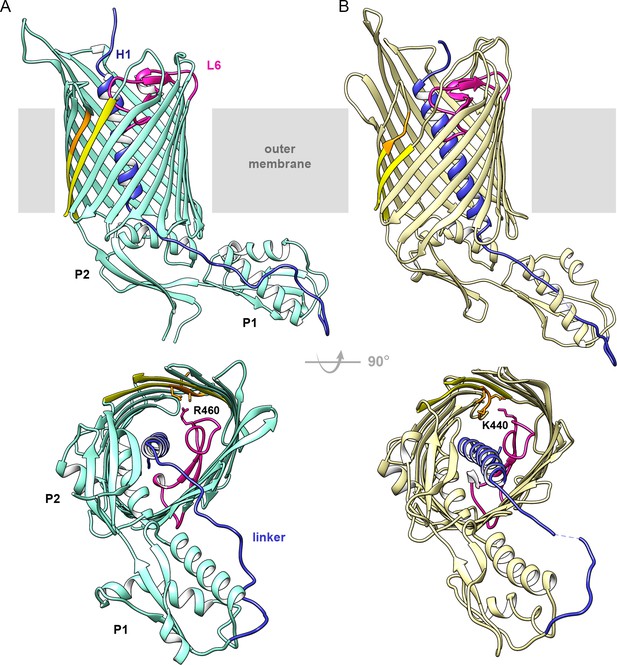
Structures of CdiB transporters.
Membrane view (upper panel) and periplasmic view (lower panel) of (A) CdiB from Acinetobacter baumannii (CdiBAb) in light teal and, (B) CdiB from Escherichia coli (CdiBEc) in pale yellow. The first POTRA domain is indicated by P1, and the second POTRA domain, closer to the β-barrel, by P2. Inside the β-barrel, the N-terminal helix H1 is shown in blue and Loop 6 (L6) in magenta. The linker connecting H1 to P1 is also colored blue. The first and last β-strands from the β-barrel are shown in yellow, with the DxxG motif in orange. In the lower panels, selected sidechains from DxxG and L6 are shown as sticks, with the interacting loop 6 residue highlighted.
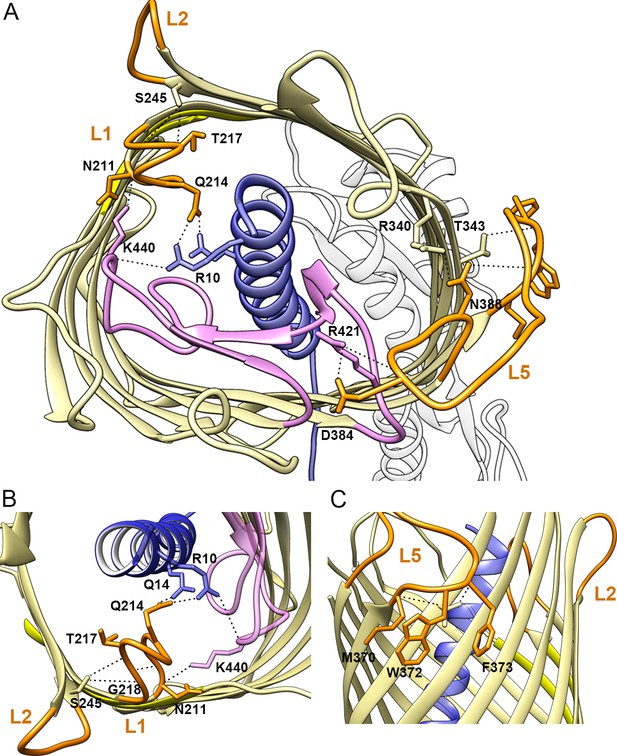
Positions of extracellular loops in the CdiBEc structure.
Extracellular loops 1, 2, and 5 from CdiBEc structure are colored in orange (indicated by L1, L2, and L5). (A) Extracellular view of the network of interaction (dashed lines) between helix H1 (blue), loop 6 (magenta) and loop 1. Lysine 440 from loop 6 stabilizes N211, the second residue of the conserved DxxG motif. The long extracellular loop 5 is stabilized by interactions with loop 6 and β8 from the β-barrel (pale yellow). (B) zoomed view of loop 1-helix H1-loop 6 interactions where Q214 forms hydrogen bonds with two residues from helix H1 (R10 and Q14), while R10 also interacts with K440 from loop 6. The loop 1 conformation is secured by an interaction between S245 on β3 and T217-G218 on loop 1. As seen in the sequence alignment (Supplementary file 1), the glycine of the DxxG motif, and T217-G218 residues are conserved, and the CdiBEc structure displays their spatial proximity. The conserved glycines complete the turn made by loop 1, where one amine group interacts with the carbonyl group of T217. (C) side view of loop 5 showing three hydrophobic residues (M370, W372, F373) that extend outside the β-barrel toward the outer membrane, potentially interacting with the membrane bilayer.
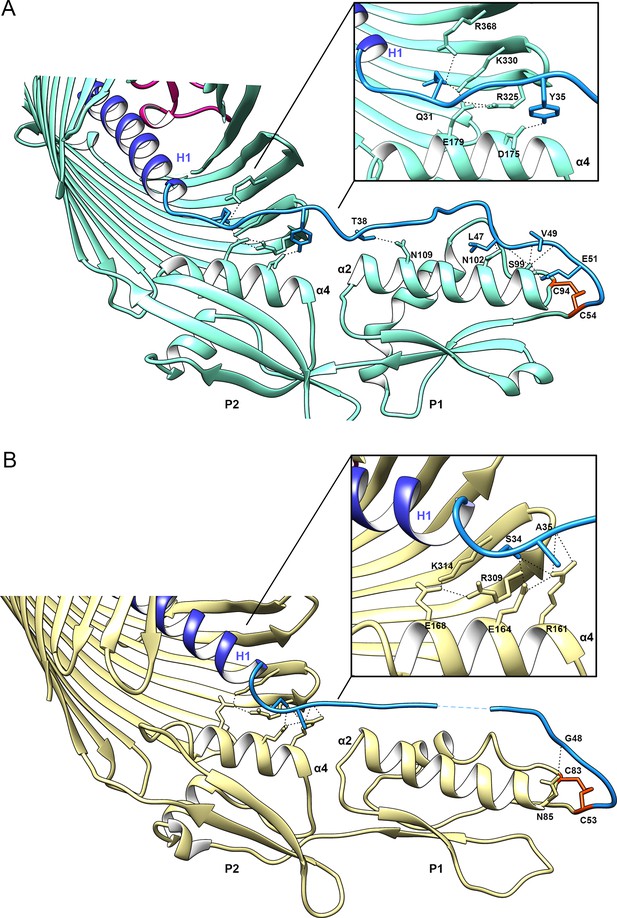
Network of interactions connecting the β-barrel, POTRA domains, and Linker.
View of the periplasmic region for (A) CdiBAb and, (B) CdiBEc. The α-helix proximal to the β-barrel lumen is labeled α4, where a conserved glutamate forms a salt bridge with an arginine and a lysine present on β6 and β7 from the β-barrel in both structures. On the same face of α4, a polar residue connects the N-terminal region of the linker (colored in blue). A zoomed view of this region is shown in the inset. The C-terminal region of the linker shows greater variation between the CdiB structures, where the linker in CdiBAb makes multiple interactions along the α2 helix of POTRA1. In both structures, the conserved cysteines (colored in red) form a disulfide bond that connects the linker to POTRA1. The two residues missing in the CdiBEc linker are represented by dashed lines.
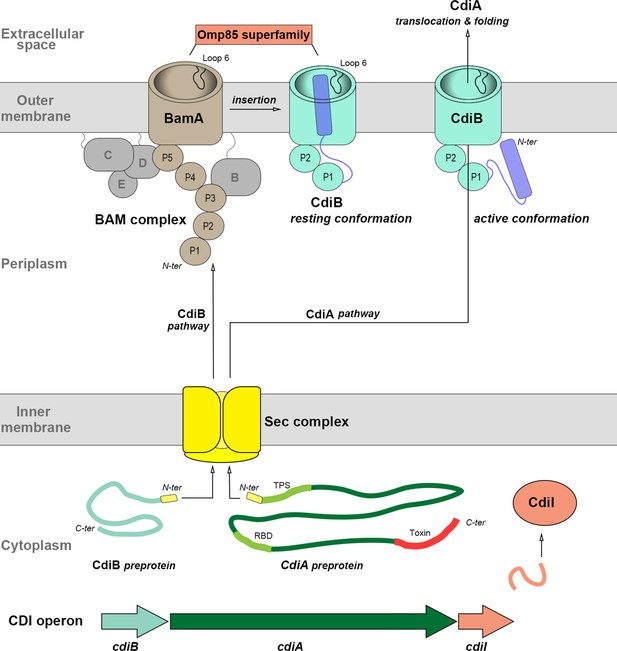
Biogenesis of CDI proteins.
In the cytoplasm, the CDI operon synthetizes CdiB and CdiA as preproteins. The N-terminal signal peptide (represented by a yellow box) is cleaved by the SEC complex (yellow) during inner membrane translocation. In the periplasm, CdiB and CdiA follow two different pathways (aided by periplasmic chaperones; not shown). As a TpsB transporter, CdiB interacts with the BAM complex, which folds and inserts CdiB into the outer membrane. BamA (brown), the central element of the BAM complex, and TpsB transporters are part of the same Omp85 superfamily sharing structural homologies (loop 6 and periplasmic POTRA domains (P) are indicated). CdiAs are large multi-domains proteins: the signaling domains TPS and RBD are represented in light green, FHA-1 and FHA-2 domains in dark green, and C-terminal toxin domain in red. In the periplasm, the N-terminal TPS domain recognizes and interacts with CdiB POTRA domains to initiate outer membrane translocation. At the surface of the cell, CdiA forms a filament folded as a β-helix that presents the RBD and toxin domains to neighboring bacteria. To prevent fratricide and auto-inhibition, the CDI operon produces a cytoplasmic immunity protein (CdiI). The boundaries of CdiB, CdiA, and BamA proteins are indicated by ‘N-ter’ and ‘C-ter’.
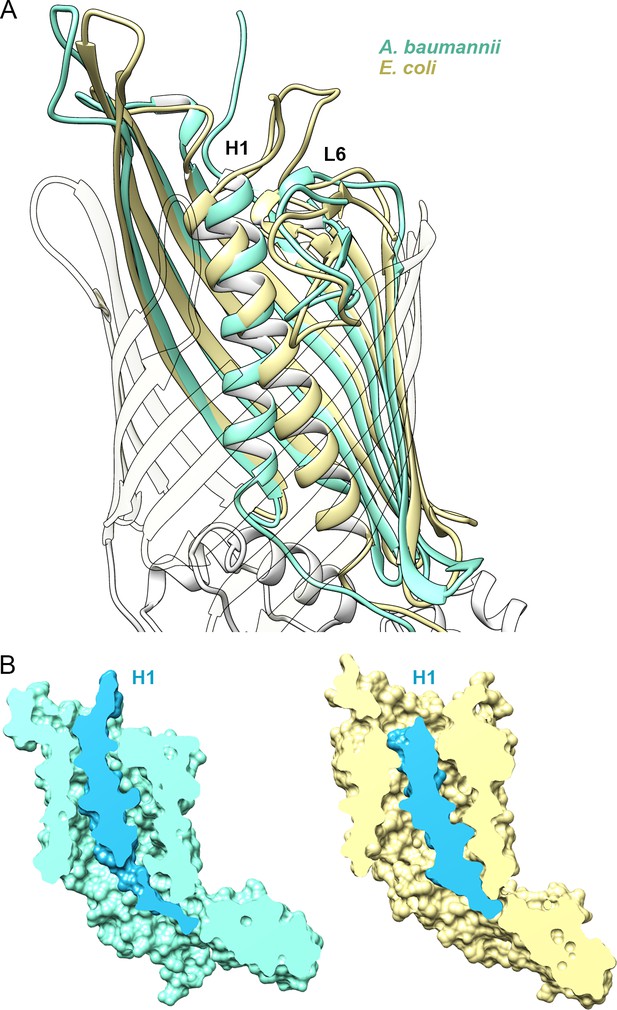
Position of Helix H1 in CdiB transporters.
(A) Membrane view of a superposition of CdiBAb (light teal) and CdiBEc (pale yellow) that illustrates conformational differences of helix H1 inside the β-barrel. To better visualize helix H1, β-strands from the front of the barrel are transparent. (B) Molecular surface cross-section of CdiBAb (light teal) and CdiBEc (pale yellow) where helix H1 and linker are shown in blue.
-
Figure 2—source data 1
List of interactions involving helix H1 for CdiBAb and CdiBEc.
List of amino acid interactions between β-barrel-helix H1, and helix H1-loop 6 in the crystal structures of CdiB A.baumannii (left) and CdiB E. coli (right). H-bond and salt-bridge interactions are labeled ‘h ‘and ‘s‘, respectively. *Interaction networks between R10-Q14 and Q214-K440 from CdiBEc are also depicted in Figure 1—figure supplement 1.
- https://cdn.elifesciences.org/articles/58100/elife-58100-fig2-data1-v2.docx
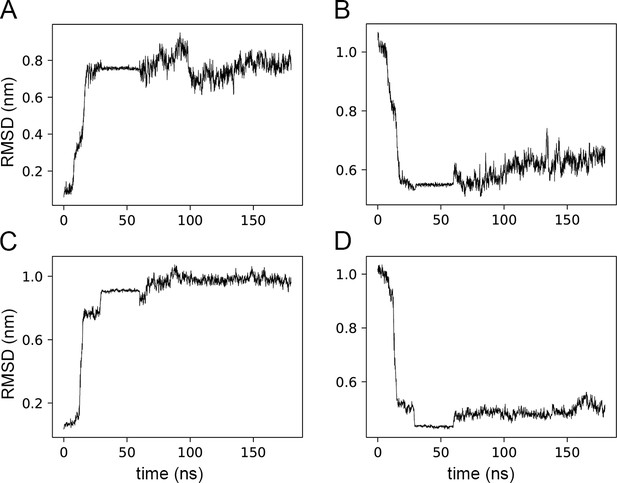
RMSD comparison from targeted MD simulations on H1.
Root-Mean-Square Deviation (RMSD) of the position of helix H1 for the two CdiB models generated by Targeted Molecular Dynamics (TMD) simulations. Top panels, CdiBAb structure where the helix H1 is moved from its initial position toward the position of the CdiBEc helix H1. Bottom panels, CdiBEc structure where the helix H1 is moved from its initial position toward the position of the CdiBAb helix H1. TMD was run for 30 ns, followed by 30 ns of H1 restrained to the new position, and then 120 ns of free equilibration. (A) CdiBAb H1 RMSD comparison with the initial frame (CdiBAb crystal structure). (B) CdiBAb H1 RMSD comparison with the targeted model (CdiBEc H1 position). (C) CdiBEc H1 RMSD comparison with the initial frame (CdiBEc crystal structure). (D) CdiBEc H1 RMSD comparison with the targeted model (CdiBAb H1 position).
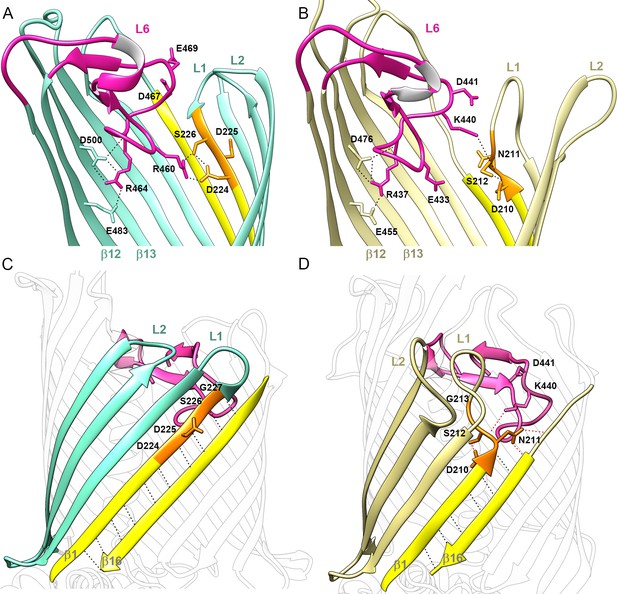
Position of Loop 6 and the DxxG motif in CdiB transporters.
Zoomed view of loop 6 (L6, magenta) in the lumen of the β-barrel of (A) CdiBAb (light teal) and, (B) CdiBEc (pale yellow). Sidechains involved in the network of interactions between L6 (VRGF/Y motif), β12 and β13 (F/GxDxG motif) and the DxxG motif (orange) are indicated by black dashed lines. β1 and β16 are colored in yellow, and extracellular loops 1 and 2 are indicated by ‘L1' and ‘L2', respectively. A conformational difference is shown between CdiB structures: in CdiBAb, R460, located before the VRG(F/Y) motif, interacts with the first and third residues of the DxxG motif, while In CdiBEc, K440 located after VRG(F/Y), interacts with the second residue of DxxG. Membrane view of the β-barrel of (C) CdiBAb and, (D) CdiBEc where β2–4 are highlighted in light teal and pale yellow respectively. β1 and β16 are highlighted in yellow, and DxxG motif in orange with important residues numbered. At the β1–β16 interface, mainchain interactions are shown as black dashed lines, and sidechain interactions as red dashed lines. In CdiBEc the two sidechains from L6 pointing toward β1–β16 are shown.
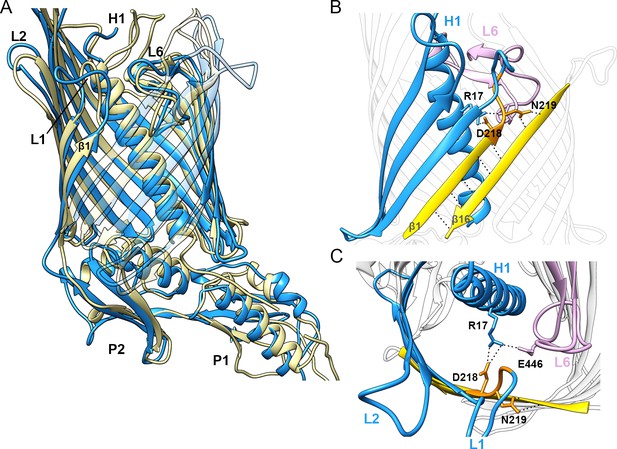
Superposition of CdiBEc and FhaC structures.
Superposition of CdiBEc (pale yellow) and FhaC (blue, PDB: 4QKY). Helix H1 lies at the same angle inside the barrel and the β1–β16 interface is shorter, where the second residue of DxxG (N219) stabilizes β16 and starts loop 1. Loop 1 is flexible and twisted inward, whereas loop 2 is oriented outward. (A) Membrane view illustrates conformational similarities between the CdiBEc (pale yellow) and FhaC (blue) structures. N-terminal helix H1, loop 1, loop 2, loop 6, and the first β-strand are indicated by H1, L1, L2, L6, and β1, respectively. For clarity, the front of the β-barrel is transparent. (B) Membrane view of the β-barrel of FhaC for comparison with Figure 3D. Helix H1 and strands β2 - β4 are highlighted in blue. β1 and β16 are highlighted in yellow, with the DxxG motif in orange. Interactions between β1 and β16 are shown by black dashed lines. (C) Extracellular view of DxxG-H1-L6 in FhaC structure. R17 from helix H1 (blue) forms a network of interactions between E446 on loop 6 (pink) and the conserved aspartate D218 from the DxxG motif (orange).
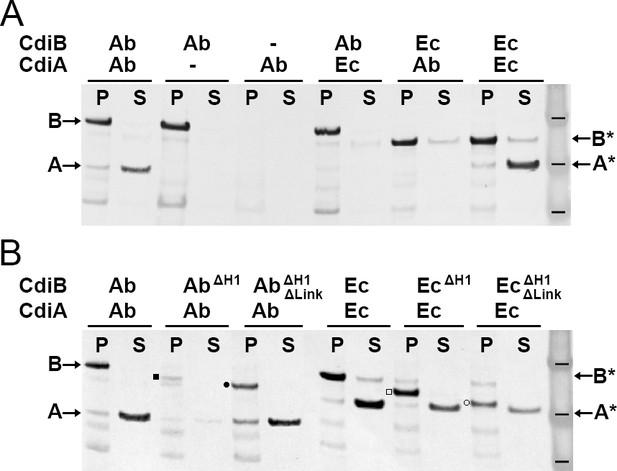
Functional analysis of CdiB transporters.
Immunoblot analyses of Escherichia coli C41 cultures co-expressing CdiB and CdiA-Nt. (A) After induction, cell pellets ‘P’ and culture supernatants ‘S’ are separated and analyzed to detect the presence of CdiB and CdiA-Nt from Acinetobacter baumannii ACICU ‘Ab’ or E. coli EC93 ‘Ec’, respectively. Dash indicates that cdi genes are present on the plasmid but not induced. CdiA-NtAb 74-472 and CdiA-NtEc 29-416 are only detected in the supernatant when their respective CdiB transporters are present. (B) CdiB variants where helix H1 ‘ΔH1’ and Linker ‘ΔLink’ are genetically deleted and co-expressed with their cognate CdiA-Nt substrates from CDI systems of A. baumannii ‘Ab’ and E. coli ‘Ec’. CdiB and CdiA-Nt wildtype proteins detected in immunoblot are indicated by B and A for A. baumannii ACICU and B* and A* for E. coli. Detection of CdiBΔH1 and CdiBΔH1ΔLink variants are indicated respectively, by a black square and a black circle for A. baumannii; and a white square and a white circle for E. coli. Protein ladder bands indicate 62, 49, 38 kDa respectively.
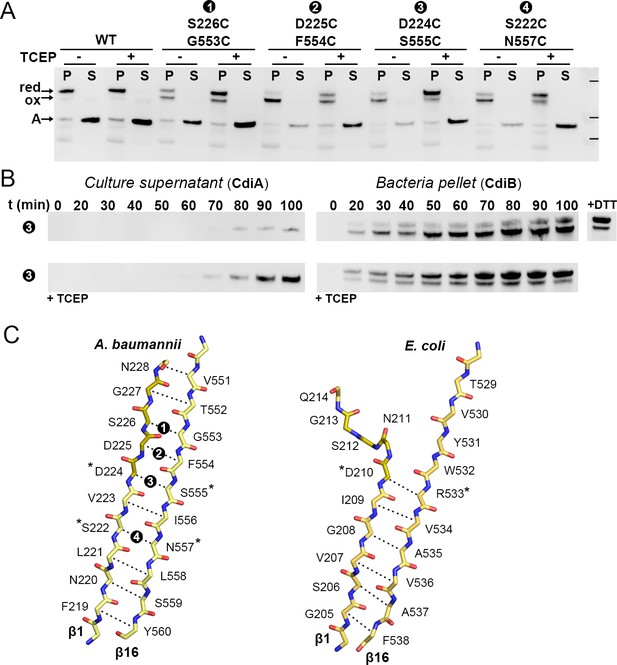
β1–β16 disulfide crosslinking in CdiBAb.
(A) CdiBAb cysteine variants are co-expressed with CdiA-Nt substrate from Acinetobacter baumannii in presence + or absence - of reducing agent ‘TCEP’ in the culture. After induction, pellet ‘P’ and supernatant ‘S’ are separated and analyzed by western blot to detect the presence of CdiA-Nt ‘A’ and, CdiB without cross-linking ‘red’, or CdiB with β1–β16 crosslinked ‘ox’. Protein ladder bands indicate 70, 50, 40 kDa, respectively. (B) After induction (t0) the secretion of CdiA-Nt and the production of CdiBAb double cysteine D224C-S555C are monitored at 10 min intervals. Samples from culture supernatant and bacterial pellet are separated, analyzed by western blot, and the bands corresponding to CdiA-Nt (left) and CdiBAb (right) displayed. For comparison, reducing agent was added to the pellet fraction t100 (‘+DTT’) or incubated with the culture at t0 (‘+TCEP’). (C) Mainchain representation of β1–β16 of CdiB from A. baumannii (left) and E. coli (right) colored in yellow, or orange for the DxxG motif. Adjacent sidechains oriented in the same direction are indicated by dashed lines, where * indicate sidechains involved in H-bonding in the crystal structure. Black circles with white numbers indicate engineered disulfides.
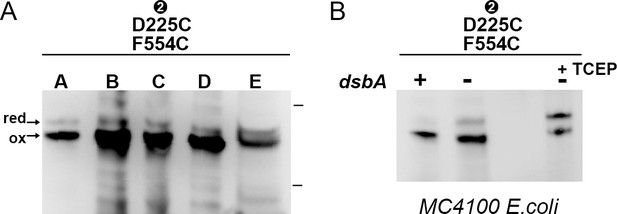
Expression of CdiBAb double cysteine variant.
(A) Western blot of isolated membrane fraction from CdiB D225C/F554C double cysteine variant (represented by bold letters A to E). Fraction A, bacterial pellet fraction (‘P ‘lane from Figure 5A). Fraction B, bacterial lysate fraction homogenized three times. Fraction C, supernatant fraction after centrifugation at 7000 g (where inclusion bodies and unlysed cells are removed). Fraction D, pellet from ultracentrifugation at 160,000 g to isolate membrane fractions after a 2% Triton wash (which removes soluble and inner membrane proteins). Membrane fractions are resuspended in 50 mM Tris-HCl, pH7.4, 200 mM NaCl, and solubilized by constant stirring in 5% Elugent. Fraction E, supernatant of a second ultracentrifugation at 220,000 g containing solubilized membrane proteins. As seen in Figure 5, β1–β16 are mostly crosslinked in the D225C-F554C mutant where ‘ox’ and ‘red’ indicate the oxidized and reduced form, respectively. Protein ladder bands indicate 70 and 50 kDa. (B) Western blot analysis of bacterial pellet from CdiBAb D225C/F554C variant expressed in MC4100 E. coli parent cell (dsbA ‘+’) and MC4100 E. coli dsbA:ChloroR strain (dsbA ‘-‘), in absence (second lane) or presence of TCEP (‘+TCEP’).
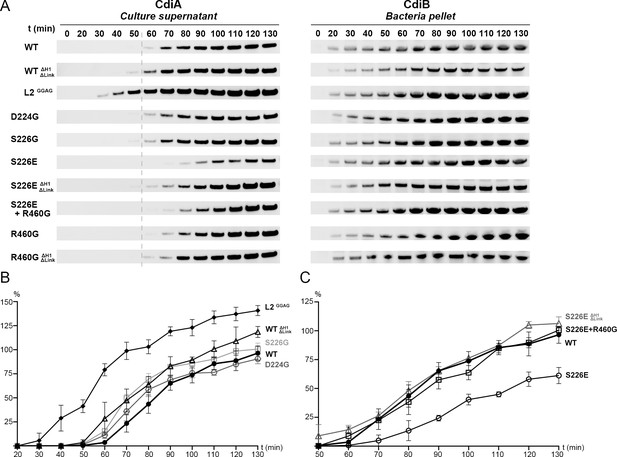
Analysis of CdiBAb secretion activity.
(A) Secretion of CdiA-NtAb 74-472 and expression of CdiBAb are monitored after induction (t0) up to 130 mins. Samples from culture supernatant and bacterial pellet were separated by centrifugation, analyzed by western blot, and the bands corresponding to CdiA-Nt (left) and CdiBAb variants (right) displayed. A gray dashed line at 60 min after induction indicates the reference point for the W.T where enough CdiA-Nt is present in the culture supernatant for immunoblot detection. (B), (C) Secretion activity was assessed by the levels of CdiA-NtAb 74-472 detected in culture supernatants normalized to the wildtype at t130 min (100%), repeated three times independently where error bars represent the standard error of the mean. (B), Secretion activity comparison from 20 to 130 min for CdiBAb wildtype (WT, ●), CdiB where H1 and Linker are genetically deleted (WTΔH1ΔLinker, Δ), Loop 2 substitutions GGAG (L2GGAG,♦) and single substitution variants D224G ( , gray), S226G (□, light gray). (C), Secretion activity comparison from 50 to 130 min between CdiBAb wildtype (WT, ●), single substitution S226E (○), S226E without H1 and Linker (S226EΔH1ΔLinker, Δ gray), and double substitution S226E+R460G (□).
, gray), S226G (□, light gray). (C), Secretion activity comparison from 50 to 130 min between CdiBAb wildtype (WT, ●), single substitution S226E (○), S226E without H1 and Linker (S226EΔH1ΔLinker, Δ gray), and double substitution S226E+R460G (□).
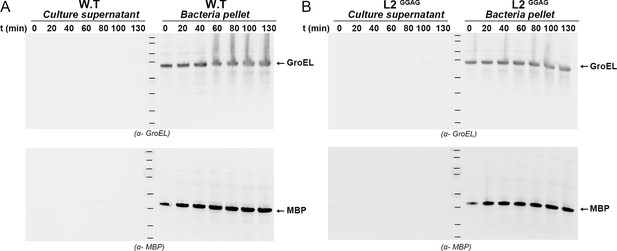
Western blot analysis of GroEL and MBP in wild type and L2GGAG cultures.
Western blot detection of GroEL and MBP from culture supernatant and bacterial pellet over 130 min after induction of CdiA-NtAb with CdiBAb wild type (A) or CdiBAb L2GGAG variant (B). Protein ladder bands indicate 180, 130, 100, 70, 55, 40, 35 and 25 kDa.
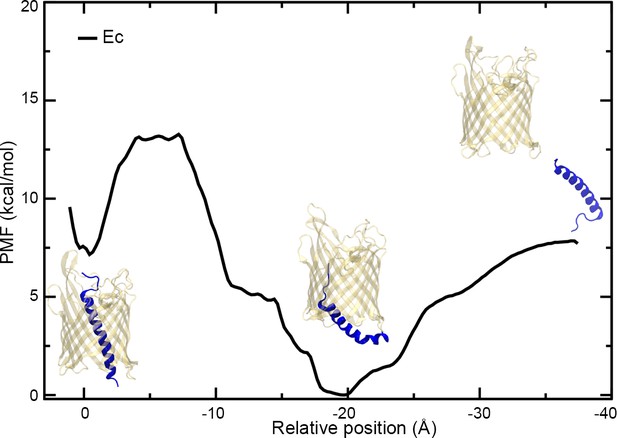
PMF of the extraction of H1 from the CdiBEc β-barrel.
Plot of the potential of mean force (PMF) measured as a function of the distance between the centers-of-mass of the α-helix H1 and β-barrel of CdiBEc. Three images of CdiBEc have been added to the plot representing different states, from the left to right: resting conformation, intermediate extraction, and H1 out of the pore.
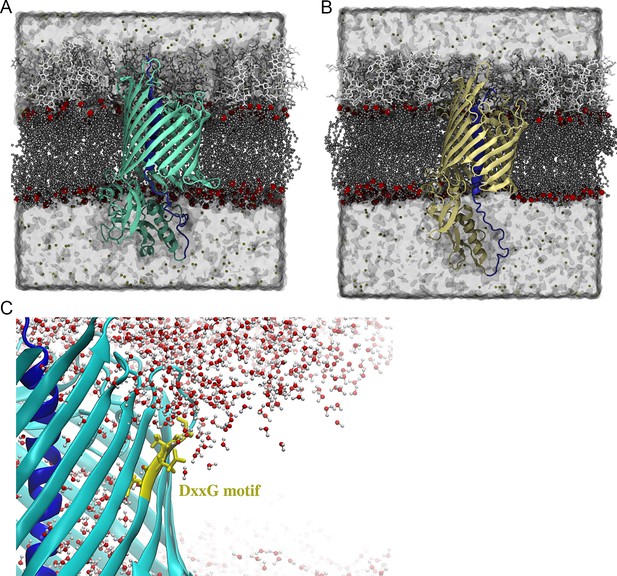
Initial states of the CdiBAb and CdiBEc simulations.
(A) CdiBAb was inserted into a bilayer that mimics the A. baumannii outer membrane. The outer leaflet is composed of type 1 and type 2 Lipid A with a ratio of 1:1 and the R1 core. (B) CdiBEc was inserted into an asymmetric bilayer that mimics the E. coli outer membrane. The outer leaflet is composed of type1 Lipid A with the R1 core. The inner leaflets of both membranes are composed of PPPE, PVPG and PVCL with a ratio of 15:4:1. The protein-membrane systems are surrounded by TIP3P water. The system size for the E. coli simulation is 110 × 115×125 Å3 and ~170,000 atoms, while the system size for the A. baumannii simulation is 115 × 125×125 Å3 and ~200,000 atoms. (C) Zoomed-in view of the DxxG motif from CdiBAb inserted into the outer membrane bilayer. Water molecules (in red and white) penetrate into the membrane to keep DxxG solvated.
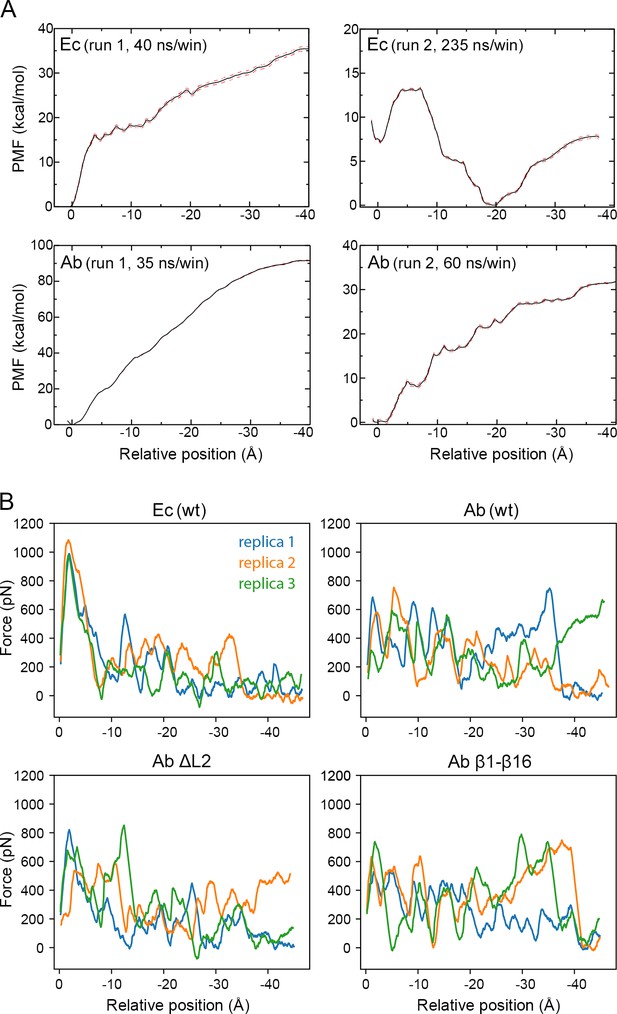
Extraction of H1 from the CdiB β-barrel.
(A) Plots of the results of four independent PMF calculations measured as a function of the distance between helix H1 and β-barrel of CdiBEc (top panels) and CdiBAb (lower panels). Statistical error (+ / - 1 standard deviation) is represented as a red dashed line. CdiBEc run two is also presented in Figure 7. (B) Force vs. position plots of the steered molecular dynamics simulations for H1 extraction from CdiBEc and CdiBAb structures (wt), from CdiBAb where Loop 2 is in-silico deleted (Ab ΔL2), and from CdiBAb double cysteine variants (Ab β1–β16). For each system, three independent replicas are presented (blue, orange, green). The main differences observed between the CdiBAb replicas is due to the formation of an ion-bridged interaction between H1 and D224 (DxxG) and D261 (loop 2). D261 is removed in the ΔL2 mutant, although we still see an ion-bridged interaction between H1 and E469 (loop 6) in one out of the three trajectories.
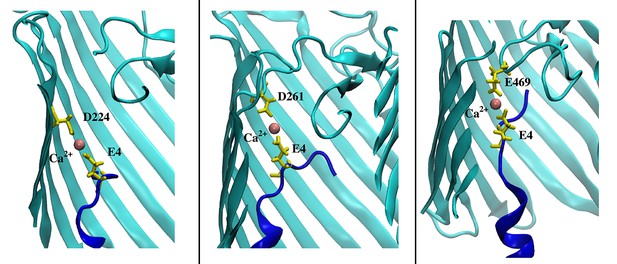
Examples of ion-bridged interactions in CdiBAb.
Left and middle images are taken from CdiBAb WT trajectories and show interactions between E4 and D224 (DxxG) and E4 and D261 (loop 2), respectively. The image on the right is taken from simulation of CdiBAb ΔL2 and shows an interaction between E4 and E469 (loop 6).
Videos
Morph of CdiBAb and CdiBEc crystal structures.
Linear interpolation morph of CdiBAb and CdiBEc crystal structures highlights the conformational differences observed. Helix H1 adopts a different angle and is positioned at a different height in the two structures. β1 twists inward, whereas loop 2 is oriented outward. The POTRA domains are relatively rigid. We note that although the length of the small α-helix in POTRA 1 differs between CdiBAb and CdiBEc, the mobility detected in the morph movie is probably an artefact due to the length.
Targeted Molecular Dynamic simulations of H1 in CdiBAb and CdiBEc models.
From the initial state: X-ray structures of CdiBAb (left) or CdiBEc (right), the helix H1 is displaced toward the position of the helix from CdiBEc helix (left) or CdiBAb (right).
Equilibrium simulation of CdiBAb and CdiBEc.
500 ns equilibrium simulations of three CdiBAb (light teal) and three CdiBEc (pale yellow) molecules. Full length CdiB proteins are inserted into their respective species-specific outer membranes (shown in Figure 7—figure supplement 1). Strands β1 and β16 are highlighted in yellow, and helix H1 in blue. The H-bonds between G227-T552 and I229-V534 are indicated for CdiBAb and CdiBEc, respectively. The POTRA domains and linker were present for the simulations but not shown in the final movies.
Extraction of CdiBAb helix H1 by Steered Molecular Dynamics.
SMD simulations of CdiBAb (light teal) where helix H1 is pulled toward the direction of the periplasm at a constant speed (0.29 Å/ns).
Extraction of CdiBEc helix H1 by Steered Molecular Dynamics.
SMD simulations of CdiBEc (pale yellow) where helix H1 is pulled toward the direction of the periplasm at a constant speed (0.29 Å/ns).
Tables
Data collection and refinement statistics for CdiBAb and CdiBEc.
| CdiB Ab (Se) | CdiB Ab | CdiB Ec | |
|---|---|---|---|
| Data collection | |||
| λ (Å) | 0.979415 | 1.0 | 1.0 |
| Space group | P1 | P1 | P21212 |
| a, b, c (Å) | 47 49.3 86.8 | 46.9 49.3 86.8 | 45.3 112.9 183.4 |
| α, β, γ (°) | 100.7 90.6 109.9 | 100.8 90.4 109.9 | 90 90 90 |
| Resolution (Å) | 50–2.6 | 50–2.4 | 50–2.6 |
| Rsym/Rmerge†* | 0.1 (1.4) | 0.1 (1.3) | 0.1 (1.3) |
| I / σ (I)* | 13.6 (1.4) | 10.1 (1.3) | 13.9 (2.0) |
| CC (1/2) (%)* | 0.998 (0.688) | 0.99 (0.619) | 0.99 (0.9) |
| Completeness (%)* | 90.8 (90.0) | 97.0 (82.4) | 99.9 (100) |
| Ano Completeness (%)* | 90.1 (89.5) | ||
| Redundancy* | 7.7 (7.7) | 5.8 (5.6) | 13 (13.5) |
| Refinement | |||
| Resolution (Å) | 44–2.4 | 44–2.6 | |
| No. reflections | 27153 | 29889 | |
| Rwork§/Rfree¶ | 0.20/0.25 | 0.24/0.26 | |
| r.m.s. deviations | |||
| Bonds (Å) | 0.003 | 0.002 | |
| Angles (°) | 0.61 | 0.58 | |
| No. Protein atoms | 4256 | 4130 | |
| No. Ligand atoms | 52 | 90 | |
| No. Waters | 55 | 13 | |
| B-factors (Å2) | |||
| Wilson B | 53.20 | 68.29 | |
| Protein | 59.6 | 79.1 | |
| Ligands | 69.1 | 81.2 | |
| Waters | 54.4 | 58.5 | |
| Ramachandran Analysis | |||
| Favored (%) | 98.3 | 97.18 | |
| Allowed (%) | 1.7 | 2.82 | |
| Outliers (%) | 0 | 0 | |
| PDB code | 6WIL | 6WIM | |
-
†Rsym = Σhkl,j (|Ihkl-<Ihkl > |) / Σhkl,j Ihkl, where < Ihkl > is the average intensity for a set of j symmetry-related reflections and Ihkl is the value of the intensity for a single reflection within a set of symmetry-related reflections.
§R factor = Σhkl (||Fo| - |Fc||) / Σhkl|Fo| where Fo is the observed structure factor amplitude and Fc is the calculated structure factor amplitude.
-
¶Rfree = Σhkl,T (||Fo| - |Fc||) / Σhkl,T|Fo|, where a test set, T (5% of the data), is omitted from the refinement.
* Statistics for highest resolution shell shown in parentheses.
Additional files
-
Supplementary file 1
Sequence alignment of TpsB transporters.
The first two sequences correspond to CdiBAb and CdiBEc studied in this paper, where the names of strains are indicated. Amino acid sequences from CdiB to BcpB correspond to proteins involved in CDI mechanisms, whereas the last nine sequences (FhaC to LepB) represent other TpsB transporters. α-helices and β-strands are shown by cylinders and arrows respectively, from the structures of CdiBAb (light teal) and CdiBEc (pale yellow). The boundaries of POTRA domains are indicated in gray. Structural elements discussed in the text and figures are indicated in bold black, and important residues are marked with a star and label, in light teal for CdiBAb and pale yellow for CdiBEc. Omp85/TpsB conserved residues on L6, β13, β1 and cysteines (involved in disulfide bond formation) are shown in red.
- https://cdn.elifesciences.org/articles/58100/elife-58100-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58100/elife-58100-transrepform-v2.pdf





