Spatial inter-centromeric interactions facilitated the emergence of evolutionary new centromeres
Figures
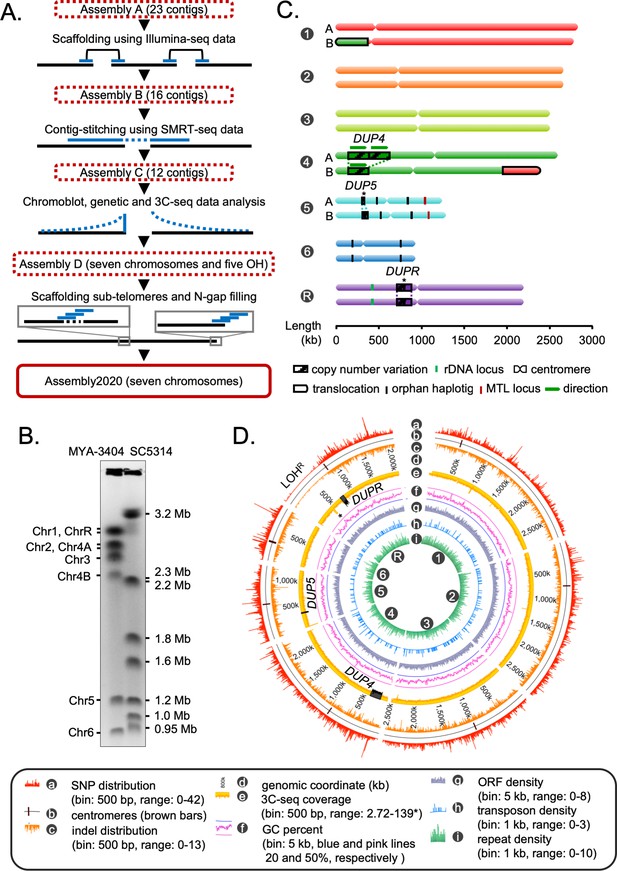
Construction of the gapless assembly of C. tropicalis type strain MYA-3404 in seven chromosomes.
(A) Schematic showing the stepwise construction of the gapless chromosome-level assembly (Assembly2020) of C. tropicalis (also see Figure 1—figure supplement 1 and Figure 1—figure supplement 2). (B) An ethidium bromide (EtBr)-stained CHEF gel image of separated chromosomes of the C. tropicalis (strain MYA-3404) and C. albicans (strain SC5314) (Materials and methods). C. albicans chromosomes are used as size markers for estimation and validation of lengths and identities of C. tropicalis chromosomes in the newly constructed Assembly2020. (C) An ideogram of seven chromosomes of C. tropicalis as deduced from Assembly2020 and drawn to scale. The genomic location of the three loci showing copy number variations (CNVs), DUP4, DUP5 and DUPR located on Chr4, Chr5 and ChrR respectively, are marked and depicted as striped box. The CNVs for which the correct homolog-wise distribution of the duplicated copy is unknown are marked with asterisks. Homolog-specific differences for Chr1 and Chr4, occurred due to an exchange of chromosomal parts in a balanced heterozygous translocation between Chr1B and Chr4B, are highlighted with black borders (also see Figure 1—figure supplement 4C). (D) A circos plot showing the genome-wide distribution of various sequence features. Very high sequence coverage at rDNA locus is clipped for more precise representation and marked with an asterisk.
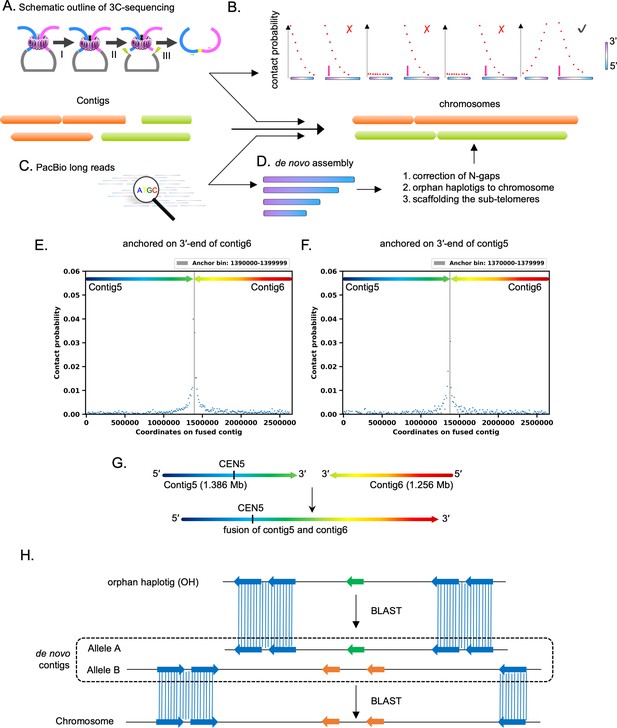
Schematic of the strategies used for construction of the gapless chromosome-level assembly of C. tropicalis.
(A - D) The outline of the steps followed for the construction of the genome assembly. (A) Major steps followed for 3C-sequencing in this study were (I) crosslinking, (II) restriction digestion, and (III) ligation, library preparation, and sequencing. (B) A cartoon explaining the use of contact probability values for establishing contiguity between two DNA fragments. Pink arrows denote coordinates of the anchor bin, with respect to which the contact probability scores (represented as the red dots) are determined. (C) Long reads generated by SMRT-seq were used to construct de novo genome assemblies using Canu as well as the FALCON pipeline. (D) Use of de novo contigs to fill N-gaps, finding the alleles of the orphan contigs in the genome and scaffolding of the sub-telomeres. (E) The 3C profile (bin size = 10 kb) of 3′-terminal bin of contig6 (anchor; gray vertical line) showing its contact probabilities (blue dots) with bins on contig5 and contig6. (F) The 3C profile (bin size = 10 kb) of 3′ terminal bin of contig5 (anchor; gray vertical line) showing its contact probabilities (blue dots) with bins on contig5 and contig6. (G) A schematic representation of chromosome 2 assembled by fusion of contig5 and contig6 in a tail-to-tail orientation based on the 3C profile results. (H) Orphan contigs OHs are mapped to the chromosomes by BLAST analysis of the ORFs, which are located on the Canu assembled de novo contigs. The allelic difference in the OH loci is depicted by color-coded ORFs (orange and green arrows).
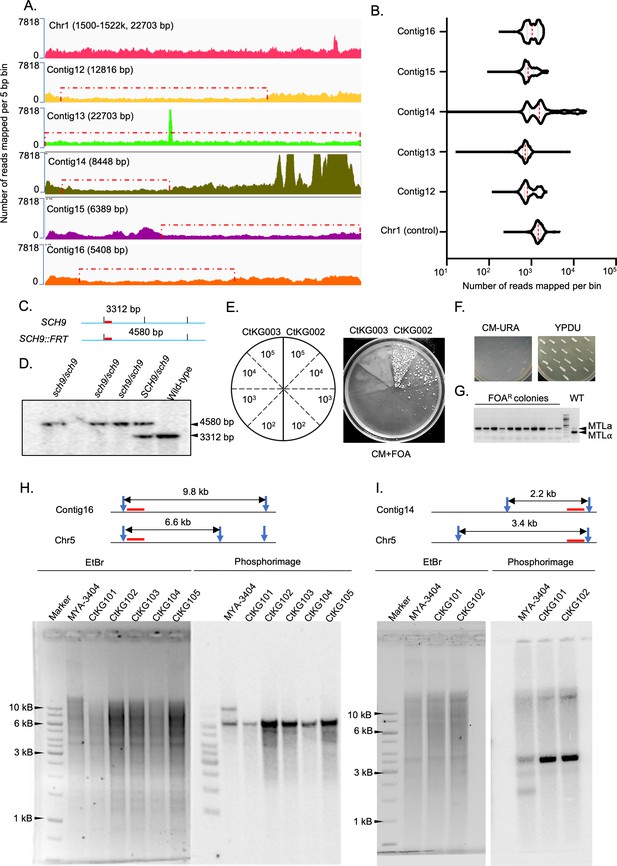
Orphan contigs are alleles in the diploid genome of C. tropicalis.
(A) IGV track images showing the coverage of 3C-seq data on the y-axis (number of reads mapped per bin for each million of the total reads) over the orphan contigs and a control locus from Chr1. (B) Violin plots showing the distribution of read coverage of 3C-seq data across the orphan contigs (bin size = 5 bp) and the control region on Chr1 generated using deepTools2 bamCoverage script. (C) Schematic showing the positions of HindIII sites (vertical black lines) and the length of the expected bands detectable by the probe used (red bars) for Southern hybridization to confirm sch9 deletion strains (CtKG001). (D) Phosphorimage of the blot for confirmation of sch9△/sch9△ mutant strains. (E) Schematic showing various number of cells (105, 104, 103, and 102) of CtKG002 (left) and CtKG003 (right) plated on CM+FOA plate. The plate image showing appearance of FOAR colonies in CtKG002 but not in CtKG003 strain. (F) The FOAR colonies thus obtained were picked up, patched on CM-URA (left), and YPDU media (right) and imaged after 48 hr of growth at 30°C. (G) The EtBr stained gel image for multiplex PCR products to detect the loss of MTLa or MTLα alleles in the FOAR colonies along with the wild-type control (Primers are listed on Supplementary file 9). (H and I) Experimental validation of the allelic nature of contig16 and contig14, respectively, using Southern blot analysis. The length of restriction fragments polymorphisms between the alleles after digesting with ClaI and EcoRI (restriction enzyme sites are indicated using blue arrowheads) are graphically represented for contig16 and contig14, respectively. The lanes in gels represent the wild-type MYA-3404 (2nd lane) and the monosomic aneuploid strains (CtKG101 - 105) where one homolog of Chr5 is absent. The probes used in this experiment are denoted using red bars.
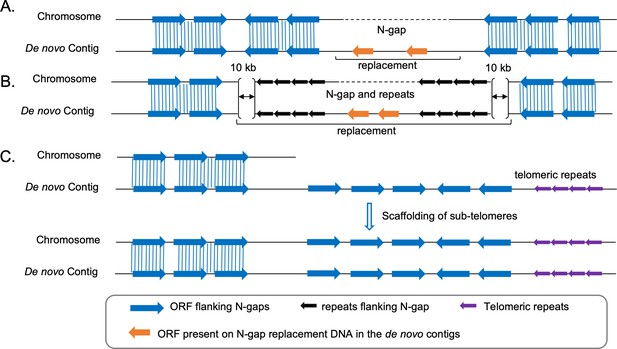
Schematic outline of the strategy followed for N-gap filling and scaffolding of sub-telomeres.
(A - B) Strategy-I and strategy-II (Materials and methods) for filling N-gaps without flanking repeats or with flanking repeats, respectively. Repeats are presented as black arrows. (C) Schematic for scaffolding of sub-telomeres using the de novo assembled contigs.
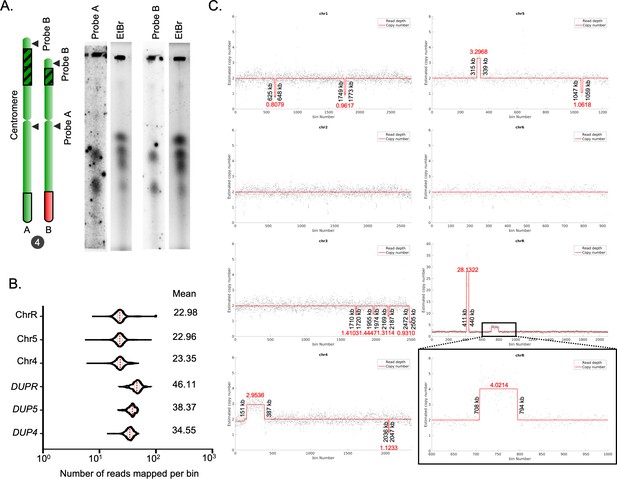
Identification of CNVs in the C. tropicalis strain MYA-3404.
(A) EtBr stained gel images and phosphorimages obtained from Southern hybridization experiments using a centromere-proximal (Probe A) and a centromere-distal probe (Probe B) from Chr4 (Supplementary file 9). (B) Violin plots for the number of reads mapped per bin (2 bp) on ChrR (excluding the rDNA locus), and other chromosomes or loci as indicated. The average number of reads mapped on the chromosomes and DUP4, DUP5, and DUPR loci are presented. (C) CNAtra output of read depth signals calculated from 3C-seq reads (black dots; bin size = 1 kb) and estimated copy numbers (red lines) of each chromosome in C. tropicalis. For regions whose estimated copy numbers are <1.5 or >2.5, their respective start and end coordinates (shown in black vertical text) as well as estimated copy numbers (shown in red horizontal text) are manually labelled. Black box represents a zoom in view of a region on ChrR (600–1000 kb) where a duplicated region shows an estimated copy number of ~4.
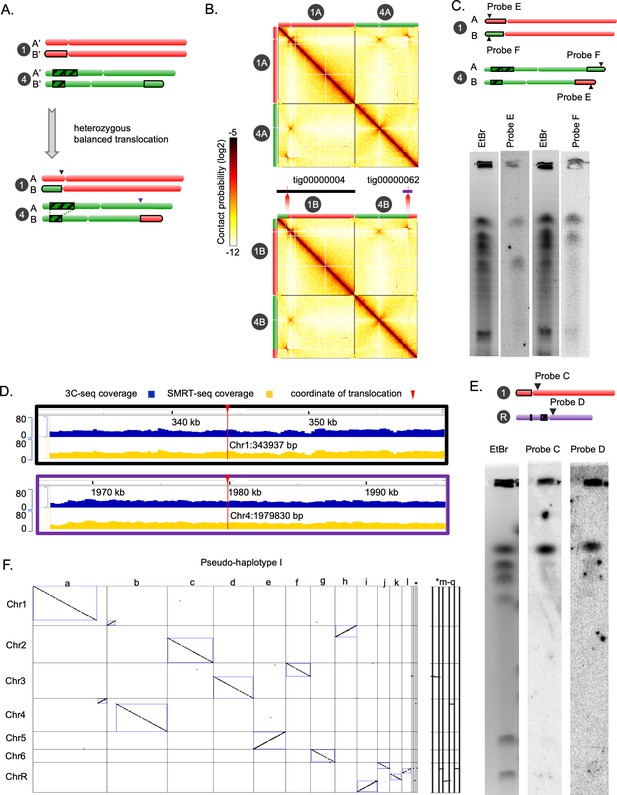
Chromoblot, sequence coverage analysis, and haplotyping for validation of the chromosome-level genome assembly of C. tropicalis.
(A) Schematic of the balanced heterozygous translocation between Chr1B and Chr4B. The DUP4 locus is highlighted with the black striped box. The junction between Chr1 and Chr4 on Chr1B and Chr4B are marked with black and purple arrows, respectively. (B) Contact probability heatmaps (bin size = 10 kb) of Chr1 and Chr4 of C. tropicalis showing a balanced translocation as evidenced by a butterfly-like pattern (chromatin contacts split into two blocks) in the interchromosomal area. The 3C-seq reads were mapped to Assembly2020 (top; original) with Chr1A and Chr4A genomic sequences (Figure 1C). We have also mapped the 3C-seq reads to an alternate assembly (bottom) with Chr1B and Chr4B sequences. Alternate assembly has been generated by exchanging the genomic sequences at the translocation breakpoint in Chr1 and Chr4. Coordinate of translocation was mapped using two de novo assembled contigs supporting the junctions. Chromosome labels and their corresponding ideograms are shown on the heatmap. Colorbar represents the contact probability in log2 scale. (E) An ethidium bromide stained gel image and phosphorimages obtained from Southern hybridization using a probe from part of Chr1, which is exchanged with Chr4 (probe F) and a second probe from part of Chr4, that is exchanged with Chr1 (Probe E) (Supplementary file 9). Black triangles point to the genomic coordinates of the probes used. (C) IGV tracks showing 3C-seq (blue) and SMRT-seq coverage (yellow) across the translocation junctions on each of the unaltered homolog of Chr1 (black border) and Chr4 (purple border), respectively. (D) An ethidium bromide stained gel image and phosphorimages obtained from Southern hybridization using centromere-proximal probes from Chr1 (Probe C) and ChrR (Probe D). (E) A synteny dot-plot comparing the colinearity between the chromosomes and the FALCON-generated contigs (labeled as a-l). Five very short contigs are denoted by an asterisk. The enlarged version of the dot plot for these contigs is shown on the right panel. The dot-plot was generated using Symap.
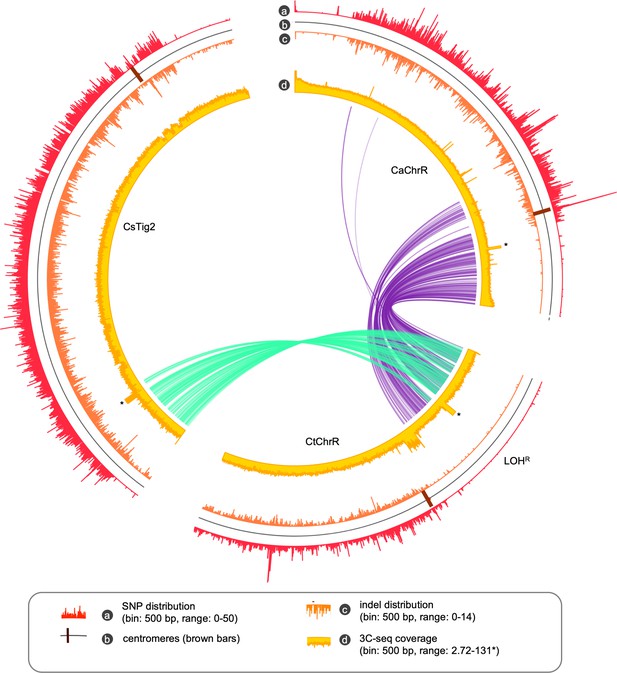
Partial conservation of a LOH block in each of the C. albicans, C. tropicalis and C. sojae genome.
The circos tracks represent the SNP density, positions of the centromeres, Indel density, Illumina sequence coverage (the sequence coverage at the rDNA loci is clipped for clearer representation and marked with an asterisk) as indicated. The ribbon plot was drawn by connecting the genomic coordinates of the conserved single copy orthologs between C. tropicalis and C. sojae (teal), and C. tropicalis and C. albicans (purple).
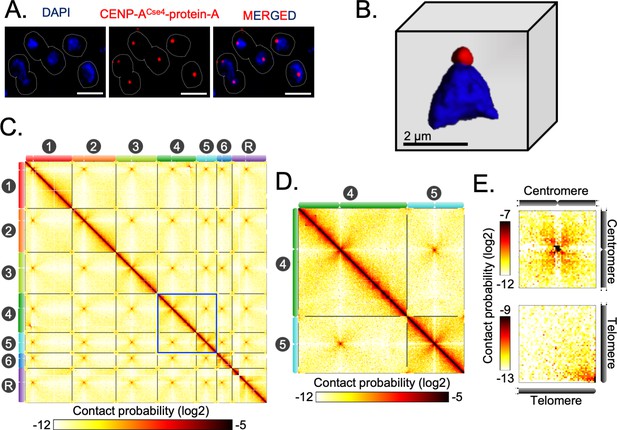
Spatial genome organization reveals centromere-centromere and telomere-telomere contacts in C. tropicalis.
(A) A representative field image of C. tropicalis (strain CtKS102) cells expressing Protein-A tagged CENP-ACse4. CENP-A signals (red) were obtained using anti-Protein A antibodies by indirect immuno-fluorescence microscopy. Nuclei of the corresponding cells were stained by DAPI (blue). The images were acquired using a DeltaVision imaging system (GE) and processed using FIJI software (Schindelin et al., 2012). Scale, 2 µm. (B) A 3D reconstruction showing clustered kinetochores marked by CENP-ACse4 (red) at the periphery of the DAPI-stained nucleus (blue) using Imaris software (Oxford Instruments) in C. tropicalis. Scale, 2 µm. (C) A genome-wide contact probability heatmap (bin size = 10 kb) generated using 3C-seq data. Chromosome labels and their corresponding ideograms are shown on the axes of the heatmap. Colorbar represents the contact probability in the log2 scale. (D) Zoom in view of heatmap showing Chr4 and Chr5 from panel C (blue box). (E) Heatmaps plotted from aggregate signal analysis of matrices (bin size = 2 kb) surrounding centromere-centromere (top) or telomere-telomere interactions (bottom). Top, genomic loci containing mid-points of centromeres are aligned at the center ; bottom, genomic loci from 5′ or 3′ ends of chromosomes are aligned at the bottom right corner.
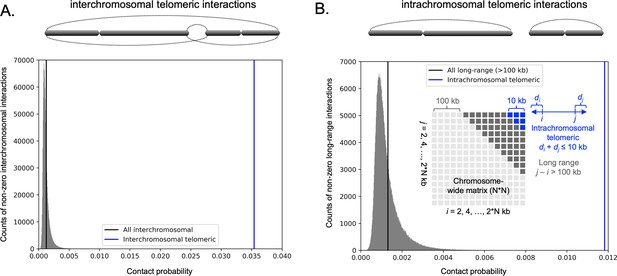
Analysis of 3C-seq data reveals interchromosomal and intrachromosomal telomeric contacts in C. tropicalis genome.
(A) Histogram of all interchromosomal interactions (excluding zero values; gray) was plotted from the 3C-seq contact probability matrix (bin size = 2 kb) of C. tropicalis. The mean value of all interchromosomal interactions is indicated by the black vertical line. A cartoon of interchromosomal telomeric interactions (dotted curves) is depicted above the histogram as the interactions between telomeres of different chromosomes. The mean value of interchromosomal telomeric interactions is indicated by the blue vertical line in the histogram. The interchromosomal telomeric interactions are significantly greater than all interchromosomal interactions (Mann-Whitney U test P value = 1.129 × 10−11). (B) Histogram of all intrachromosomal long-range (>100 kb) interactions (excluding zero values; gray) was plotted from chromosome-wide contact probability matrices (bin size = 2 kb) of C. tropicalis. A cartoon of intrachromosomal telomeric interactions (dotted curves) is depicted above the histogram as the interactions between two telomeres of the same chromosome. Inset, a cartoon depicting long-range interactions (gray blocks) and intrachromosomal telomeric interactions (blue blocks) in a chromosome-wide matrix. A long-range interaction is defined as the cis interaction between two loci separated by a distance of >100 kb. Intrachromosomal telomeric interactions are computed as cis interactions between loci whose distances to two telomeres have a sum of ≤10 kb. Note that the distances indicated in the cartoon for 100 kb and 10 kb are not drawn to scale. The mean values of all long-range and intrachromosomal telomeric interactions are indicated by black and blue vertical lines, respectively. The intrachromosomal telomeric interactions are significantly greater than all long-range interactions (Mann-Whitney U test P value = 7.374 × 10−11).
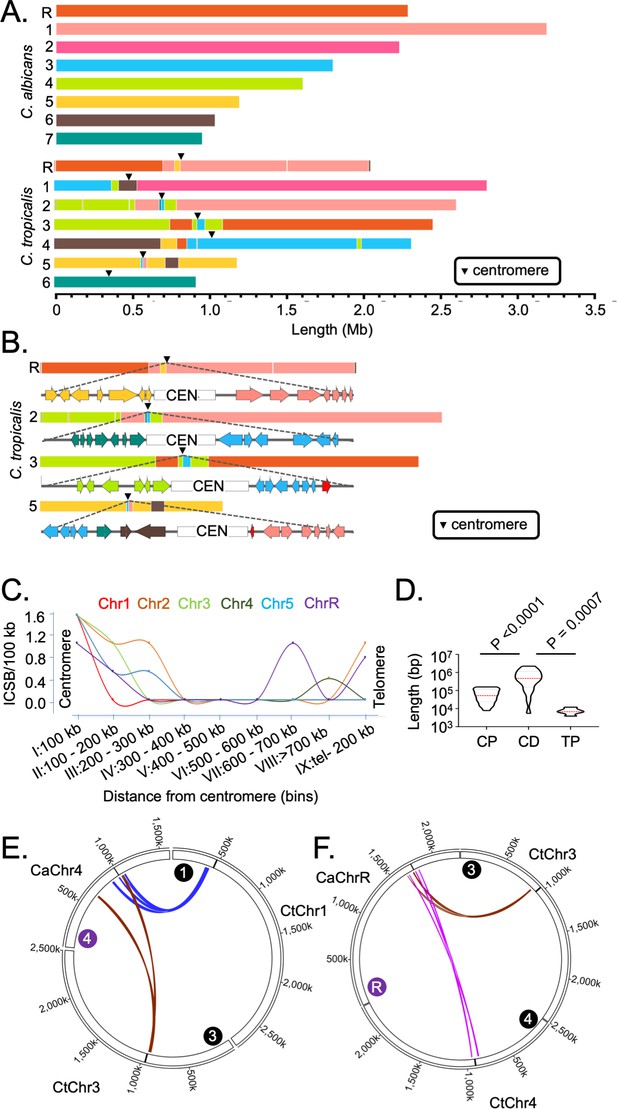
Genome-wide mapping of interchromosomal synteny breakpoints in C. tropicalis identifies a spatial cue for karyotype evolution.
(A) Scaled representation of the color-coded orthoblocks (relative to C. albicans chromosomes) and ICSBs (white lines) in C. tropicalis (Materials and methods). Orthoblocks are defined as stretches of the target genome (C. tropicalis) carrying more than two syntenic ORFs from the same chromosome of the reference genome (C. albicans). The centromeres are represented with black arrowheads. (B) Zoom in view of the C. tropicalis centromere-specific ICSBs on CEN2, CEN3, CEN5 and CENR showing the color-coded (relative to C. albicans chromosomes) ORFs flanking each centromere. C. tropicalis-specific unique ORFs proximal to CEN3 and CEN5 are shown in red. (C) A plot showing the chromosome-wise ICSB density, calculated as number of ICSBs per 100 kb of the C. tropicalis genome (y-axis), as a function of the linear distance from the centromere in nine bins. These bins are a) 0–100 kb on both sides of centromere (bin I), (b) 100–200 kb (bin II), (c) 200–300 kb (bin III), (d) 300–400 kb (bin IV), (e) 400–500 kb (bin V), (f) 500–600 kb (bin VI), (g) 600–700 kb (bin VII), (h) >700 kb to 200 kb from telomere ends (bin VIII), and i) 200 kb from the telomere ends (bin IX). Chr6 was excluded from this analysis, as it does not harbor any ICSB. (D) A violin plot comparing the distribution of lengths of orthoblocks (y-axis) at three different genomic zones: a) the centromere-proximal zone (CP), (b) the centromere-distal zone (CD), and c) telomere-proximal zone (TP). Orthoblocks, which span over more than one zone, were assigned to the zone with maximum overlap. The centromere-distal dataset was compared with the other two groups using the Mann-Whitney U test and the respective P values are mentioned. (E - F) Circos plots representing the convergence of centromere-proximal ORFs of C. tropicalis chromosomes near the centromeres (CEN4 and CEN7) of C. albicans. Chromosomes of C. tropicalis and C. albicans are marked with black and purple filled circles at the beginning of each chromosome, respectively.
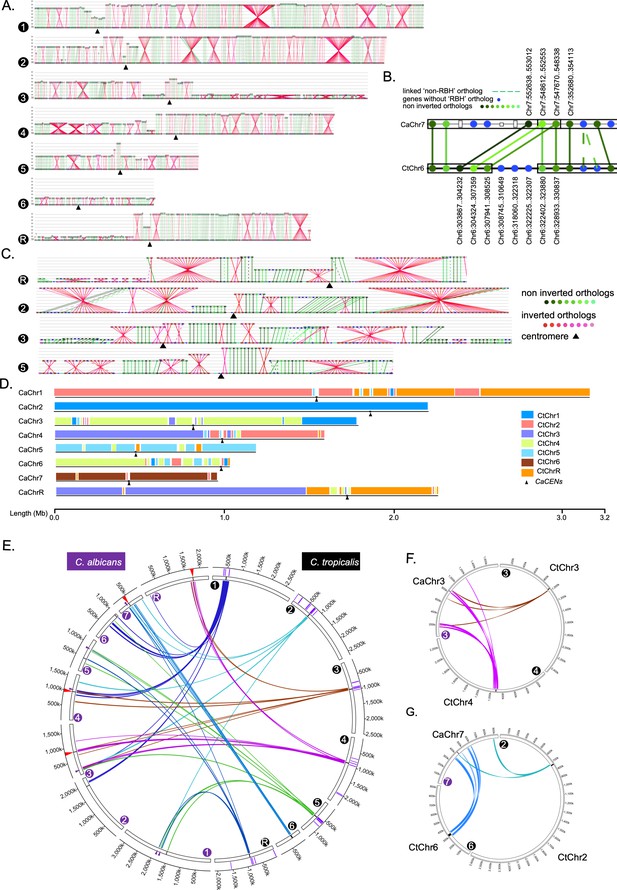
Genome-wide synteny analysis between C. albicans and C. tropicalis suggests evidence of inter-centromere translocations in the last common ancestor.
(A) Synteny maps of C. tropicalis chromosomes (the lowermost line of each panel, marked by filled black circles numbered from 1 to R), with respect to C. albicans chromosomes (lines above the C. tropicalis chromosomes), in the order of Chr1 to ChrR (top to bottom) for all panels. Centromeres, black triangles. The ORFs (represented as beads) are color-coded: inverted, red and non-inverted, green. The more conserved the reciprocal best hits (RBH) are, the darker are the shades of red/green color. (B) Zoom in view of the synteny relationship between the centromere proximal ORFs of C. tropicalis Chr6 with C. albicans Chr7. (C) The zoom in view of RBH ORFs proximal to the centromeres of C. tropicalis as indicated in the figure, where each centromere is located at an ICSB. (D) A scaled representation of the color-coded orthoblocks (relative to C. tropicalis chromosomes) and ICSBs (white lines) in C. albicans (Materials and methods). Orthoblocks are defined as stretches of the target genome (C. albicans) carrying more than two syntenic ORFs from the same chromosome of the reference genome (C. tropicalis). The centromeres are represented with black arrowheads. (E) Circos plot showing the ICSBs (purple lines on the outer-most circle) on C. tropicalis chromosomes (marked with black filled circles). The centromere proximal ORFs (10 ORFs on both sides) present in C. tropicalis are connected to their homologs present on C. albicans chromosomes (marked by purple filled circles) by color-coded lines (based on their origin). The positions of centromeres are marked with black lines of the inner-most circle in each chromosome. The genomic locations in C. albicans chromosomes showing the convergence of ORFs from at least two centromere-proximal loci of C. tropicalis are marked with red (proximal to the C. albicans centromere) and purple (a non-centromere locus) triangles. Note that all centromeres of C. albicans are proximal to ORFs, homologs of which are proximal to centromeres of C. tropicalis. (F - G) Circos plots showing the convergence of centromere proximal ORFs of C. tropicalis chromosomes near the centromeres on C. albicans chromosomes 3 (CaChr3) and 7 (CaChr7), respectively. Chromosomes of C. tropicalis and C. albicans are marked with black and purple filled circles at the beginning of each chromosome, respectively.
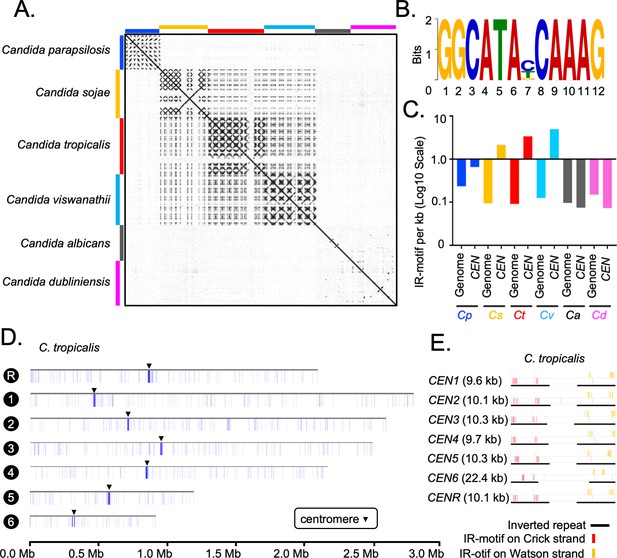
Genome-wide analysis of centromere DNA sequences across the CUG-Ser1 clade reveals the emergence of unique centromeres from an ancestral homogenized inverted repeat-associated centromere type.
(A) A dot-plot matrix representing the sequence and structural homology among species of the CUG-Ser1 clade was generated using Gepard (Materials and methods). (B) A logo plot showing the 12-bp-long IR-motif, identified using MEME-suit (Materials and methods). (C) The distribution of IR-motif density on centromere DNA sequences and across the entire genome of each species was calculated as the number of motifs per kb of DNA (Materials and methods). Note that C. albicans and C. dubliniensis centromeres that form on unique and different DNA sequences do not contain the IR-motif. (D) IGV track images showing the IR-motif density across seven chromosomes of C. tropicalis. The location of the centromere on each chromosome is marked with a black arrowhead. (E) IGV track images showing the IR-motif distribution across seven HIR-associated centromeres of C. tropicalis.
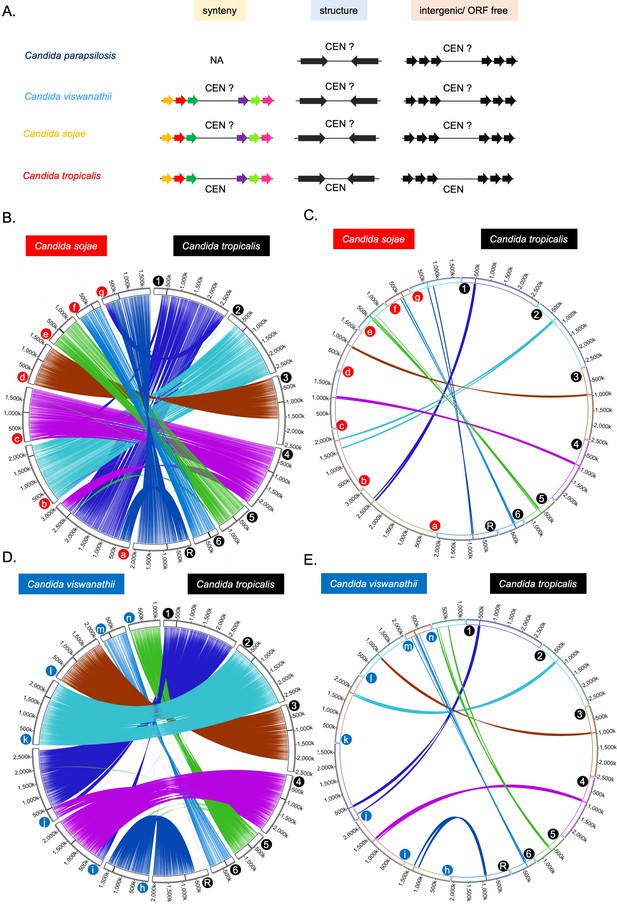
Identification of HIR-associated centromeres in the CUG-Ser1 clade.
(A) Schematic of the method used for the identification of putative centromeres in C. sojae, C. viswanathii, and C. parapsilosis. Putative centromeric loci in these species were tested for gene synteny with C. tropicalis (for C. sojae and C. viswanathii), presence of IRs, and overlap with intergenic/ORF-free regions. (B) Genome-wide synteny of conserved ortholog pairs between C. sojae and C. tropicalis. (C) Circos plot similar to that of B, showing 10 ORFs on both sides of each centromere of C. tropicalis connected to the corresponding genomic loci carrying homologs in C. sojae. (D) Genome-wide synteny between conserved ortholog pairs between C. viswanathii and C. tropicalis. (E) Circos plot similar to that of D, showing 10 ORFs on both sides of each centromere of C. tropicalis connected to the corresponding genomic loci carrying homologs in C. viswanathii. The location of the centromere on each chromosome on subpanel B, C, D, and E is marked with a black line. Chromosomes/contigs are marked at the beginning of each chromosome/contig with colored filled circles (a to g are tig00000002, tig00000008, tig00000017, tig00000038, tig00000050, tig00016100, tig00000001, for C. sojae and h to n are NW_020797881.1, NW_020797885.1, NW_020797858.1, NW_020797886.1, NW_020797884.1, NW_020797877.1 and NW_020797878.1 for C. viswanathii). Contigs that are either <100 kb in length or do not carry putative centromeres (for C. sojae) or duplicated in the genome assembly (for C. viswanathii) were excluded from this analysis. The chromosomal coordinates of the ortholog pairs are connected using color-coded lines.
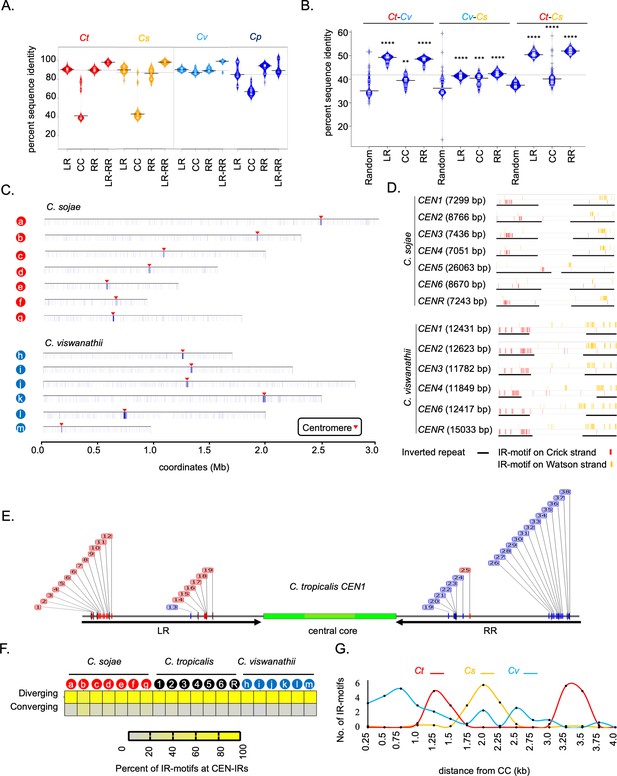
Inter-species conservation of centromere DNA sequences of closely related Candida species.
(A) Bean plots showing the distribution of the percent sequence identity among the centromeric left repeat (LR), the central core (CC), and right repeat (RR) elements in C. tropicalis (Ct), C. sojae (Cs), C. viswanathii (Cv), and C. parapsilosis (Cp). The extent of sequence identity between all possible pairs of CC, LR, RR, and LR-RR pairs were calculated in blastn analysis using Clustal Omega. The means are depicted as a horizontal black bar on each of the bean pod. (B) Bean plots showing distribution and means (horizontal black bar on each of the bean pods) of percent sequence identity values obtained from pairwise DNA sequence alignment of all possible combinations for each of seven random loci (Random), centromeric left repeats (LR), central cores (CC) and right repeats (RR) between species-pairs as indicated, using Clustal Omega. The significance of difference between percent sequence identity of centromere elements and random loci for all three species pairs were tested using the Mann-Whitney U test (p<0.05) and the P value summary for each comparison is represented with asterisks. (C) Heatmap plot across the contigs carrying the putative centromeres showing the extent of enrichment of IR-motifs in C. sojae (contigs a-g, as described in Figure 4—figure supplement 1) and C. viswanathii (contigs h-n, as described in Figure 4—figure supplement 1). The locations of the centromeres are pointed with red arrowheads. D. IGV track images showing the extent of enrichment of IR-motif on the putative centromeres of C. sojae and C. viswanathii. (E) A representative figure showing a zoom in view of the IR-motif distribution on C. tropicalis CEN1 DNA. The motifs on the Crick strand (red) and Watson strand (blue) are color coded. (F) Heatmap showing the percent of IR-motifs present in converging and diverging orientation with respect to the central core region for each of the HIR associated putative centromeres present in C. sojae, C. tropicalis, and C. viswanathii. (G) The average number of IR-motifs per 250 bp on the IRs is plotted in the y-axis as a function of the distance from the start of CC (x-axis) for C. tropicalis (red), C. sojae (yellow), and C. viswanathii (blue). Bean plots were generated using BoxplotR.
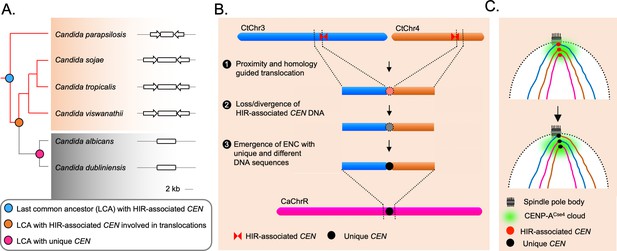
The spatial genome organization remained conserved in the CUG-Ser1 clade despite centromere type diversity.
(A) A maximum likelihood-based phylogenetic tree of closely related CUG-Ser1 species analyzed in this study. The centromere structure of each species is shown and drawn to scale. (B) A model showing possible events during the loss of HIR-associated centromeres and emergence of the unique centromere type through inter-centromeric translocations possibly occurred in the common ancestor of C. tropicalis and C. albicans. The model is drawn to show translocation events involving two C. tropicalis chromosomes (CtChr3 and CtChr4) as representatives, which can be mapped proximal to the centromere on C. albicans ChrR (CaChrR) as shown in Figure 3F. (C) Rabl-like chromosomal conformation is maintained despite inter-centromeric translocations that facilitated centromere type transition.
Additional files
-
Source data 1
Source_data_combined.
- https://cdn.elifesciences.org/articles/58556/elife-58556-data1-v3.xlsx
-
Supplementary file 1
Assembly C with 12 contigs.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp1-v3.pptx
-
Supplementary file 2
Assembly of sub-telomeres and filling up N-gaps in the genome assembly of C. tropicalis using de contigs.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp2-v3.pptx
-
Supplementary file 3
Statistics for different versions of genome the assembly of C. tropicalis (MYA-3404) generated in this study.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp3-v3.pptx
-
Supplementary file 4
A comparative analysis of Assembly A and the improved Assembly2020 of C. tropicalis.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp4-v3.pptx
-
Supplementary file 5
Features of centromere DNA elements in C. sojae.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp5-v3.pptx
-
Supplementary file 6
Features of centromere DNA elements in C. viswanathii.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp6-v3.pptx
-
Supplementary file 7
Centromere coordinates used for identifying conserved DNA sequence motifs in Candida species.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp7-v3.pptx
-
Supplementary file 8
List of strains used in this study.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp8-v3.pptx
-
Supplementary file 9
List of primers used in this study.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp9-v3.pptx
-
Supplementary file 10
List of plasmids used in this study.
- https://cdn.elifesciences.org/articles/58556/elife-58556-supp10-v3.pptx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58556/elife-58556-transrepform-v3.docx





