Homology-guided identification of a conserved motif linking the antiviral functions of IFITM3 to its oligomeric state
Figures
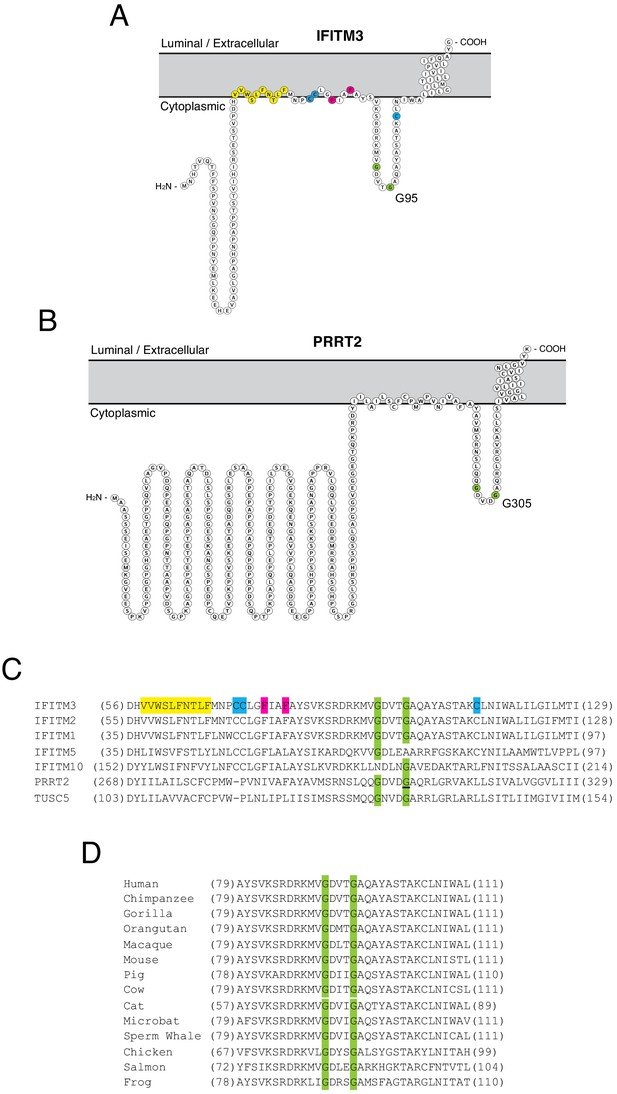
Homology-guided identification of a putative oligomerization motif within CD225 domains.
(A) Schematic representation of the membrane topology of IFITM3 made with Protter. Residues corresponding to the amphipathic helix (yellow), palmitoylated cysteines (blue), phenylalanines purported to regulate oligomerization (red), and the glycines of the GxxxG motif (green) are indicated. (B) Schematic representation of the membrane topology of PRRT2 made with Protter. Residues corresponding to the glycines of the GxxxG motif (green) are indicated. (C) A partial amino acid alignment of CD225 domains from IFITM proteins, PRRT2, and TUSC5. Color codes are included as in (A and B). The position of the polymorphic glycine in PRRT2 associated with neurological disease (G305W) is underlined. (D) A partial amino acid alignment of IFITM3 orthologs in vertebrates. Conserved glycines in the GxxxG motif (green) are indicated.
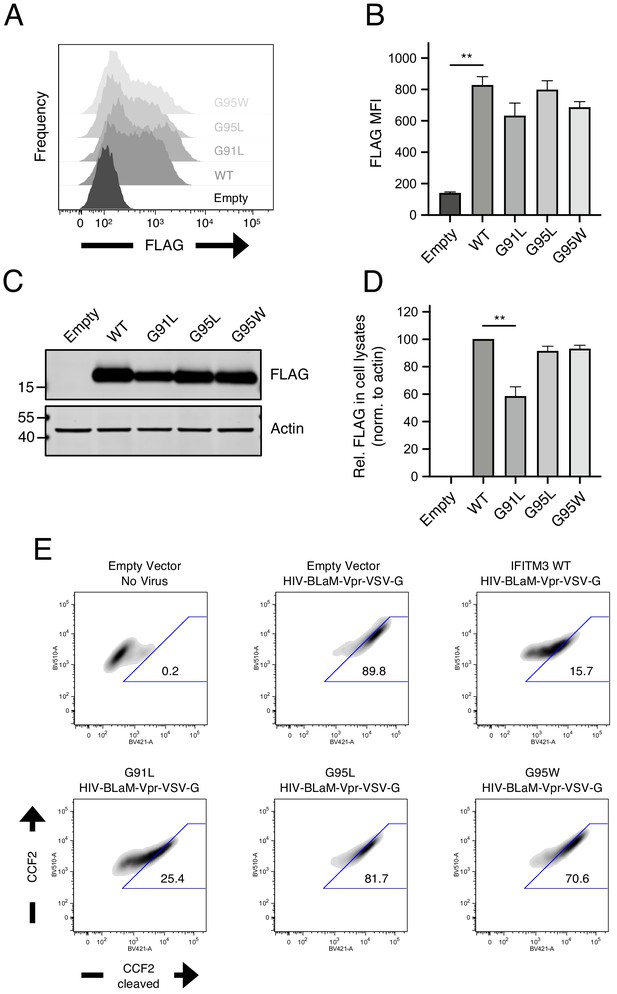
Quantitative measurement of IFITM3 construct expression following transient transfection and flow cytometric analysis of virus-cell fusion.
(A) HEK293T were transiently transfected with 1.5 µg of empty pQCXIP, IFITM3 WT-FLAG, or the indicated mutants and cells were fixed 48 hr later, stained with anti-FLAG, and assessed by flow cytometry. (B) Mean fluorescence intensity measurements of FLAG staining in (A) are shown for three independent experiments. (C) SDS-PAGE and western blot analysis of whole cell lysates produced from HEK293T transiently transfected with 1.5 µg empty pQCXIP, WT IFITM3-FLAG, or mutants and lysed 48 hr later. Immunoblotting was performed with anti-FLAG. Actin served as a loading control. Number and tick marks indicate size (kilodaltons) and position of protein standards in ladder. (D) FLAG signal was quantified by measuring fluorescence of DyLight-conjugated secondary antibody and was normalized to that of Actin in three independent experiments. Normalized FLAG signal was shown relative to IFITM3 WT (set to 100). (E) An example of flow cytometry dot plots from a single experiment of data summarized in Figure 2E. Error bars indicate standard error. Rel., relative. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. MFI, mean fluorescence intensity. Rel., relative. Norm., normalized.
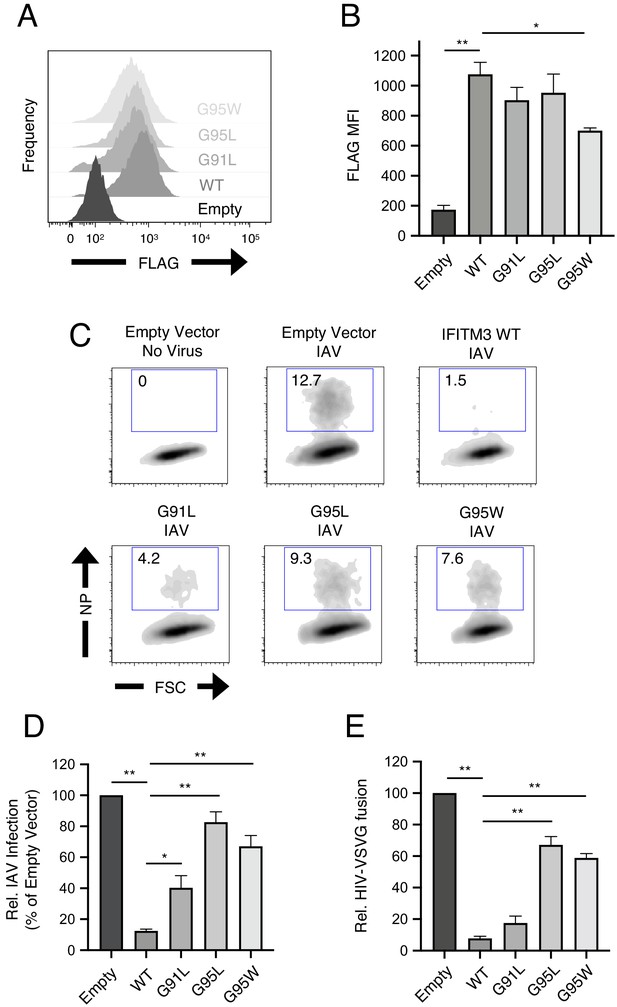
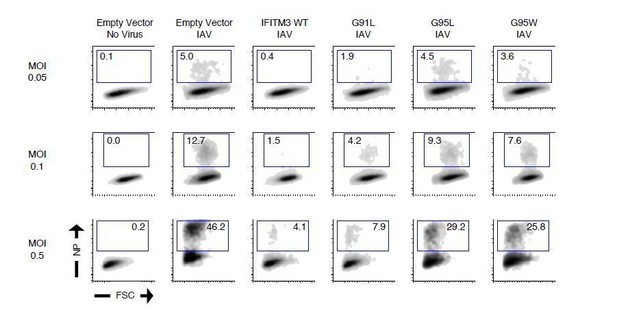
91GxxxG95 is important for restriction of virus entry by IFITM3.
(A) HEK293T cells stably transfected with empty pQCXIP, IFITM3 WT-FLAG, or the indicated mutants were fixed, stained with anti-FLAG antibody and assessed by flow cytometry. FLAG levels are displayed as histograms. (B) Mean fluorescence intensity measurements of FLAG staining in (A) are shown for three independent experiments. (C) HEK293T cells stably transfected with empty pQCXIP, IFITM3 WT-FLAG, or the indicated mutants were challenged with IAV PR8 strain (MOI of 0.1), fixed at 18 hr post-infection, stained with an anti-nucleoprotein antibody, and assessed by flow cytometry. (D) Mean infection results representing 5–8 independent experiments are normalized to empty vector (set to 100%). (E) HEK293T cells stably transfected with empty pQCXIP, IFITM3 WT-FLAG, or the indicated mutants were challenged with replication-incompetent HIV-1 incorporating BlaM-Vpr and pseudotyped with VSV glycoprotein. Virus-cell fusion was assessed at 2.5 hr post-virus addition using the beta lactamase assay and flow cytometry. Results represent the mean of three independent experiments and are normalized to empty vector (set to 100%). Error bars indicate standard error. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. MFI, mean fluorescence intensity. Rel., relative.
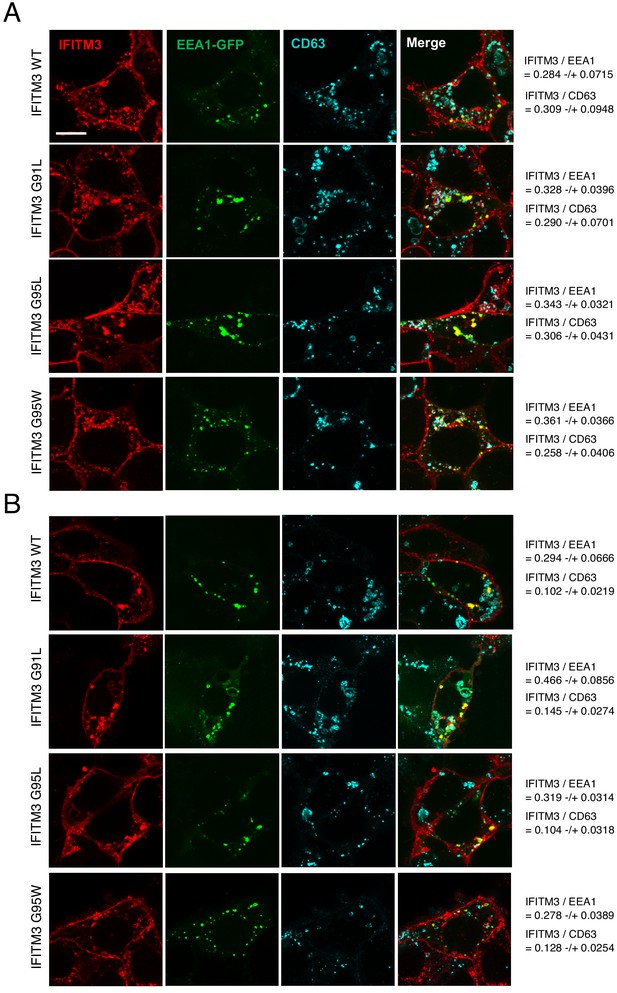
The subcellular localization of IFITM3 WT and mutants as measured by confocal immunofluorescence microscopy.
(A) HEK293T cells stably expressing IFITM3 constructs were transfected with 0.2 micrograms of EEA1-GFP and cells were fixed at 48 hr post-transfection, permeabilized, and immunostained with anti-IFITM3 and anti-CD63 and cells were analyzed by immunofluorescence confocal microscopy. Scale bars, 10 μm. (B) HEK293T cells were transiently transfected with 0.2 µg of pQCXIP-IFITM3 WT-FLAG or the indicated mutants and 0.2 µg of EEA1-GFP and cells were fixed at 48 hr post-transfection, permeabilized, and immunostained with anti-IFITM3 and anti-CD63 and cells were analyzed by immunofluorescence confocal microscopy. All images represent mean intensity averages (Z-stacks) from five consecutive medial sections. Scale bars, 10 μm. Colocalization between IFITM3 and EEA1-GFP and IFITM3 and CD63 was measured using Imaris software. The mean Pearson’s correlation coefficient values and standard deviations were calculated in each condition from at least 10 cells displaying EEA1-GFP signal.
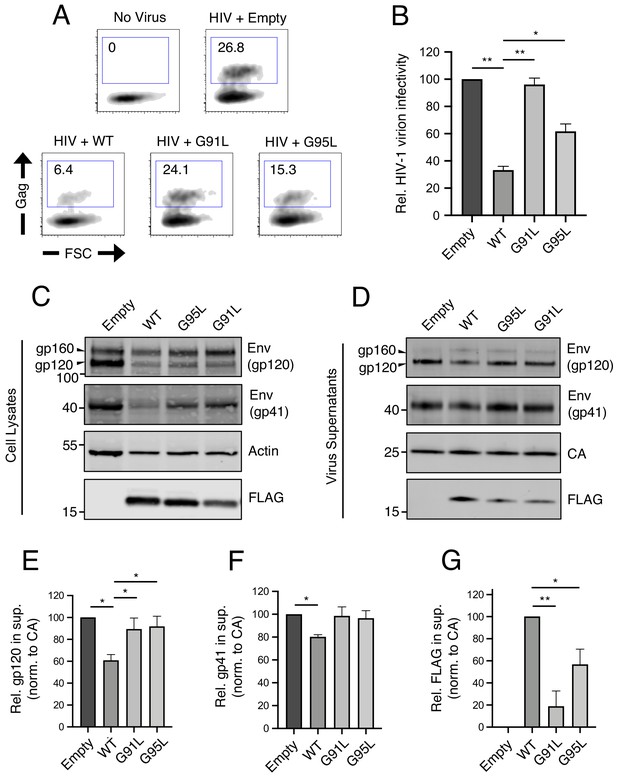
91GxxxG95 is important for restriction of HIV-1 virion infectivity by IFITM3.
(A) HEK293T were co-transfected with HIV-1 molecular clone pNL4.3 and empty pQCXIP, IFITM3 WT-FLAG, or the indicated mutant. Virus-containing supernatants were harvested at 48 hr post-transfection and subjected to ultracentrifugation over sucrose pellets. Virion content was quantified by p24 CA ELISA and 50 ng p24 equivalent was added to TZM.bl cells for infectivity measurements. TZM.bl were fixed at 48 hr post-infection, stained with anti-CA antibody, and infection was assessed by flow cytometry. (B) Mean infection results of TZM.bl using virus derived from three independent transfections of HEK293T are shown and normalized to empty vector (set to 100%). (C) Whole cell lysates and (D) virus-containing supernatants were collected from HEK293T co-transfected with the HIV-1 molecular clone pNL4.3 and empty pQCXIP, IFITM3 WT-FLAG, or the indicated mutant at 48 hr post-transfection. Virus-containing supernatants were ultracentrifuged through sucrose cushions. Both lysates and concentrated, purified virus-containing supernatants (50 ng p24 equivalent) were subjected to SDS-PAGE and Western blot analysis. Immunoblotting was performed with anti-gp120, anti-gp41, anti-CA, anti-actin, and anti-FLAG. (E) Virion-associated levels of gp120 Env were quantified by measuring fluorescence of DyLight-conjugated secondary antibody and were normalized to levels of CA in three independent experiments. (F) Virion-associated levels of gp41 Env and (G) IFITM3-FLAG were quantified similarly. For anti-Env immunoblotting, the amount of gp120 or gp41 in virions was presented relative to empty vector (set to 100%). For anti-FLAG immunoblotting, the amount of IFITM3 WT in virions was set to 100%. Error bars indicate standard error. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. Rel., relative. Norm., normalized.
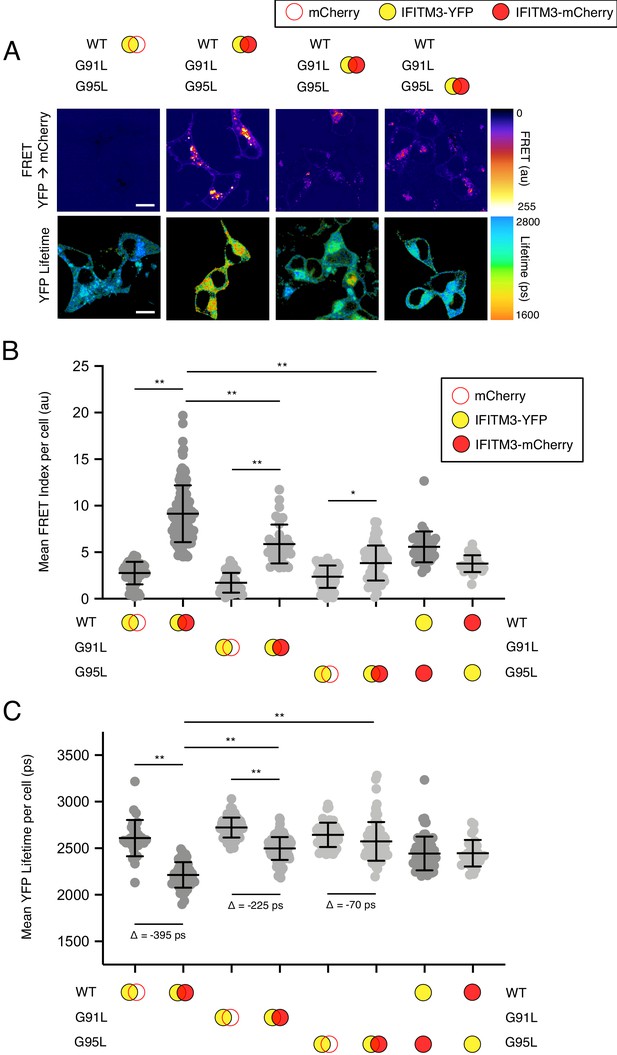
91GxxxG95 regulates oligomerization of IFITM3 in living cells.
(A) HEK293T were transiently co-transfected with IFITM3-YFP and mCherry or IFITM3-YFP and IFITM3-mCherry. Constructs encoded IFITM3 WT, IFITM3 G91L, or IFITM3 G95L. FRET-FLIM measurements were made, and images of FRET signal and YFP lifetimes are representative of 12–20 captured images per condition. (B) Whole-cell FRET analysis was performed on a minimum of 50 cells per condition and the results of three independent experiments were pooled. Dots correspond to individual cells. (C) Whole-cell YFP lifetimes were measured on a minimum of 50 cells per condition and the results of three independent experiments were pooled. Dots correspond to individual cells. A mean delta (Δ) value is indicated to represent the drop in YFP lifetime resulting from the pairing of IFITM3-YFP and mCherry versus the pairing of IFITM3-YFP and IFITM3-mCherry. Empty red circles are used to depict mCherry, filled red circles are used to depict IFITM3-mCherry (either WT, G91L, or G95L), and filled yellow circles are used to depict IFITM3-YFP (either WT, G91L, or G95L). Error bars indicate standard deviation. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. Scale bars, 10 μm. Ps, picoseconds. Au, arbitrary units.
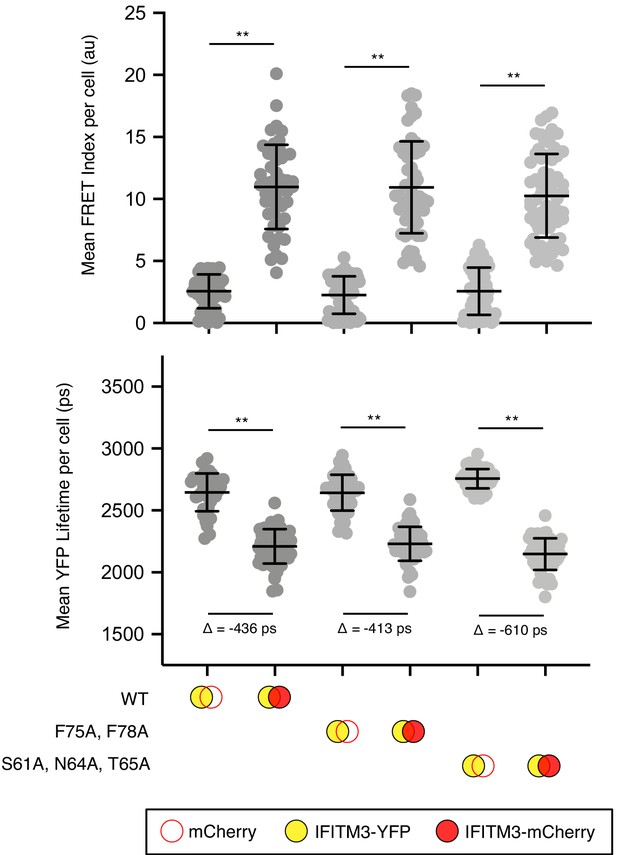
Assessment of additional IFITM3 mutations on oligomerization measured by FRET-FLIM.
HEK293T were transiently co-transfected with IFITM3-YFP and mCherry or IFITM3-YFP and IFITM3-mCherry (fluorescent proteins were tagged at the amino-terminus). Constructs encoded IFITM3 WT, IFITM3 F75A/F78A, or IFITM3 S61A/N64A/T65A. FRET-FLIM measurements were made, and images of FRET signal and YFP lifetimes are representative of 12–20 captured images per condition. (Top) Whole-cell FRET analysis was performed on a minimum of 50 cells per condition and the results of three independent experiments were pooled. Dots correspond to individual cells. (Bottom) Whole-cell YFP lifetimes were measured on a minimum of 50 cells per condition and the results of three independent experiments were pooled. Dots correspond to individual cells. A mean delta (Δ) value is indicated to represent the drop in YFP lifetime resulting from the pairing of IFITM3-YFP and mCherry versus the pairing of IFITM3-YFP and IFITM3-mCherry. Empty red circles are used to depict mCherry, filled red circles are used to depict IFITM3-mCherry and filled yellow circles are used to depict IFITM3-YFP. Error bars indicate standard deviation. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. Au, arbitrary units. Ps, picoseconds.
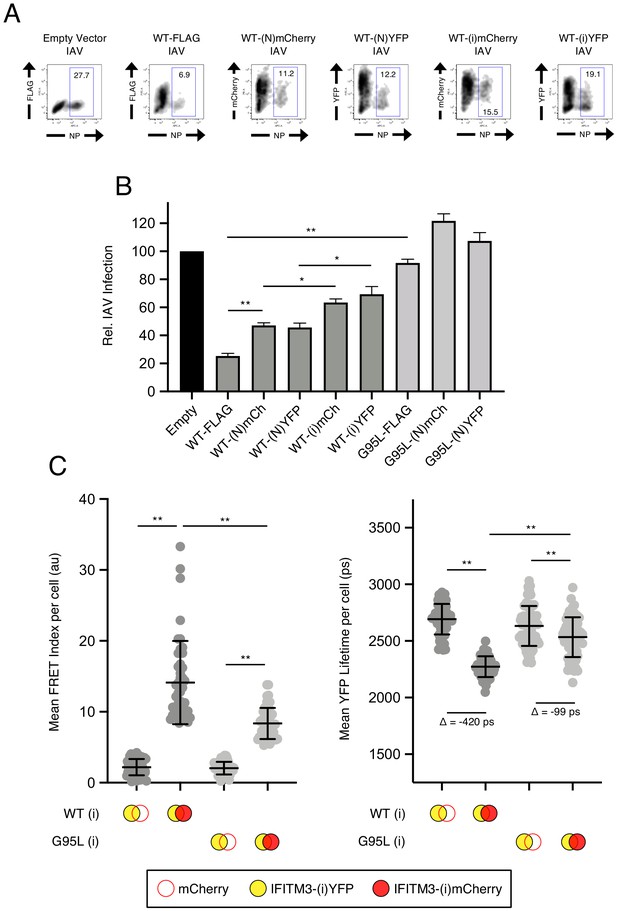
Assessing the functional impact of fluorescent protein placement within IFITM3 fusion proteins.
(A) HEK293T were transiently transfected with 1.5 micrograms of empty pQCXIP, or pQCXIP encoding IFITM3-FLAG (amino-terminal FLAG), IFITM3-(N)YFP (amino-terminal YFP), IFITM3-(N)mCherry (amino-terminal mCherry), IFITM3-(i)YFP (internal YFP), IFITM3-(i)mCherry (internal mCherry) or the indicated constructs encoding G95L. 48 hr after transfection, cells were challenged with IAV PR8 strain at a MOI of 0.2. Cells were fixed at 18 hr post-infection, stained with an anti-nucleoprotein antibody, and assessed by flow cytometry. (B) Mean infection results representing five independent experiments are normalized to empty vector (set to 100%). Error bars indicate standard error. (C) HEK293T were transiently co-transfected with IFITM3-(i)YFP and mCherry or IFITM3-(i)YFP and IFITM3-(i)mCherry. Constructs encoded IFITM3 WT or IFITM3 G95L. Whole-cell FRET (left) and whole-cell YFP lifetimes were measured on a minimum of 50 cells per condition and the results of three independent experiments were pooled. Dots correspond to individual cells. A mean delta (Δ) value is indicated to represent the drop in YFP lifetime resulting from the pairing of IFITM3-YFP and mCherry versus the pairing of IFITM3-YFP and IFITM3-mCherry. Empty red circles are used to depict mCherry, filled red circles are used to depict IFITM3-(i)mCherry (either WT or G95L), and filled yellow circles are used to depict IFITM3-(i)YFP (either WT or G95L). Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. Au, arbitrary units. Ps, picoseconds. Rel., relative.
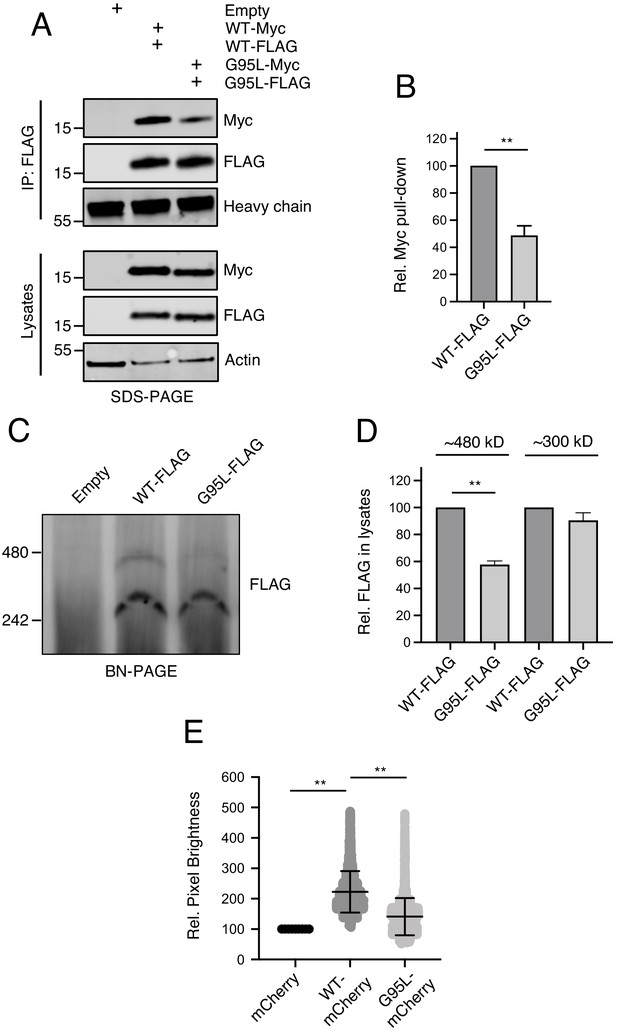
Glycine-95 regulates oligomerization of IFITM3 in denaturing and non-denaturing conditions.
(A) HEK293T were transiently transfected with empty pQCXIP or the following pairs: IFITM3 WT-FLAG and IFITM3 WT-myc or G95L-FLAG and G95L-myc. Whole cell lysates were produced under mildly denaturing conditions and immunoprecipitation (IP) using anti-FLAG antibody was performed. IP fractions and volumes of whole cell lysates were subjected to SDS-PAGE and Western blot analysis. Immunoblotting was performed with anti-FLAG and anti-myc. Heavy chain IgG and Actin served as loading controls in the IP fraction and lysates fraction, respectively. Number and tick marks indicate size (kilodaltons) and position of protein standards in ladder. (B) Levels of IFITM3-myc (either WT or G95L) co-immunoprecipitated by anti-FLAG IP were quantified in (A) and represented as the mean of three independent experiments. Error bars indicate standard error. (C) HEK293T were transiently transfected with empty pQCXIP, IFITM3 WT-FLAG or G95L-FLAG. Cell lysates were produced with 1% digitonin and blue native PAGE was performed, followed by immunoblotting with anti-FLAG. Number and tick marks indicate size (kilodaltons) and position of protein standards in ladder. (D) Levels of IFITM3-FLAG (either WT or G95L) corresponding to ~480 kd and ~300 kD were quantified in (C) and represented as the mean of three independent experiments. Error bars indicate standard error. (D) Number and Brightness analysis was performed on monomeric mCherry and IFITM3-mCherry (either WT or G95L) as described in the Materials and methods. Statistical analysis was performed using student’s T test. *, p<0.05; **, p<0.001. Rel., relative.
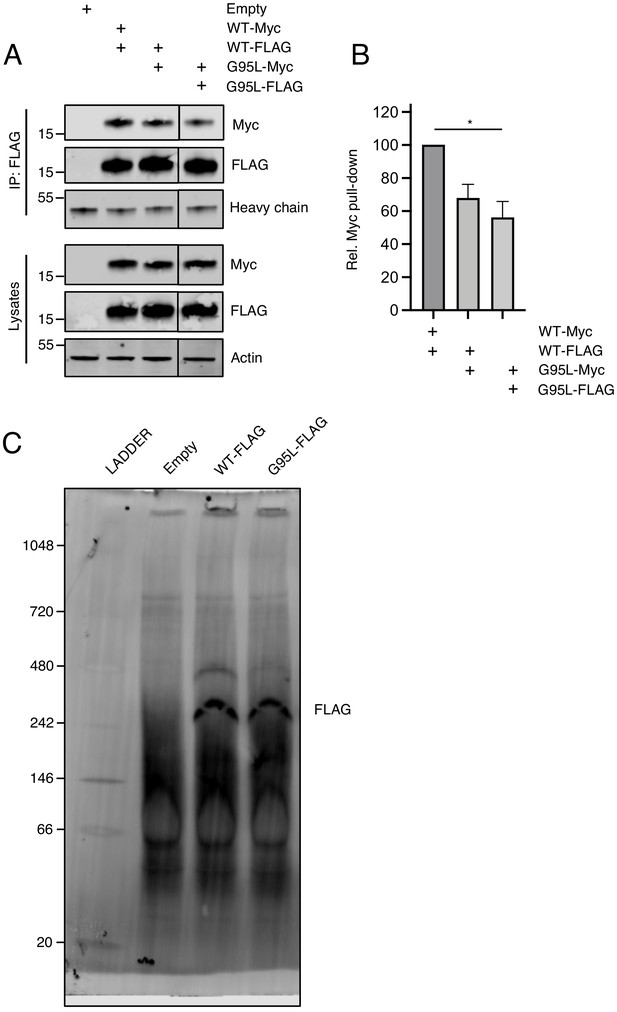
Blue native PAGE of IFITM3 and assessment of heteromultimerization between IFITM3 WT-FLAG and G95L-myc.
(A) HEK293T were transiently transfected with empty pQCXIP or the following pairs: IFITM3 WT-FLAG and IFITM3 WT-myc, IFITM3 WT-FLAG and G95L-myc, or G95L-FLAG and G95L-myc. Whole cell lysates were produced under mildly denaturing conditions and immunoprecipitation (IP) using anti-FLAG antibody was performed. IP fractions and volumes of whole cell lysates were subjected to SDS-PAGE and Western blot analysis. Immunoblotting was performed with anti-FLAG and anti-myc. Heavy chain IgG and Actin served as loading controls in the IP fraction and lysates fraction, respectively. Number and tick marks indicate size (kilodaltons) and position of protein standards in ladder. (B) Levels of IFITM3-myc (either WT or G95L) co-immunoprecipitated by anti-FLAG IP were quantified in (A) and the mean of three independent experiments is shown. Error bars indicate standard error. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. (C) A full scan of the membrane depicted in Figure 5C is shown. Immunoblotting was performed with anti-FLAG. Number and tick marks indicate size (kilodaltons) and position of protein standards in ladder. Rel., relative.
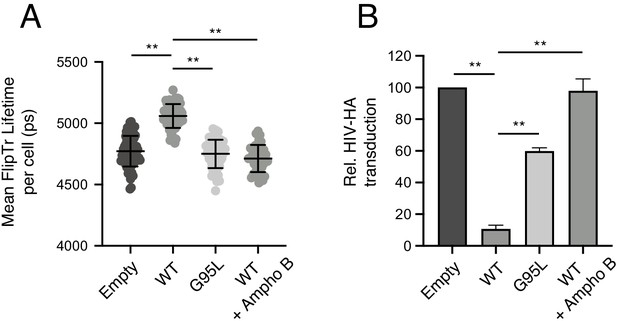
Membrane order is increased by IFITM3 oligomers in an Ampho B-sensitive manner.
(A) HEK293T cells stably transfected with empty pQCXIP, IFITM3 WT-FLAG, or G95L-FLAG were stained with 1 μM FliptR for 5 min and imaged by FLIM. In the condition indicated, 1 μM Amphotericin B was added to cells for 1 hr and washed away prior to addition of FliptR and imaging. The whole-cell mean fluorescence lifetime (τ), in addition to individual component lifetimes (long and short lifetimes, τ1 and τ2), was calculated using Symphotime for a minimum of 40 cells per condition and τ1 from three independent experiments were pooled and plotted. Dots correspond to individual cells. Error bars indicate standard deviation. (B) HEK293T cells stably transfected with empty pQCXIP, IFITM3 WT-FLAG, or G95L-FLAG were challenged with HIV pseudotyped with hemagglutinin (HA) from IAV WSN strain at a MOI of 1. In the condition indicated, 1 μM Amphotericin B was added to cells for 1 hr prior to virus addition. Cells were fixed at 48 hr post-infection, and infection was scored by GFP expression using flow cytometry. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. Ps, picoseconds. Rel., relative.
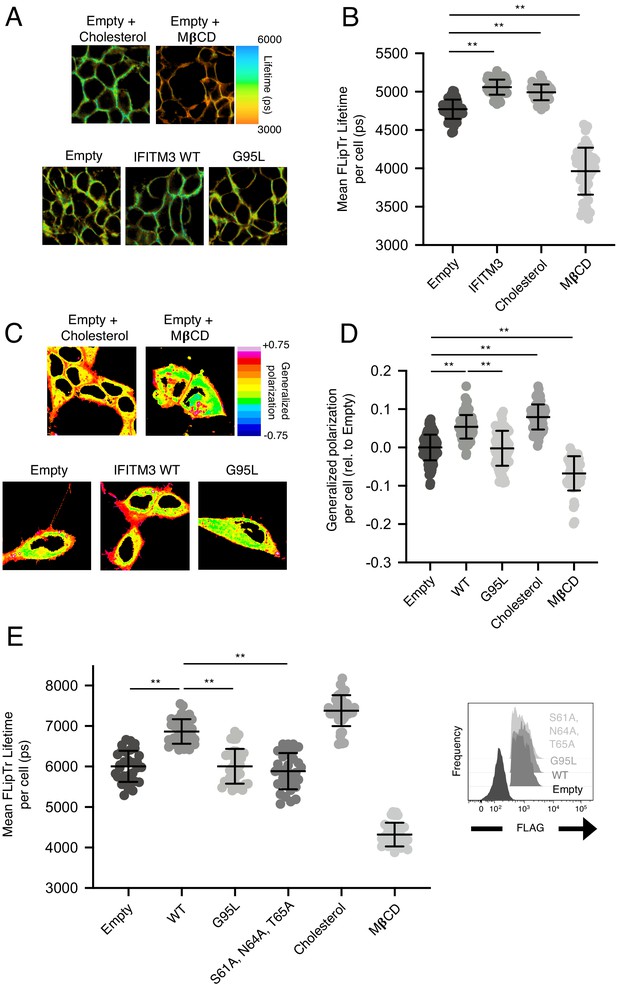
Assessment of cholesterol addition and cholesterol depletion on membrane order by FliptR and Laurdan.
(A) Examples of FliptR images from data plotted in Figure 6A. In addition, soluble cholesterol (100 μg/mL) or methyl-beta-cyclo-dextrin (5 mM) were added to untransfected HEK293T cells for 1 hr and washed away prior to addition of 1 μM FliptR for 5 min and cells were imaged by FLIM. (B) HEK293T cells stably transfected with empty pQCXIP or IFITM3 WT-FLAG were compared to cholesterol-treated and cholesterol-depleted cells. FliptR τ1 was measured for a minimum of 40 cells per condition and the pooled results of three independent experiments are shown. Dots correspond to individual cells. Error bars indicate standard deviation. (C) HEK293T cells stably transfected with empty pQCXIP, IFITM3 WT-FLAG, or G95L-FLAG were incubated with 1.8 μM Laurdan for 1 hr, rinsed with PBS, and imaged. Where indicated, soluble cholesterol (100 μg/mL) or methyl-beta-cyclo-dextrin (5 mM) were added to untransfected HEK293T cells for 1 hr and washed away prior to addition of Laurdan. Examples of images depicting generalized polarization values are shown. (D) Generalized polarization values were calculated for at least 40 cells per condition, and the pooled results from three independent experiments are shown. Dots correspond to individual cells. Error bars indicate standard deviation. (E) HEK293T were transiently transfected with empty pQCXIP, IFITM3 WT-FLAG, G95L-FLAG, or S61A/N64A/T65A-FLAG and stained with 1 μM FliptR for 5 min followed by imaging by FLIM. In the conditions indicated, soluble cholesterol (100 μg/mL) or methyl-beta-cyclo-dextrin (5 mM) were added to untransfected HEK293T cells for 1 hr and washed away prior to addition of FliptR. The whole-cell mean fluorescence lifetime (τ), in addition to individual component lifetimes (long and short lifetimes, τ1 and τ2), were calculated using Symphotime for a minimum of 40 cells per condition and τ1 from three independent experiments were pooled and plotted. Dots correspond to individual cells. Error bars indicate standard deviation. Representative flow cytometry histograms indicate the expression of IFITM3 WT-FLAG, G95L-FLAG, or S61A/N64A/T65A-FLAG following transient transfection and immunostaining with anti-FLAG. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. Ps, picoseconds. Rel., relative.
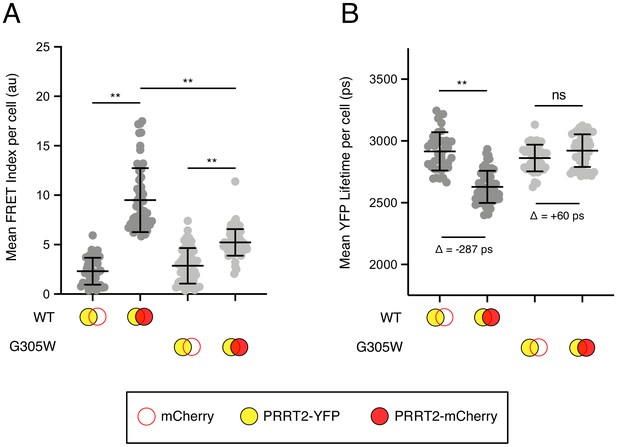
Disease-associated G305W disrupts the oligomerization of PRRT2 in living cells.
(A) HEK293T were transiently co-transfected with PRRT2-YFP and mCherry or PRRT2-YFP and PRRT2-mCherry. Constructs encoded PRRT2 WT or PRRT2 G305W. Whole-cell FRET analysis was performed on a minimum of 50 cells per condition, and the results of three independent experiments were pooled. Dots correspond to individual cells. (B) Whole-cell YFP lifetimes were measured on a minimum of 50 cells per condition and the results of three independent experiments were pooled. Dots correspond to individual cells. A mean delta (Δ) value is indicated to represent the drop in YFP lifetime resulting from the pairing of PRRT2-YFP and mCherry versus the pairing of PRRT2-YFP and PRRT2-mCherry. Empty red circles are used to depict mCherry, filled red circles are used to depict PRRT2-mCherry (either WT or G305W), and filled yellow circles are used to depict PRRT2-YFP (either WT or G305W). Error bars indicate standard deviation. Statistical analysis was performed using one-way ANOVA. *, p<0.05; **, p<0.001. Au, arbitrary units. Ps, picoseconds.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo-sapiens) | HEK293T | ATCC | CRL-3216 RRID:CVCL_0063 | |
| Cell line (Homo-sapiens) | TZM-bl | NIH AIDS Reagent Resource | 8129–442 RRID:CVCL_B478 | |
| Strain, strain background (Influenza A Virus) | A/PR/8/34 (H1N1) | Charles River Laboratories | 10100781 | Clarified allantoic fluid |
| Recombinant DNA reagent | pNL4-3 (plasmid) | NIH AIDS Reagent Resource | 114 | |
| Recombinant DNA reagent | pNL4-3ΔEnv (plasmid) | Eric O. Freed | ||
| Recombinant DNA reagent | pMD2.G (VSV-G) (plasmid) | Addgene | 12259 RRID:Addgene_12259 | |
| Recombinant DNA reagent | pCMV4-BlaM-Vpr (plasmid) | Addgene | 21950 RRID:Addgene_21950 | |
| Recombinant DNA reagent | pReceiver-YFP-IFITM3 and mutants (plasmid) | This paper | YFP fused to IFITM3 at amino terminus | |
| Recombinant DNA reagent | pReceiver-mCherry-IFITM3 and mutants (plasmid) | This paper | mCherry fused to IFITM3 at amino terminus | |
| Recombinant DNA reagent | pReceiver-PRRT2-YFP and mutants (plasmid) | This paper | YFP fused to PRRT2 at carboxy terminus | |
| Recombinant DNA reagent | pReceiver-PRRT2-mCherry and mutants (plasmid) | This paper | mCherry fused to PRRT2 at carboxy terminus | |
| Recombinant DNA reagent | pQCXIP-FLAG-IFITM3 and mutants (plasmid) | Compton et al., 2014 and this paper | FLAG fused to IFITM3 at amino terminus | |
| Recombinant DNA reagent | EEA1-GFP (plasmid) | Addgene | 42307 RRID:Addgene_42307 | |
| Commercial assay or kit | LiveBLazer FRET-B/G Loading Kit with CCF2-AM | Thermo Fisher | K1032 | |
| Chemicalcompound, drug | Amphotericin B | Sigma | C4951 | |
| Chemical compound, drug | FliptR | Spirochrome | CY-SC020 | |
| Chemical compound, drug | Laurdan | Invitrogen | D250 | |
| Chemical compound, drug | Cholesterol, water-soluble | Sigma | A2942 | |
| Chemical compound, drug | Methyl-beta-cyclo-dextrin | Sigma | C4555 | |
| Antibody | Anti-IAV NP mouse monoclonal | Abcam | AA5H | 1:500 (flow) |
| Antibody | Anti-p24 CA mouse monoclonal | NIH AIDS Reagent Resource | 3537 | 1:1000 (WB) |
| Antibody | Anti-p24 CA [KC57-FITC] mouse monoclonal | BD | CO6604665 | 1:500 (flow) |
| Antibody | Anti-CD63 [MX-49.129.5] mouse monoclonal | Santa Cruz Biotechnology | sc-5275 | 1:400 (IF) |
| Antibody | Anti-FLAG [M2] mouse monoclonal | Sigma | F1804 | 1:1000 (WB) 1:500 (flow) |
| Antibody | Anti-IFITM3 [EPR5242] rabbit monoclonal | Abcam | ab109429 | 1:1000 (WB) 1:200 (IF) |
| Antibody | Anti-Env gp120b sheep polyclonal | NIH AIDS Reagent Resource | 288 | 1:1000 (WB) |
| Antibody | Anti-Env gp41 [2F5] human monoclonal | NIH AIDS Reagent Resource | 1475 | 1:1000 (WB) |
| Antibody | Anti-Actin [C4] mouse monoclonal | Santa Cruz Biotechnology | sc-47778 | 1:1000 (WB) |
| Antibody | Anti-c-Myc rabbit monoclonal | Sigma | C3956 | 1:1000 (WB) |