Molecular rationale for antibody-mediated targeting of the hantavirus fusion glycoprotein
Figures
Composition and neutralization potency of recombinantly-derived bank vole mAb P-4G2.
(A) Composition of the complementarity-determining regions (CDRs) of the mAb P-4G2 antigen-binding fragment (Fab) heavy (VH) and kappa (VK) chains. For the full sequence of Fab P-4G2 variable regions, please see Figure 1—figure supplement 1. (B) A hantavirus-pseudotyped VSV-ΔG RFP neutralization assay shows that recombinantly produced mAb P-4G2 neutralizes Puumala virus- and Andes virus-pseudotyped VSV (black and blue traces, respectively), but not Hantaan virus-pseudotyped VSV (gray trace). Each neutralization assay was carried out three times in duplicate. A representative experiment is shown. Error bars represent the range of the value for the experiment performed in duplicate.
Sequence alignment of antibody variable regions from bank vole mAb P-4G2 and a representative mouse antibody.
Fab P-4G2 variable regions with variable regions from representative mouse (Mus musculus) antibodies. Mouse homologues used in the alignment were identified by NCBI BLAST (Altschul et al., 1990; Johnson et al., 2008). The P-4G2 heavy chain variable region (VH) was aligned with the VH sequence from mouse IgG (GenBank: BAN14007.1). The P-4G2 light chain variable region (VK) was aligned with the VK sequence from mouse IgG (GenBank: ATI98427.1). Complementarity-determining regions (CDRs) of P-4G2 are annotated under the alignment.
Crystal structure of neutralizing antibody P-4G2 in complex with Puumala virus (PUUV) Gc.
(A) Crystal structure of Fab P-4G2−PUUV Gc complex at 3.5 Å resolution. PUUV Gc, a class II fusion protein, comprises domains I−III (colored red, yellow, and blue, respectively), a Gc C-terminal tail (light blue) and the viral fusion loop at Ser771–Thr785 (orange). Fab P-4G2, comprised of a heavy chain (dark gray) and a light chain (white), is observed bound at the junction of domains I and II on PUUV Gc. A domain schematic of the PUUV glycoprotein precursor with the signal peptide (SP), transmembrane domains (TM), intra-virion domain (IV), and WAASA signal peptidase cleavage site is shown alongside the construct used in crystallization (schematic was produced using the DOG software [Ren et al., 2009]). N-linked glycosylation sequons are shown in the domain schematic as pins and the N-linked glycan observed at Asn937 is rendered as sticks. There was no evidence of glycosylation at Asn898. (B) A close-up view of the central portion of the Fab P-4G2-PUUV Gc interface around paratope residue Arg100 of the Fab P-4G2 CDRH3. Residues contributing to a putative hydrogen bonding network (indicated by dashed lines) are rendered as sticks and were identified using the PDBePISA server (Krissinel and Henrick, 2007). (C) Site-directed mutagenesis of Arg100 to Ala reduces the neutralizing potency of mAb P-4G2, confirming the importance of this residue in the interaction interface. We compared the neutralization potency of mAb P-4G2 R100A mutant (magenta trace) to that of the wild-type mAb P-4G2 (black trace) against PUUV pseudovirus in a hantavirus-pseudotyped VSV-ΔG RFP neutralization assay. The virus neutralization assay presented in panel C was conducted as a part of the same experiment as the neutralization assay presented in Figure 1B. Each neutralization assay was carried out three times in duplicate. A representative experiment is shown. Error bars represent the range of the value for the experiment performed in duplicate.
Electron density at the Fab P-4G2−Puumala virus (PUUV) Gc interface.
Stereo-view of the interaction between PUUV Gc and CDRH3 of Fab P-4G2, with the simulated annealing composite omit electron density map shown contoured at 1σ. The structure is shown in stick representation, with nitrogen atoms colored blue, oxygen atoms red, and carbon atoms yellow for Gc, salmon for Fab P-4G2 CDRH2, and magenta for Fab P-4G2 CDRH3, respectively. Arginine 100 of CDRH3 and residues Ile726 and Glu725 of Gc, which are predicted by the PDBePISA server to form hydrogen bonds and a salt bridge with Arg100, are labeled.
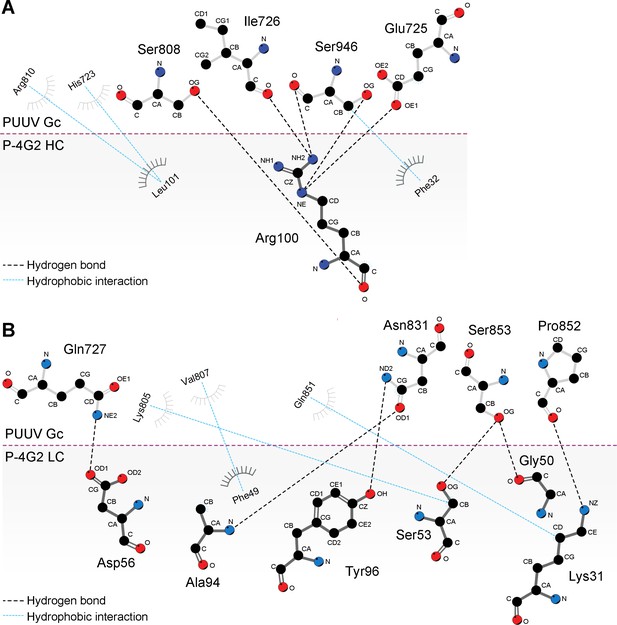
Key interactions at the Fab P-4G2−Puumala virus Gc complex interface.
Paratope and epitope residues involved in hydrogen bonding (dashed black lines) for contacts made by the heavy (A) and light (B) chains were identified using the PDBePISA server (Krissinel and Henrick, 2007). Residues involved in hydrophobic interactions (dashed blue lines) were identified with LigPlot+ (Laskowski and Swindells, 2011). The interface was visualized using LigPlot+. Residues contributing to hydrophobic interactions are shown as curved lines with dashes. Atoms in residues participating in hydrogen bonding are depicted as circles, with oxygen in red, carbon in black, and nitrogen in blue.

Sequence conservation at the mAb P-4G2 epitope.
A sequence alignment of Gc glycoproteins from Puumala virus (PUUV; CAB43026.1), Andes virus (ANDV; AAO86638.1), and Hantaan virus (HTNV; AIL25319.1) numbered according to PUUV Gc. In the alignment, fully conserved residues are shown white on a red background, partially conserved residues red on a white background, and variable residues are rendered black and blue. Residues comprising the mAb P-4G2 epitope were identified using the PDBePISA server (Krissinel and Henrick, 2007) and are demarcated by dark gray bars above the alignment. The relatively high level of similarity between PUUV and ANDV is in line with the ability of mAb P-4G2 to cross-neutralize ANDV-pseudotyped VSV (Figure 1).
The epitope of antibody P-4G2 denotes a key antigenic site at the hantaviral surface.
(A) Puumala virus (PUUV) Gc from the Gc-4G2 complex displays a domain III conformation distinct from that observed in (B) post-fusion PUUV Gc (Willensky et al., 2016). (C) Fab P-4G2 binding precludes the rearrangement of domain III (blue arrow) to the post-fusion conformation. PUUV Gc is represented as a surface, with the epitope of Fab P-4G2 outlined dark gray. Fab P-4G2 is shown in ribbon representation, with heavy and light chains colored gray and white, respectively. (D) Top view of the P-4G2 epitope shows that it overlaps with the neutralization evasion mutation sites reported for PUUV-neutralizing human antibody 1C9 (Hörling and Lundkvist, 1997; Lundkvist et al., 1993a) and HTNV-neutralizing mouse antibodies HC02 and 16E6 (Arikawa et al., 1989; Wang et al., 1993). Furthermore, the epitope overlaps with the binding site of domain III in the post-fusion conformation (medium blue). Interfacing residues were identified using the PDBePISA server (Krissinel and Henrick, 2007).
Overlay analysis of viral class II fusion proteins with Puumala virus (PUUV) GcGc-P-4G2 indicates that PUUV Gc has crystallized in a conformation similar to the pre-fusion conformation.
(A) The conformation of PUUV GcGc-P-4G2 is distinct from (B) the previously reported PUUV Gc post-fusion structure (PDB 5J9H) (Willensky et al., 2016), and from (C) the intermediate configuration of HTNV Gc (PDB 5LJY) (Guardado-Calvo et al., 2016), and aligns more closely with (D) the recently published prefusion Andes virus (ANDV) Gc, crystallized in complex with cognate ANDV Gn (PDB 6Y5F) (Serris et al., 2020), and (E) the experimentally confirmed pre-fusion conformations of RVFV Gc (PDB 4HJ1) (Dessau and Modis, 2013) and (F) Zika E (PDB 5IRE) (Sirohi et al., 2016). Cartoon representations of the structure are colored according to domain boundaries (red, yellow, and blue for domains I, II, and III, respectively). Dotted arrows highlight conformational differences between overlaid structures.
Treatment of Puumala virus (PUUV) virus-like particles (VLPs) with Fab P-4G2 results in additional density and is associated with loss of continuous lattice at the VLP surface.
Cryo-ET reconstructions of the Fab P-4G2-treated PUUV VLP surface, derived from (A) regions of continuous lattice (14.3 Å) and (B) regions of incomplete lattice (13.4 Å). While both reconstructions show the canonical Gn−Gc architecture (density colored white) and the viral lipid bilayer (light blue), additional density is observed in the latter reconstruction (dark gray). (C) The hantaviral surface carries tetragonal (Gn−Gc)4 spikes that can organize in patches of ordered lattice. (D) A box plot describing the frequency of (Gn−Gc)4 spikes that have a given number of lattice compatible neighbors, from zero to a maximum of eight, shows that treatment with Fab P-4G2 alters the presentation of the (Gn−Gc)4 spike assemblies at the VLP surface.
The Puumala virus (PUUV) virus-like particle (VLP) surface displays ordered regions of glycoprotein lattice and is congruent with previously published reconstructions.
Size-distribution plot of the VLPs (upper right corner) shows that the pleomorphic particles vary in size, with most particles measuring between 80 and 100 nm in the longest dimension. Despite the variable resolutions of the present reconstructions of hantaviral surfaces, the tetragonal lattice organization observed in PUUV VLP (13.9 Å) and in PUUV VLP treated with P-4G2 (14.3 Å) is similar to that of Tula virus (TULV, 15.6 Å, EMDB-4867) (Li et al., 2016) and Hantaan virus (HTNV, 25 Å, EMDB-2056) (Battisti et al., 2011). A top view of each viral surface reconstruction is shown, and densities corresponding to viral membrane and envelope glycoproteins are rendered in light blue and shades of gray and white, respectively.

Treatment with Fab P-4G2 alters the presentation of the hantaviral glycoprotein lattice at the surface of Puumala virus (PUUV) virus-like particles (VLPs).
Tomographic slices of the selected PUUV VLPs (left) and corresponding Mercator projections (right) describing the positions of (Gn−Gc)4 spikes on VLP surfaces are shown (A) in the absence and (B) presence of Fab P-4G2. The subset of spike positions that belong to regular patches (determined using the PatchFinder script; please see main Methods) are displayed in black and the remainder in gray. Units shown are in nanometers and scale bars are 50 nm.
Fitting of the Fab P-4G2−Puumala virus (PUUV) Gc crystal structure into the cryo-ET reconstruction confirms the P-4G2 epitope in the context of the viral surface and supports the hypothesis that the observed Gc conformation constitutes a pre-fusion state.
(A) Side view and (B) top view of the Fab P-4G2-treated PUUV virus-like particle (VLP) spike at 13.4 Å resolution. Crystal structures of Fab P-4G2−PUUV, along with PUUV Gn (PDB id 5FXU), were fitted in the cryo-ET reconstruction as rigid bodies and display excellent conformity with the cryo-ET derived envelope. (C) A zoom-in of the fit of domain III of Gc into the cryo-ET reconstruction presented from two points of view, which are related by a 180° rotation. The goodness of fit supports the hypothesis that domain III has crystallized in a conformation that closely resembles the pre-fusion state presented in the mature hantaviral spike. (D) A close-up of Fab P-4G2 similarly presents a high-correlation fit. A simplified look at the composition of the hantaviral spike, where fitted Gc and Gn are represented as surfaces, is shown from side view (E) and top view (F).
Fitting of Puumala virus (PUUV) Gn and Gc into the cryo-ET-derived reconstruction of PUUV virus-like particles (VLPs) shows that lattice formation is mediated by Gc homo-dimers.
(A) Top- and side views of the PUUV lattice. Crystal structures of PUUV Gc from the Fab P-4G2 complex, and PUUV Gn (PDB id: 5FXU) are shown fitted into a cryo-ET reconstruction of the PUUV VLP surface at 13.9 Å resolution. PUUV Gn and Gc are shown in cartoon representation and colored as presented at the bottom of panel C. (B) A zoom-in of an individual spike reveals that contacts between spikes are mediated by Gc. (C) The fitting identifies the topology of Gc dimers formed between neighboring spikes. Gc domain I is indicated to be involved in the formation of the dimer interface, corroborating the model reported by Bignon et al. (Bignon et al., 2019) based on a crystallographically observed HTNV Gc homo-dimer (Guardado-Calvo et al., 2016).
Domain I mediates dimer contacts at the Gc–Gc interface.
(A) Gc homodimer interface generated by fitting Puumala virus (PUUV) GcGc-P-4G2 into a cryo-ET reconstruction of PUUV virus-like particle (VLP) surface at 13.9 Å resolution (see also Figure 6C). Within the constraints of low-resolution fitting, the interface matches with the homodimerization interface from the (B) crystallographically observed HTNV Gc dimer (Guardado-Calvo et al., 2016). The twofold symmetry axis is indicated by an oval.
Fab P-4G2 epitopes from neighboring Puumala virus (PUUV) Gc proteins within the Gn−Gc lattice are in close proximity.
Top (panel A) and side (panel B) view representations of the PUUV Gn–Gc surface at the Fab P-4G2 epitope are shown. The surface of PUUV Gn and Gc are colored according to the legend at the bottom of the figure. Neighboring Fab P-4G2 epitope residues were identified using the PDBePISA server (Krissinel and Henrick, 2007) and are highlighted in red (‘epitope a.’) and blue (‘epitope b.’). A ribbon representation of Fab P-4G2 bound to epitope b is shown in the side view representation and indicates that lattice-incorporated, neighboring Gc glycoproteins are unlikely to be able to sterically accommodate more than one Fab molecule in this region.
A representative example of a hantaviral lattice “break point” displayed as a Mercator projection.
Points colored blue represent points that were manually picked and aligned to generate the map in Author response image 2. This figure is adapted from Figure 4—figure supplement 2.

A reconstruction of a hantaviral spike using particles manually picked from “break points” in the lattice.
The resulting map displays an empty patch of membrane where an 8th neighbour would normally be expected as indicated with a dashed circle. Additional density that likely corresponds to fab P-4G2 is indicated with an asterisk. This reconstruction is generated from 160 manually picked and aligned particles and displayed at a resolution of 50 Å for clarity.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Puumala orthohantavirus) | Glycoprotein precursor (GPC); used in recombinant Gc production | GenBank | CAB43026.1 | Synthetic cDNA was produced by GeneArt, Life Technologies |
| Gene (Puumala orthohantavirus) | Glycoprotein precursor (GPC); used in PUUV VLP production | GenBank | CCH22848.1 | Synthetic cDNA was produced by GeneArt, Life Technologies |
| Gene (Hantaan orthohantavirus) | Glycoprotein precursor (GPC) | GenBank | AIL25321.1 | Synthetic cDNA was produced by GeneArt, Life Technologies |
| Gene (Andes orthohantavirus) | Glycoprotein precursor (GPC) | GenBank | AAO86638.1 | Synthetic cDNA was produced by GeneArt, Life Technologies |
| Strain, strain background (Escherichia coli) | Subcloning Efficiency DH5α Competent Cells | Thermo Fisher Scientific | Cat#: 18265017 | Competent cells |
| Cell line (Homo-sapiens) | Human embryonic kidney HEK 293T | ATCC | CRL-3216 | |
| Cell line (Homo-sapiens) | Human embryonic kidney HEK293F | Thermo Fisher Scientific | Cat#: R79007 | |
| Biological sample (Myodes Glareolus, Mus musculus) | Bank vole-mouse heterohybridoma producing mAb P-4G2 | Lundkvist and Niklasson, 1992 | ||
| Biological sample (Indiana vesiculovirus) | VSV-ΔG RFP | Reynard and Volchkov, 2015 | ||
| Recombinant DNA reagent | pHLsec plasmid | Aricescu et al., 2006 | ||
| Recombinant DNA reagent | pHLsec-8H-SUMO-1D4 plasmid | Chang et al., 2015 | ||
| Recombinant DNA reagent | pgk-φC31/pCB92 plasmid | Chen et al., 2011 | ||
| Recombinant DNA reagent | pURD plasmid | Zhao et al., 2014 | ||
| Recombinant DNA reagent | Tim1/pCAGGs plasmid | Watt et al., 2014 | ||
| Recombinant DNA reagent | pCAGGS plasmid | Niwa et al., 1991 | ||
| Recombinant DNA reagent | Mouse IgG1 plasmid | von Boehmer et al., 2016 | ||
| Recombinant DNA reagent | Mouse IgK plasmid | von Boehmer et al., 2016 | ||
| Recombinant DNA reagent | Fab P-4G2 light chain synthetic DNA fragment | This study | Synthetic cDNA was produced by GeneArt, Life Technologies | |
| Recombinant DNA reagent | Fab P-4G2 heavy chain synthetic DNA fragment | This study | Synthetic cDNA was produced by GeneArt, Life Technologies | |
| Sequence-based reagent | Mouse IgG sequencing primers | von Boehmer et al., 2016 | List of primer sequences is provided in the referenced study | |
| Sequence-based reagent | PUUV Gc ectodomain cloning primer, forward | This study | CGCACCGGTGAGACACAGAACCTGAACAGCGGC | |
| Sequence-based reagent | PUUV Gc ectodomain cloning primer, reverse | This study | GCGGTACCCTCGCCGGACTTGGTGAACC | |
| Sequence-based reagent | Fab P-4G2 HC R100A mutagenesis primer, forward | This study | GTATTACTGTACAAGAGATGCATTAGGCCCTTTTGA | |
| Sequence-based reagent | Fab P-4G2 HC R100A mutagenesis primer, reverse | This study | TCAAAAGGGCCTAATGCATCTCTTGTACAGTAATAC | |
| Commercial assay or kit | Phusion High-Fidelity PCR Master Mix with HF Buffer | New England Biolabs | Cat#: M0531S | |
| Commercial assay or kit | SuperScript III reverse transcriptase | Thermo Fisher Scientific | Cat#: 18080093 | |
| Commercial assay or kit | Quick Ligation kit | New England Biolabs | Cat#: M2200S | |
| Chemical compound, drug | PEI Max 40K | Polysciences, Inc | Cat#: 24765–1 | |
| Chemical compound, drug | PEI | Polysciences, Inc | Cat#: 23966–1 | |
| Chemical compound, drug | Lipofectamine 2000 Transfection reagent | Thermo Fisher Scientific | Cat#: 11668027 | |
| Chemical compound, drug | Kifunensine | Cayman Chemical | Cat#: 10009437 | |
| Software, algorithm | XIA2 | Winter, 2010 | ||
| Software, algorithm | CCP4 | Potterton et al., 2003 | ||
| Software, algorithm | SWISS-MODEL | Waterhouse et al., 2018 | ||
| Software, algorithm | Coot | Emsley and Cowtan, 2004 | ||
| Software, algorithm | REFMAC | Murshudov et al., 1997 | ||
| Software, algorithm | PHENIX | Adams et al., 2002 | ||
| Software, algorithm | Molprobity | Davis et al., 2007 | ||
| Software, algorithm | IMGT/V-QUEST server | Brochet et al., 2008 | ||
| Software, algorithm | GraphPad Prism | GraphPad Software, San Diego, CA, USA | ||
| Software, algorithm | PDBePISA server | Krissinel and Henrick, 2007 | ||
| Software, algorithm | PyMOL | The PyMOL Molecular Graphics System, Schrödinger, LLC | ||
| Software, algorithm | UCSF Chimera | Pettersen et al., 2004 | ||
| Software, algorithm | LigPlot+ software | Laskowski and Swindells, 2011 | ||
| Software, algorithm | Motioncor2 | Zheng et al., 2017 | ||
| Software, algorithm | CTFFIND4 | Rohou and Grigorieff, 2015 | ||
| Software, algorithm | tomo_preprocess script | This study | ||
| Software, algorithm | IMOD | Mastronarde and Held, 2017 | ||
| Software, algorithm | Dynamo | Castaño-Díez et al., 2012 | ||
| Software, algorithm | PatchFinder script | This study | ||
| Other | Chromatography column, Superdex 200 10/300 Increase | Cytiva | Cat#: 28990944 | |
| Other | Chromatography column, HisTrap FF Crude 5 ml | Cytiva | Cat#: 17528601 | |
| Other | 1-MDa cut-off dialysis membrane | Spectrum Chemical | ||
| Other | Holey carbon grids, 2 μm hole diameter | Protochips |
Additional files
-
Supplementary file 1
Table S1: Crystallographic data collection and refinement statistics for Fab P-4G2−Puumala virus Gc.
Table S2. Cryo-EM tomography data collection, sub-tomogram reconstruction, and fitting statistics.
- https://cdn.elifesciences.org/articles/58242/elife-58242-supp1-v1.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58242/elife-58242-transrepform-v1.docx





