How cells determine the number of polarity sites
Figures
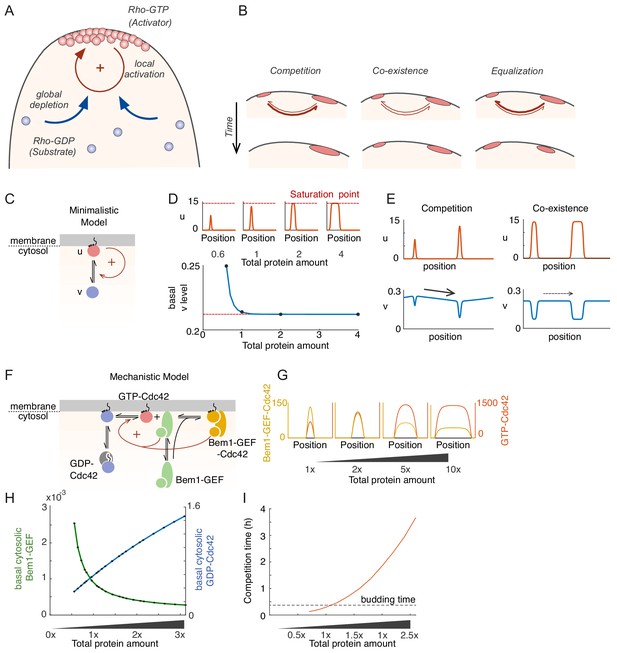
Competition in a mechanistic Rho-GTPase model.
(A) Rho-GTPase polarity circuits can be modeled as mass-conserved reaction-diffusion systems where membrane-bound Rho-GTP (red) is a slow-diffusing activator and cytosolic Rho-GDP (blue) is a fast-diffusing substrate. Such systems polarize Rho-GTPase to spatially confined polarity sites based on positive feedback (+) via local activation that recruits substrate from the cytoplasm, leading to global substrate depletion. (B) A starting condition with two unequal peaks of activator can evolve in three ways. Competition occurs if the larger peak recruits substrate better than the smaller; coexistence occurs if both peaks recruit substrate equally well; and equalization occurs if the smaller peak recruits substrates better than the larger. (C) Schematic of the minimalistic MCAS model with one Rho-GTPase converting between activator (u: Rho-GTP, red) and substrate (v: Rho-GDP, blue) forms. Arrows depict reactions. The positive feedback is highlighted in red (see text for details). (D) In the minimalistic model, as total protein amount in the system increases, peak u concentration (red) approaches a saturation point while basal v concentration (blue) declines to a limit. Basal here refers to the concentration at the edge of the peak. 1x protein amount equivalent to starting uniform v concentration of 2. (E) Snapshots of simulations starting from two single peak steady states placed next to each other in the same domain. With peaks far from saturation, the larger peak depletes more substrate than the smaller (Left panel, starting protein amounts 0.6x, 1x), leading to a net flux of substrate (arrow) that results in competition. With peaks close to saturation, substrate depletion is similar for both (Right panel, starting protein amounts 2x, 4x), leading to little flux and therefore coexistence. (F) Schematic of a mechanistic model of the yeast polarity circuit: arrows depict reactions assumed to occur with mass-action kinetics. The positive feedback is highlighted in red. (G) Increasing total protein amount saturates the peaks similarly to D. (H) Basal cytosolic level of the limiting species Bem1-GEF declines to a limit while that of Cdc42 increases with increased total protein amount. (I) Competition time increases as peaks saturate. Competition was started from two insulated 1-peak steady states each containing 60% and 40% of the total proteins. Competition time is defined by the time it takes to evolve from 70%:30% (relative Cdc42T amounts between two peaks) to 99%:1%.
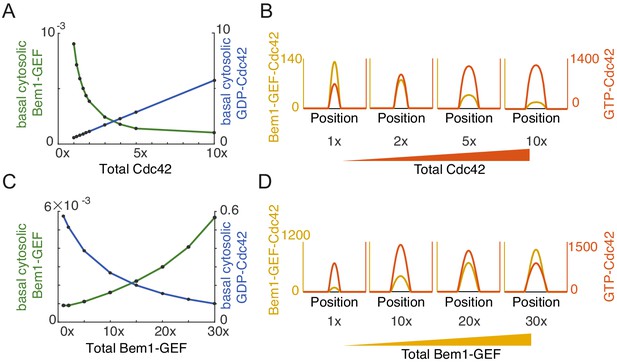
Effects of increasing the abundance of individual polarity factors (Cdc42 or Bem1-GEF) in mechanistic polarity model.
Effects of increasing the total amount of Cdc42 (A, B) and Bem1-GEF (C,D) in the system. Note that concentration profiles at the membrane are 1D representations of 2D spots. (A) Basal concentrations of the cytoplasmic substrates GDP-Cdc42 and Bem1-GEF. (B) Concentration profiles of the membrane-associated activators GTP-Cdc42 and Bem1-GEF-Cdc42. Note that the concentration of GTP-Cdc42 in the peak is the sum of unbound (GTP-Cdc42) and bound (Bem1-GEF-Cdc42) protein. Whereas Bem1-GEF is the limiting substrate whose concentration plateaus when Cdc42 is increased (A), GDP-Cdc42 becomes the limiting substrate whose concentration plateaus when Bem1-GEF is increased (C). Saturation can lower the peak Bem1-GEF-Cdc42 concentration when Bem1-GEF is the limiting substrate (B), and saturation can lower the peak GTP-Cdc42 concentration when GDP-Cdc42 is the limiting substrate (D). Total protein amount: 1x Cdc42 corresponds to a mean concentration of 1 µM in the cell; 1x Bem1-GEF corresponds to a mean concentration of 0.017 µM in the cell.
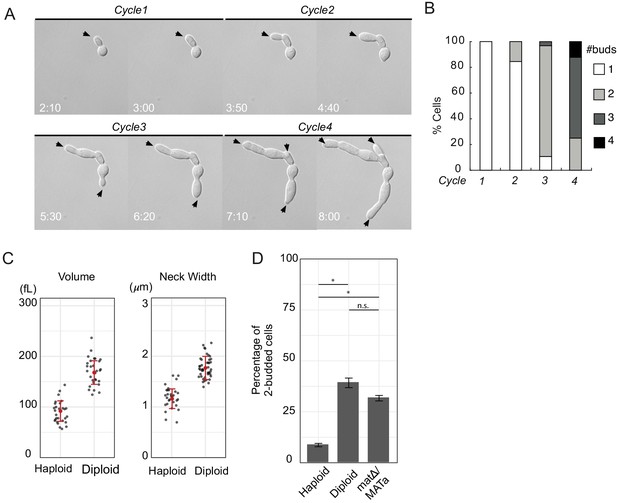
Large yeast cells can generate multiple buds simultaneously.
(A) DIC time lapse movie of a haploid cytokinesis-defective cdc12-6 mutant (DLY20240) over four budding cycles at restrictive temperature (37°C). Black arrows indicate growing buds. Time in hr:min. (B) The number of buds generated in each cell cycle was scored for N = 35 cells. (C) Volume and neck width of diploid (DLY20569) and haploid (DLY9455) cdc12-6 cells in the second cell cycle at restrictive temperature. Red dot and intervals indicate mean and standard deviation. (D) Percentage of two-budded cells observed in the second cell cycle at restrictive temperature for diploid (DLY20569) and haploid (DLY9455) cdc12-6 cells (Fisher’s exact test p<10−5). matΔ/MATa cells (DLY22887) are diploid but have a haploid mating type (Fisher’s exact test p=0.0431 compared to diploid; p<10−5 compared to haploid). Asterisks indicate p<10−5 and n.s. indicates p>0.01. Error bars indicate SEM.
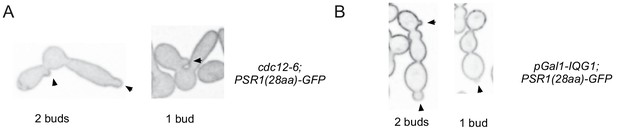
Multi-lobed cdc12-6 and Iqg1-shutoff cells have continuous cytoplasms.
(A) Medial-plane images of one- and two-budded cells after shift of cdc12-6 cells (DLY22915) to restrictive temperature for 4 hr. Membrane probe linking first 28 residues of Psr1 to GFP reveals connected cytoplasm between all mothers and buds. Arrowheads indicate buds. (B) Medial-plane images of one- and two-budded cells after IQG1 shut-off for 9 hr (DLY22875). The same membrane probe reveals contiguous cytoplasm.
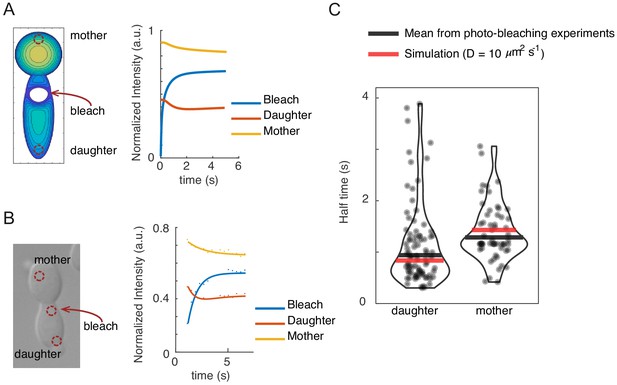
The effect of the mother-bud neck on cytoplasmic diffusion.
(A) Simulated 3D cells were bleached at a spot in the daughter compartment immediately adjacent to the neck. The schematic shows the 2D sum projection of the simulation. Dynamics of fluorescence intensity in the simulated cell were measured in the mother and daughter compartments equidistal to the bleach site (red dashed outline circles) to match experimental geometry. (B) Cells expressing cytoplasmic GFP (DLY22957) were bleached at the indicated spot and experimental recovery data (dots) were fitted to exponential curves (right). (C) Half-times extracted from photo-bleaching experiments (each dot is one photo-bleaching experiment, black line is median) and simulation data (red line). The excellent fit between simulation and experiment suggests that the neck geometry is sufficient to explain the slight difference in recovery time between mother and daughter compartments.
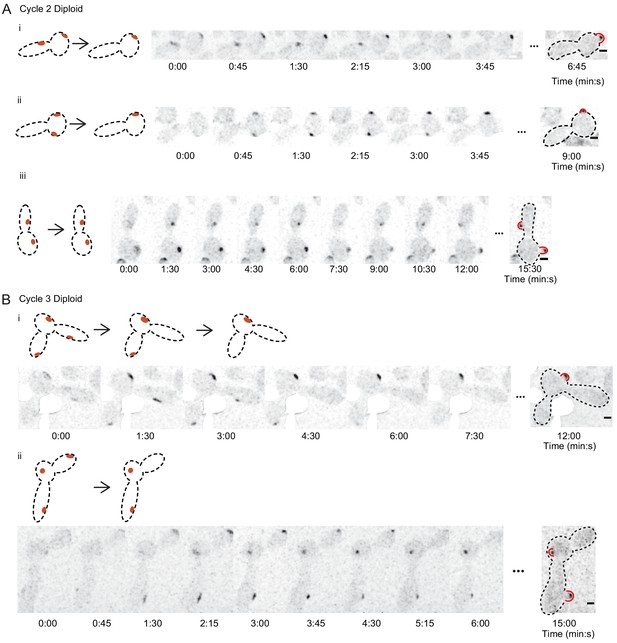
Polarity sites in large cdc12-6 cells.
(A) Cells that start with two polarity sites can show competition or coexistence. Example cells demonstrating typical Bem1 behaviors in cdc12-6 rsr1 Δ diploids (DLY15376) in the second G1 phase after switching to restrictive temperature. Cartoons (left) depict the dynamics of polarity sites (red). Red bulges (right) indicate buds (note that buds can emerge in directions that are not within the focal plane). Cells i and ii: competition yields one bud. Cell iii: coexistence yields two buds. (B) Example cells from the third G1 phase after switching to restrictive temperature. These cells initially had three polarity sites but made only one (cell i) or two (cell ii) buds. Scale bar = 2 µm. White regions cover auto-fluorescent dead cells.
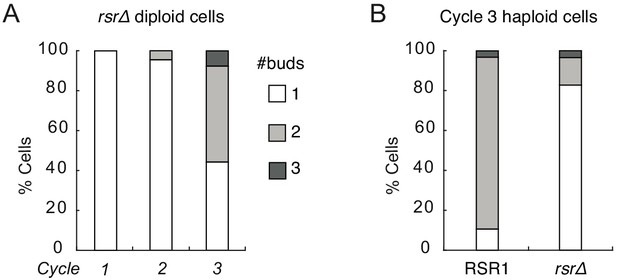
Large rsr1Δ cells can generate multiple buds similar to RSR1 cells.
(A) The number of buds generated simultaneously in each cell cycle by diploid cdc12-6 rsr1Δ cells (DLY15376) after switching to restrictive temperature. (B) rsr1Δ cells (DLY9453) generate fewer multi-budded cells compared to similar-size RSR1 cells (DLY20240).
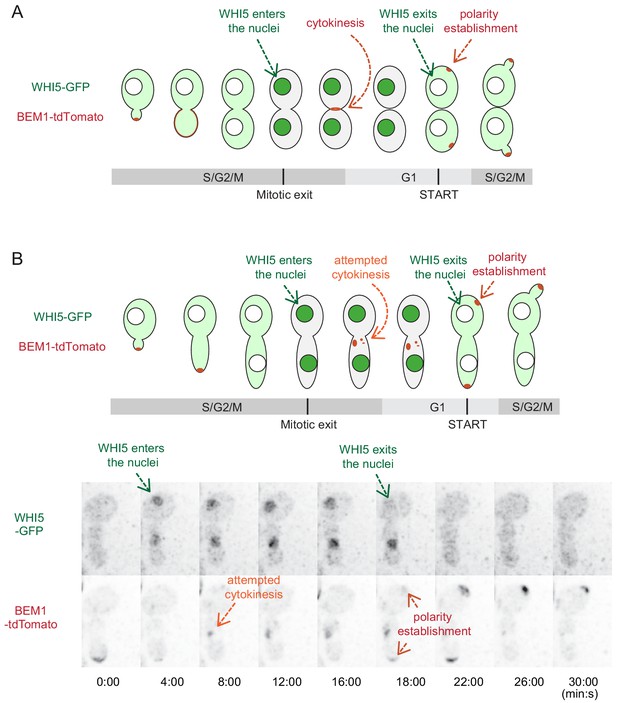
Timing of Bem1 localization in the cell cycle distinguishes abortive cytokinesis sites from polarity sites.
(A) In wild-type (not cytokinesis-defective) cells, Bem1 and Cdc42 localize to the mother-bud neck during cytokinesis and polarize at presumptive bud sites during late G1. To understand the timing of polarity events, we used the localization of Whi5 as a visual indicator of cell cycle stages. Whi5 localizes to the nucleus from late mitosis until early G1, but is cytoplasmic from late G1 through metaphase (Costanzo et al., 2004; de Bruin et al., 2004; Skotheim et al., 2008). Polarity establishment occurs at late G1 which coincides with Whi5 exit (Lai et al., 2018; Moran et al., 2019). (B) Time-lapse imaging of cell cycle (Whi5-GFP) and polarity (Bem1-tdTomato) probes in cdc12-6 cells at restrictive temperature (DLY22920). Green arrows indicate Whi5 nuclear entry (marking mitotic exit) and Whi5 nuclear exit (marking start of the next cell cycle). The first round of Bem1 localization occurred after Whi5 entered the nuclei (orange arrow, i.e. at the normal time of cytokinesis), and the second round occurred after Whi5 exited the nuclei (red arrows, that is at the normal time of polarization). Given its timing, the first round of localization likely represents a failed attempt at cytokinesis. Time in min:s.
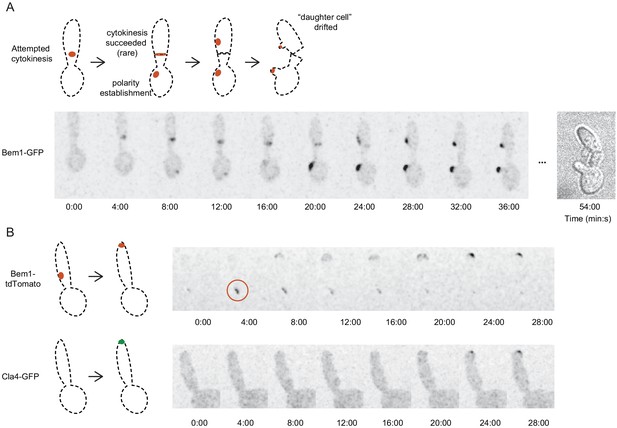
Bem1 but not Cla4 localizes to abortive cytokinesis sites as well as polarity sites in cdc12-6 mutants.
(A) A rare successful cytokinesis in cdc12-6 cells (DLY16767) at restrictive temperature. Bem1-GFP localized to the cytokinesis site and then the polarity site. The ‘daughter cell’ later drifted off, confirming cytokinesis. (B) The neck-localized Cdc42 is thought to be predominantly inactive during cytokinesis (Atkins et al., 2013), and several effectors of Cdc42 (including the PAK Cla4) do not co-localize with Cdc42 and Bem1 at the neck (Holly and Blumer, 1999). Consistent with the view that the first round of Bem1 localization in cdc12-6 represents attempted cytokinesis, Cla4 does not localize to cytokinesis site. Bem1-tdTomato localized to cytokinesis site (red circle) and then polarity site, while Cla4-GFP only localized to the polarity site (DLY23359).
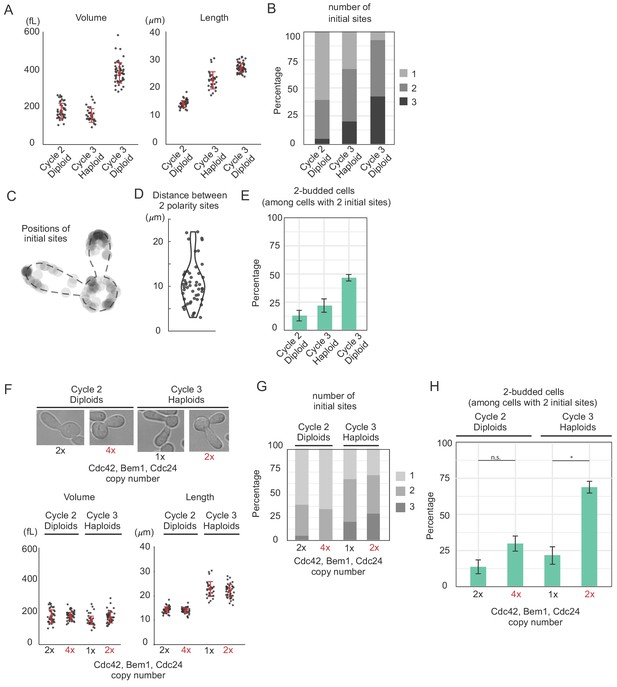
Increasing abundance of polarity proteins enhances the frequency of multipolar outcomes.
(A) Cell volume and length in three populations of cdc12-6 rsr1Δ cells at restrictive temperature: diploids (DLY15376) in the second or third cell cycle and haploids (DLY9453) in the third cell cycle. Red dot and interval indicate mean and standard deviation. (B) The number of initial polarity sites in each population. (C) Initial sites form at non-random positions. N = 123 initial polarity sites in cycle three diploids were mapped onto a generic cell outline. (D) The distance between polarity sites is highly variable. (E) Percentage of two-budded cells (bipolar outcomes) among those that established two initial sites. Larger cells show more frequent multipolar outcomes (13.3%, 21.4%, 46.2%). Error bars indicate SEM from three experiments. (F) Diploid and haploid cdc12-6 rsr1Δ cells with normal (DLY15376, DLY9453) or double (DLY23308, DLY23302) the gene dosage of CDC42, BEM1 and CDC24 show that cell size is unaffected by polarity gene dosage. (G) The number of initial polarity sites does not vary systematically as a function of polarity gene dosage (strains as in F). (H) The percentage of two-budded cells within the subpopulation that established two initial polarity sites (strains as in F). Similar sized cells expressing more polarity proteins display more frequent multipolar outcomes (13.3% vs 29.4% in cycle two diploids, although not significant, Fisher's exact test p=0.25. 21.4% vs 68.4% in cycle three haploids, Fisher’s exact test p=0.0094). Error bars indicate SEM from three experiments.
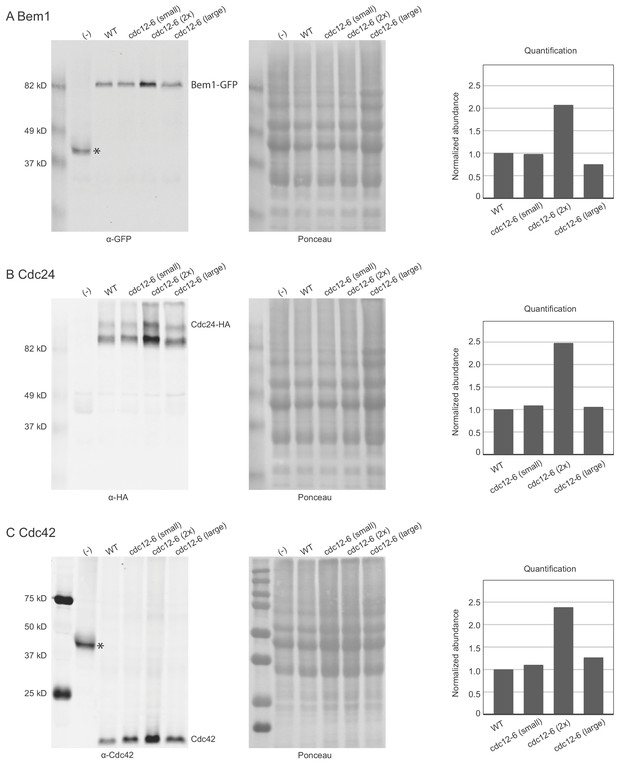
Polarity protein abundance as a function of strain and cell size.
Polarity protein abundance is comparable between wild-type and cdc12-6 cells (lanes 2 and 3) and between small and large cells (lanes 3 and 5), which are cdc12-6 cells before (small) or after (large) 4 hr incubation at 37°C. Doubling gene dosage in cdc12-6 cells (2x) causes a doubling of the abundance of the corresponding polarity protein (lanes 3 and 4). Protein amounts of key polarity proteins in lysates from negative control (lane 1, DLY13030), wild-type (lane 2, DLY22980), cdc12-6 (lanes 3 and 5, DLY22993), and cdc12-6 2x (lane 4, DLY23269). Left panels show western blots, middle panels show Ponceau S stain of the corresponding blots to control for protein loading, and right panels show quantification of abundance (Western blot band divided by Ponceau S stain of same lane) normalized to the wild-type strain. (A) Bem1-GFP abundance detected with anti-GFP antibody. The negative control strain has no tag on Bem1 but carries GFP-Cdc42 and shows a corresponding smaller band (asterisk) (B) Cdc24-HA abundance detected with anti-HA antibody. The negative control strain has no tag on Cdc24. (C) Cdc42 abundance detected with anti-Cdc42 antibody. The negative control strain has no endogenous Cdc42 but carries GFP-Cdc42 and shows a corresponding larger band (asterisk).
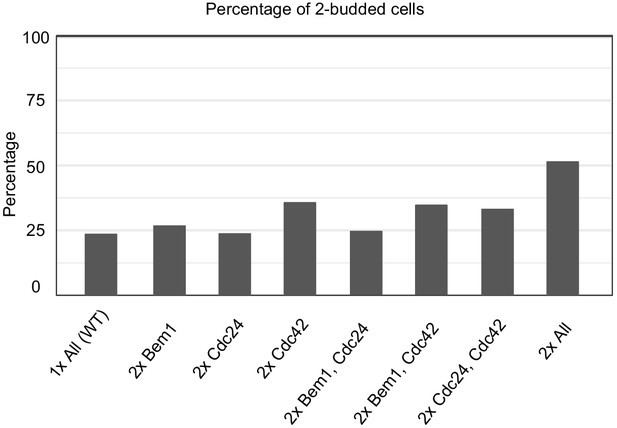
Effect of increased polarity gene dosage on the frequency of multi-polar outcomes.
The percentage of two budded cells in haploid cdc12-6 rsr1Δ strains with either one or two copies of the genes (CDC42, CDC24, BEM1) encoding the key polarity regulators. Cells were switched to 37°C for 5 hr prior to fixation and subsequent DIC imaging in order to capture the number of buds produced during the third cell cycle. ‘All’ indicates either one copy (1x All) or two copies (2x All) of all three genes. Other strains carry two copies of the indicated regulators and one copy of the others. Strains: (1x All DLY9453; 2x All DLY23302; 2x Bem1 DLY23829; 2xCdc24 DLY23830; 2xCdc42 DLY23831; 2x[Bem1 Cdc24] DLY23843; 2x[Bem1, Cdc42] DLY23833; 2x[Cdc24, Cdc42] DLY23832).
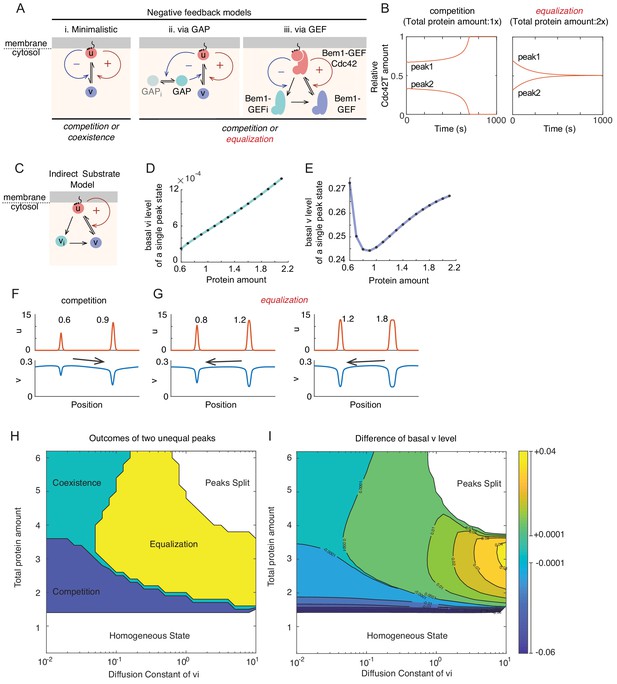
Basis for equalization in more complex models.
(A) Schematic of models incorporating negative feedback. (i) In the minimalistic model, negative feedback adds a term in which u promotes conversion of u to v in a non-linear manner. (ii) This model incorporates conversion of an inactive GAP (GAPi) to an active GAP in a manner stimulated by u. The GAP promotes conversion of u to v, providing negative feedback. (iii) Simplified scheme of a mechanistic model where positive feedback occurs via recruitment of a substrate (Bem1-GEF) to become an activator (Bem1-GEF-Cdc42). Negative feedback occurs because the activator promotes inhibitory phosphorylation of the Bem1-GEF, generating inactive Bem1-GEFi. For details of the full model see Figure 5—figure supplement 1F and Materials and methods. (B) With the GEF negative feedback model, two peaks can compete (protein amount 0.5x, 1x) or equalize (protein amount 1x, 2x). (C) Schematic of the indirect substrate model. In addition to the reactions from minimalistic MCAS model in Figure 1C,u (red) can be converted into the indirect cytoplasmic substrate vi (teal), which itself can be converted to the substrate v (blue). (D) The basal level of indirect substrate (vi) increases as the amount of total protein in the system goes up. (E) The basal substrate level (v) first decreases but then increases as the amount of total protein in the system goes up. (F) Competition occurs when the larger peak depletes cytoplasmic substrates more than the smaller. Protein amount 0.6x:0.9x. (G) Equalization occurs when the smaller peak depletes cytoplasmic substrates more than the larger. Protein amount 0.8x:1.2x and 1.2x:1.8x. (H) Outcomes vary depending on the diffusion constant of vi and total protein amount. Simulations were initiated with two single peak steady states containing unequal total protein amounts 0.4:0.6. Outcomes were classified as follows: Competition, simulations evolve to a single peak steady state within t < 2000; Coexistence, unequal peaks remain unequal (±1%) within t < 2000; Equalization, simulations evolve to a two-equal-peak steady state within t < 2000; Peaks split, simulations evolve to split a starting peak into two smaller peaks; Homogeneous, simulations evolve to a homogeneous steady state. (I) Contour plot of the difference in basal v concentration between the two initial peaks (vP0.6 - vP0.4). Contour lines are at 0.01 intervals except when basal levels are very close, when we indicate ±0.0001 contours. The starting difference in basal substrate between the peaks predicts the outcome.
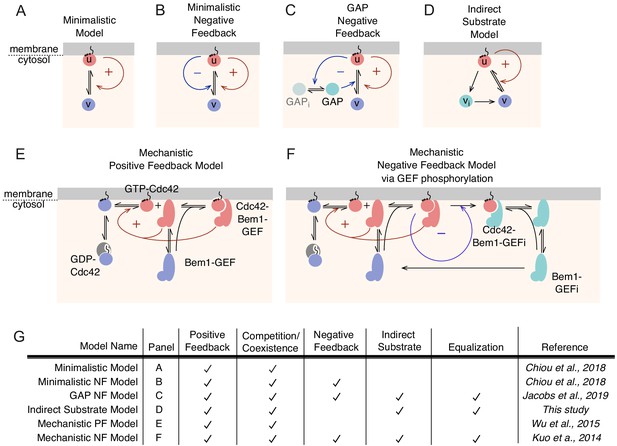
Summary of model behaviors.
(A-D) Schematic of MCAS models from Figure 1A and Figure 5B-D. (E,F) Schematic of mechanistic models without (E, as in Figure 1F) or with (F, shown in simplified form in Figure 5Aiii) negative feedback. In addition to the reactions from the positive-feedback model in E, Bem1-GEF-Cdc42 complexes contain a kinase that phosphorylates and inactivates the GEF in other Bem1-GEF-Cdc42 complexes, leading to negative feedback (F, blue arrow). Phosphorylated species (teal) lack GEF activity, but can still undergo reversible binding to Cdc42-GTP and exchange between membrane and cytoplasm. Dephosphorylation of inactive Bem1-GEFi is assumed to occur only in the cytoplasm. (G) Summary of model features and behaviors. Panel refers to schematics above. References describe first report of the models.
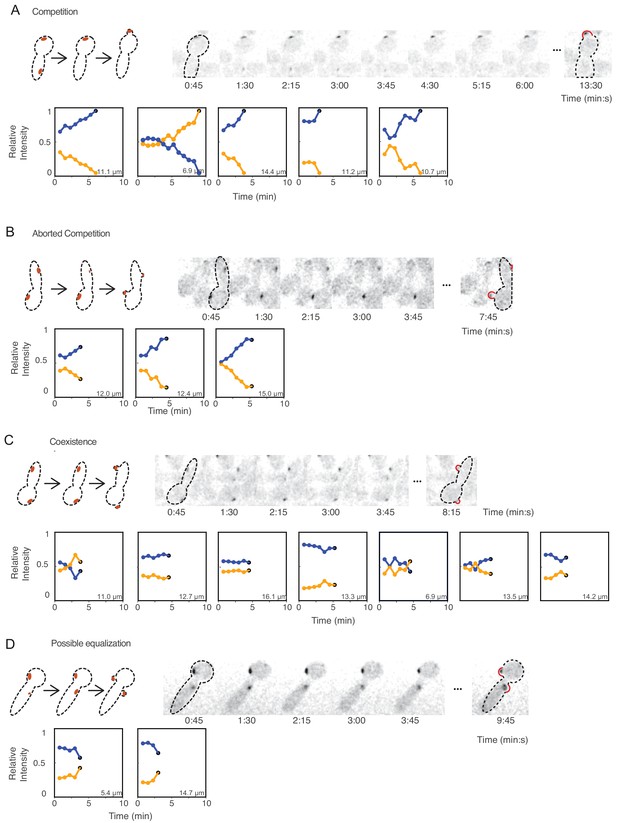
Competition, coexistence, and equalization in yeast.
Cells that exhibit (A) competition, (B) aborted competition, (C) coexistence, or (D) possible equalization. Cartoons depict the dynamics of polarity sites (red) in the accompanying montages. Red bulges (right) indicate buds, which sometimes emerge away from the focal plane (note that polarity sites in small buds were always at the bud tip, but the appearance in 2D maximum projections may not convey that when buds grow away from the focal plane). The amounts of Bem1 in each site were quantified from sum intensity traces from z-stacks of cdc12-6 rsr1Δ CDC2438A cells (DLY21100) that had two initial polarity sites. We plotted the relative amounts of Bem1 at the two polarity sites from when both sites had grown until the time of budding, with blue and orange dots representing the initially more (blue) or less (orange) intense polarity sites. Polarity sites that eventually led to bud-emergence are indicated by black dots at the last time point. Inset numbers denote the distance between the two initial polarity sites. Timelapse montages show the first cell in each category. Additional traces are shown in Figure 6—figure supplement 1.
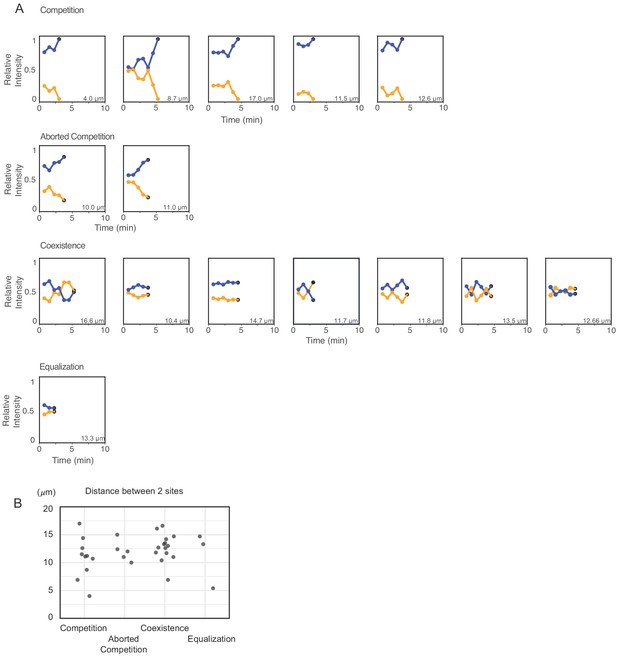
Competition, coexistence, and equalization outcomes are not correlated with the distance between initial polarity sites.
(A) Additional traces of cells with two initial polarity sites that show competition, aborted competition, coexistence, and equalization (as in Figure 6). (B) The distribution of distances between the two initial sites in each category.
Videos
Large yeast cells can generate multiple buds simultaneously.
Time-lapse DIC images of a pair of cdc12-6 cells (DLY20240) growing at restrictive temperature for four-cell cycles. The mother-daughter pair of cells were shifted to 37°C when the video begins. Time stamp indicates hr:min.
Tables
Strains.
| Strain | Relevant genotype | Source |
|---|---|---|
| DLY9453 | a; cdc12-6; rsr1::HIS3; BEM1-GFP:LEU2 | This study |
| DLY9455 | a; cdc12-6; BEM1-GFP:LEU2 | This study |
| DLY13030 | a; cdc42::TRP1; rsr1::TRP1; URA3:GFP-CDC42 | This study |
| DLY15376 | a/α; cdc12-6/cdc12-6; rsr1::HIS3/rsr1::HIS3; BEM1-GFP:LEU2/BEM1-GFP:LEU2 | This study |
| DLY16767 | a; cdc12-6; rsr1::TRP1; BEM1-GFP:LEU2; CDC2438A | This study |
| DLY20240 | a; cdc12-6; BEM1-GFP:LEU2; HTB2-mCherry:natR | This study |
| DLY20569 | a/α; cdc12-6/cdc12-6; BEM1-GFP:LEU2/BEM1-GFP:LEU2 | This study |
| DLY21100 | a/α; cdc12-6/cdc12-6; BEM1-GFP:LEU2/BEM1-GFP:LEU2; CDC2438A/CDC243838A | This study |
| DLY22875 | a; cdc12-6; WHI5-GFP:SpHIS5; BEM1-GFP:LEU2; pTEF1-PSR1-mCherry-tADH1:LEU2; rsr1::TRP1 | This study |
| DLY22887 | a/matαΔ::natR; cdc12-6/cdc12-6; BEM1-GFP:LEU2/BEM1-GFP:LEU2 | This study |
| DLY22915 | a/α; WHI5-GFP:SpHIS5/WHI5; rsr1::TRP1/RSR1; BEM1-tdTomato:HIS3/BEM1-tdTomato:HIS3; pGAL1-IQG1:LEU2/pGAL1-IQG1:LEU2; pTEF1-PSR1-GFP-tADH1:LEU2/leu2; Gal4BD-hER-VP16:URA3/Gal4BD-hER-VP16:URA3 | This study |
| DLY22920 | a/α; cdc12-6/cdc12-6; WHI5-GFP:SpHIS5/WHI5-GFP:SpHIS5; BEM1-tdTomato:HIS3/BEM1-tdTomato:HIS3 | This study |
| DLY22957 | a/α; cdc12-6/cdc12-6; BEM1-tdTomato::HIS3/BEM1; pTEF1-GFP:LEU2/pTEF1-GFP:LEU2 | This study |
| DLY22980 | α; rsr1::TRP1; BEM1-GFP:LEU2; CDC24-HA:kanR | This study |
| DLY22993 | α; cdc12-6; BEM1-GFP:LEU2; CDC24-HA:kanR | This study |
| DLY23302 | a; cdc12-6; CDC24-HA:kanR; BEM1-GFP:TRP1:BEM1-GFP:LEU2; ura3::CDC24-HA:URA3; CDC42:TRP1:CDC42; rsr1::TRP1 | This study |
| DLY23308 | a/α; cdc12-6/cdc12-6; CDC24-HA:kanR/CDC24-HA:kanR; BEM1-GFP:TRP1:BEM1-GFP:LEU2/BEM1-GFP:TRP1:BEM1-GFP:LEU2; ura3::CDC24-HA:URA3/ura3::CDC24-HA:URA3; CDC42:TRP1:CDC42/CDC42:TRP1:CDC42; rsr1::TRP1/rsr1::TRP1 | This study |
| DLY23359 | a/α; cdc12-6/cdc12-6; CLA4-GFP::HIS3/CLA4; BEM1-tdTomato::HIS3/BEM1 | This study |
| DLY23269 | α; cdc12-6; rsr1::TRP1; BEM1-GFP:TRP1:BEM1-GFP:LEU2; CDC24-HA:kanR; ura3::CDC24-HA:URA3; CDC42:TRP1:CDC42 | This study |
| DLY23829 | a; cdc12-6; rsr1::HIS3; BEM1-GFP:TRP1:BEM1-GFP:LEU2 | This study |
| DLY23830 | a; cdc12-6; rsr1::TRP1; BEM1-GFP:LEU2; CDC24-HA:kanR; ura3::CDC24-HA:URA3 | This study |
| DLY23831 | a; cdc12-6; rsr1::HIS3; BEM1-GFP:LEU2; CDC42:TRP1:CDC42 | This study |
| DLY23832 | a; cdc12-6; rsr1::TRP1; BEM1-GFP:LEU2; ura3::CDC24-HA:URA3; CDC42:TRP1:CDC42 | This study |
| DLY23833 | α; cdc12-6; rsr1::HIS3; BEM1-GFP:TRP1:BEM1-GFP:LEU2; CDC24-HA:KAN; CDC42:TRP1:CDC42 | This study |
| DLY23843 | α; cdc12-6; rsr1::HIS3; BEM1-GFP:TRP1:BEM1-GFP:LEU2; CDC24-HA:KAN; ura3::CDC24-HA:URA3 | This study |
Additional files
-
Source code 1
Mechanistic negative feedback model.
The source code files used to simulate the 2D mechanistic negative feedback model based on GEF phosphorylation (Figure 6Aiii, B) and 2D mechanistic positive feedback model (Figure 1GHIJ) if k8 is set to zero.
- https://cdn.elifesciences.org/articles/58768/elife-58768-code1-v2.zip
-
Source code 2
Indirect substrate model.
The source code files used to simulate 1D conceptual ‘Indirect substrate model’ in Figure 5.
- https://cdn.elifesciences.org/articles/58768/elife-58768-code2-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58768/elife-58768-transrepform-v2.docx





