S-phase-independent silencing establishment in Saccharomyces cerevisiae
Figures
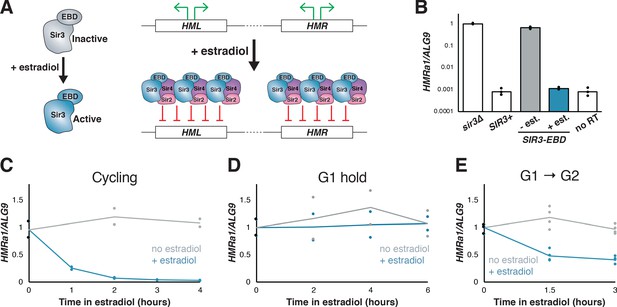
Silencing establishment using SIR3-EBD required S-phase progression.
(A) Schematic for SIR3-EBD activation. When estradiol is absent, SIR3-EBD is kept inactive and HML and HMR are expressed. Upon addition of estradiol, SIR-EBD is activated and HML and HMR are repressed. (B) RT-qPCR of mRNA from sir3∆ (JRY12168), SIR3 (JRY12171), and SIR3-EBD (JRY12170) cells grown with ethanol (solvent control) or estradiol (N = 3 for each condition). Also plotted is the non-reverse-transcribed (no RT) value HMRa1no RT/ALG9 for SIR3 cells , to demonstrate that SIR3 cells and SIR3-EBD cells grown with estradiol silenced HMRa1 to essentially the limit of detection. (C) SIR3-EBD cultures (JRY12169, JRY12170) were grown to mid-log phase, then split and grown in medium with either estradiol or ethanol added. Silencing was monitored by RT-qPCR in a time course after estradiol addition. t = 0 represents the point of estradiol addition for this and subsequent experiments. (D) SIR3-EBD cultures (JRY12169, JRY12170) were arrested in G1 with α factor, then split, with either ethanol or estradiol added. The arrest was maintained for 6 hr, and silencing was assayed by RT-qPCR throughout. (E) SIR3-EBD cultures (JRY12169, JRY12170; 2 replicates of each genotype) were arrested in G1 with α factor, then split and released to G2/M by addition of protease and nocodazole in the presence of either ethanol or estradiol. In this and all subsequent figures, dots represent biological replicates, and the bars/lines represent the averages of biological replicates.
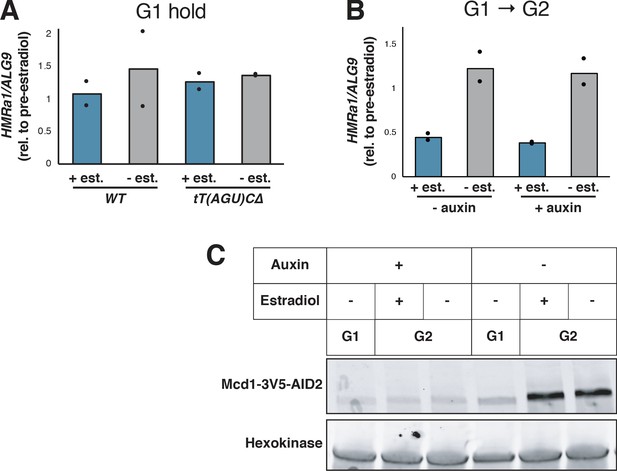
Effects of cohesin depletion and tT(AGU)C deletion on silencing establishment.
(A) SIR3-EBD strains (JRY12269, JRY12270) and SIR3-EBD strains with seamless deletion of tT(AGU)C (JRY12267; JRY12268) were arrested in G1 with α factor, then split, with half the culture receiving estradiol and half receiving ethanol. Samples were collected after 3 hr for RT-qPCR, with each sample normalized to its own pre-estradiol value. (B) Cells with MCD1-AID (JRY12560, JRY12561) were arrested in G1 with α factor, then split, with half receiving auxin and the other half receiving DMSO (solvent control). After 30 min, each culture was further split, with half receiving estradiol and the other half ethanol. All cultures were released to G2/M by addition of protease and nocodazole. Cells were collected after 3 hr for RT-qPCR, with each sample normalized to its own pre-estradiol value. (C) Immunoblot analysis showing Mcd1-AID depletion for experiment described in (B).
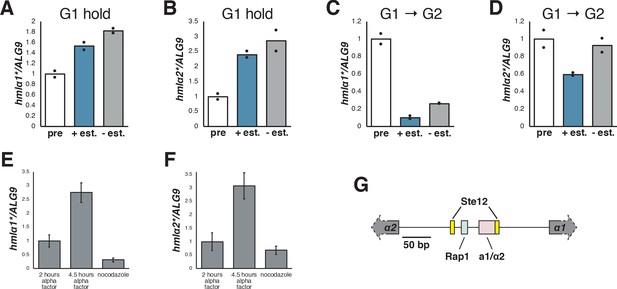
Silencing establishment required S phase at HML*.
(A) Cells with SIR3-EBD (JRY12169) were arrested in G1 with α factor, then split, with one sub-culture receiving estradiol and the other receiving ethanol. Cells were collected after 6 hr in estradiol and analyzed by RT-qPCR for hmlα1*. (B) RT-qPCR for hmlα2* in the same cells described in (A). (C) Cells were arrested in G1 with α factor, then split and released to G2/M with protease and nocodazole, with one sub-culture receiving estradiol and the other receiving ethanol. Cells were collected after 3 hr in estradiol and analyzed by RT-qPCR for hmlα1*. The pre-estradiol samples for this experiment were the same as those described in (A). (D) RT-qPCR for hmlα2* in the same cells described in (C). (E) Cells lacking SIR3 (JRY11966) were arrested in α factor for 2 hr, then split, with half staying in α factor and the other being released to G2/M by addition of protease and nocodazole. Cells were analyzed by RT-qPCR for hmlα1*, with error bars representing standard deviation among technical replicates (N = 1 biological sample). (F) RT-qPCR for hmlα2* in the cells described in (E). (G) Map of the α1/α2 promoter, showing newly-identified Ste12 motifs (TGAAACA) along with previously-identified binding sites for Rap1 and a1/α2.
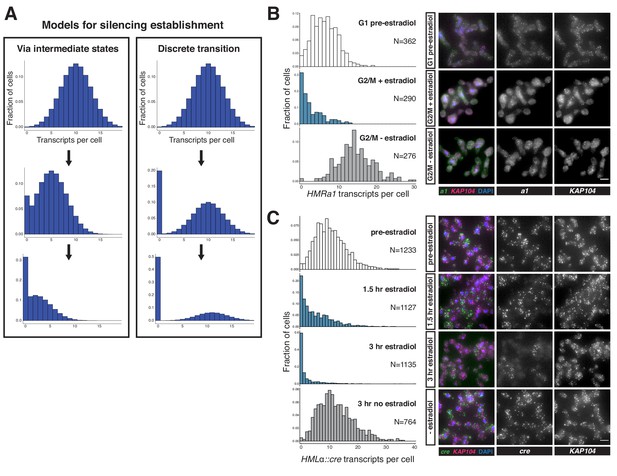
Silencing establishment proceeded via gradual repression in individual cells.
(A) Potential models for silencing establishment. Before silencing establishment (top), mRNA transcripts are present as a distribution around a mean. If silencing establishment occurred via intermediate states (left), the mean number of transcripts per cell would decrease over time, with complete silencing, that is zero transcripts per cell, occurring as the probability distribution shifted toward the y axis. If silencing establishment occurred via discrete transitions (right), an increasing fraction of cells would have zero transcripts over time, but the distribution of cells with >0 transcripts would retain the same shape. (B) smRNA-FISH for HMRa1 during silencing establishment after 1 s phase. A SIR3-EBD culture (JRY11762) was arrested in G1 with α factor (‘G1 pre-estradiol’), then split and released to G2/M by addition of protease and nocodazole in the presence of either estradiol or ethanol. Samples were collected 2 hr after estradiol addition. (C) smRNA-FISH for HMLα::cre during silencing establishment in cycling cells. A SIR3-EBD strain bearing the HMLα::cre reporter (JRY12514) was grown to mid-log phase (‘pre-estradiol’), then the culture was split in two, with one sub-culture receiving estradiol and the other receiving ethanol. Samples were collected for smRNA-FISH at t = 1.5 hr and t = 3 hr after estradiol addition. For both (B) and (C), the images displayed are representative maximum-intensity Z-projections. The data shown in (B) and (C) each represent one of two replicate experiments, for which the other replicate is shown in Figure 2—figure supplements 1 and 2, respectively. Scale bars = 5 µm.
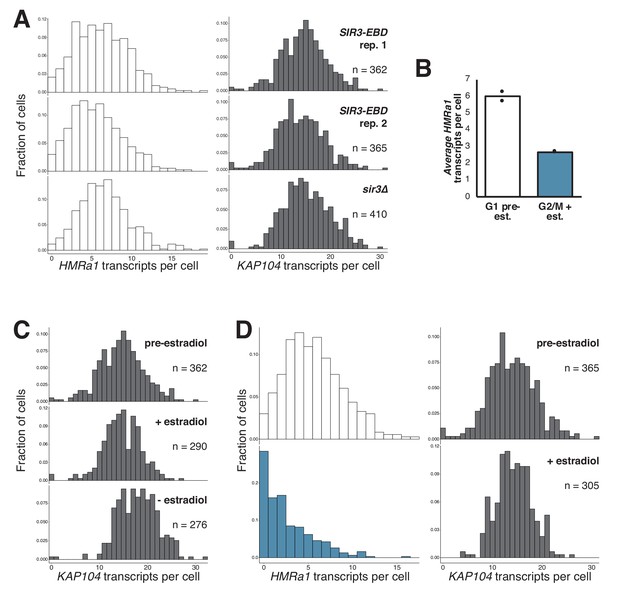
Gradual silencing establishment at HMR.
(A) Quantification of HMRa1 and KAP104 transcripts in G1-arrested SIR3-EBD (JRY11762, JRY11763) and sir3∆ (JRY11966) cells. The SIR3-EBD data are the same as displayed in D and Figure 2B. (B) Average number of HMRa1 transcripts per cell before and after silencing establishment, quantified from data shown in D and Figure 2B. Compare changes in mRNA levels to values from bulk measurement in Figure 1E. (C) Quantification of KAP104 transcripts per cell for experiment described in Figure 2B. (D) Replicate experiment to that shown in Figure 2B with isogenic cells (JRY11763).
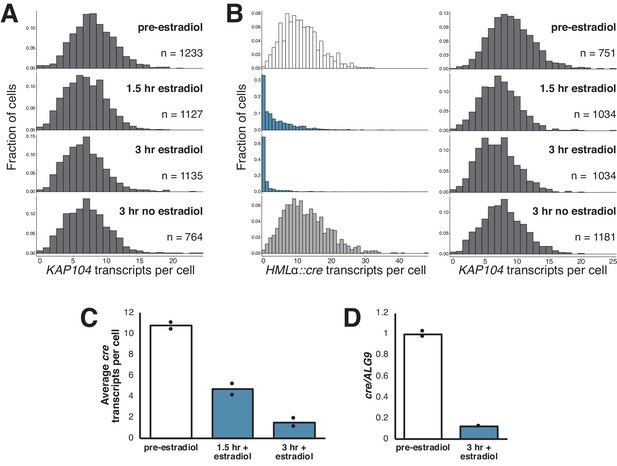
Gradual silencing establishment at HMLα::cre.
(A) Quantification of KAP104 transcripts per cell for experiment described in Figure 2C. (B) Replicate experiment to that shown in Figure 2B with isogenic cells (JRY12513). (C) Average number of cre transcripts per cell before and during silencing establishment, quantified from data shown in B and Figure 2C. (D) Bulk measurement RT-qPCR for cre during silencing establishment in an analogous experiment to that described in Figure 2C.
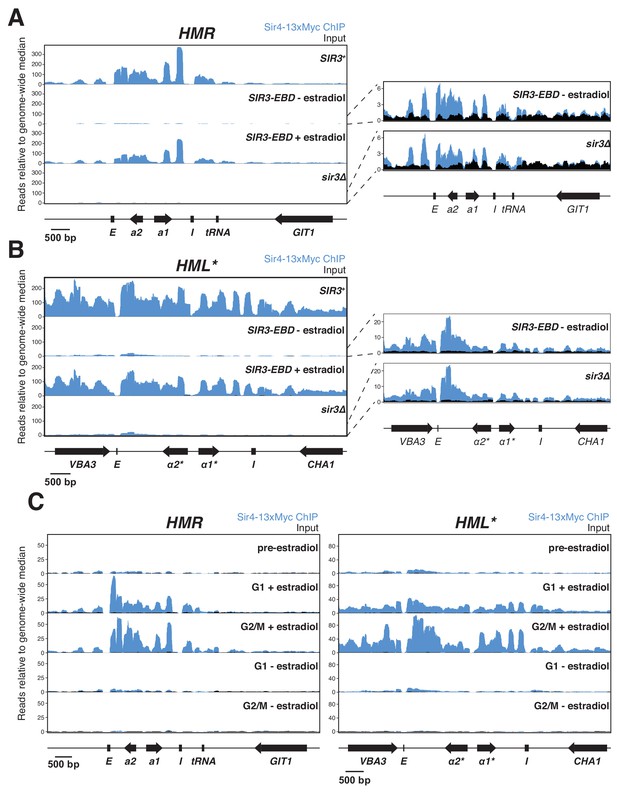
Sir protein binding and silencing were separable phenomena.
All panels show Sir4-13xMyc ChIP-seq signal in blue and input in black. Read counts were normalized to the non-heterochromatin genome-wide median. IP and input values are plotted on the same scale. (A) Left, ChIP-seq for Sir4-13xMyc at HMR in strains with SIR3 (JRY12172), sir3∆ (JRY12168), and SIR3-EBD (JRY12170) grown with or without estradiol and fixed for 60 min in formaldehyde. Right, same data as the left panel for sir3∆ and SIR3-EBD without estradiol, enlarged to show IP levels above input. (B) Same as (A), but showing data from HML*. (C) ChIP-seq for Sir4-13xMyc during silencing establishment at HMR (left) and HML* (right). Cultures of SIR3-EBD cells (JRY12169) were arrested in G1 with α factor (‘pre-estradiol’), then split four ways. Two sub-cultures were maintained in G1 in medium with estradiol or ethanol (‘G1 + estradiol’ and ‘G1 - estradiol’). The other two sub-cultures were released to G2/M by addition of protease and nocodazole; and received either estradiol or ethanol (‘G2/M + estradiol’ and ‘G2/M - estradiol’). After 3 hr in medium with estradiol or ethanol, cultures were fixed in formaldehyde for 15 min and collected for ChIP-seq. Data shown represent one of two replicates, with the other shown in Figure 3—figure supplement 1C.
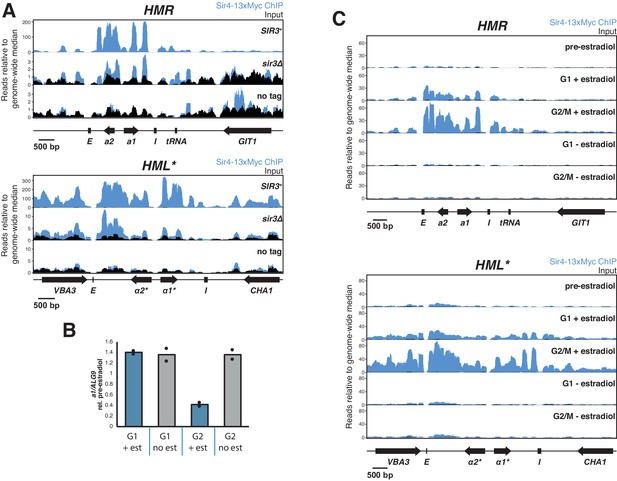
Silencing establishment ChIP-seq.
All ChIP-seq panels show Sir4-13xMyc in blue and input in black. Read counts were normalized to the non-heterochromatin genome-wide median. IP and input values are plotted on the same scale. (A) ChIP-seq for Sir4-13xMyc at HMR (top) and HML* (bottom) in strains with SIR3 (JRY12171) and sir3∆ (JRY12167). Also shown is an equivalent experiment in cells with untagged Sir4 (JRY12269). Cells were grown to mid-log phase and fixed for 15 min in formaldehyde. (B) RT-qPCR analysis of silencing establishment at HMRa1 from samples that were used for ChIP-seq described in C and Figure 3C. Values are scaled to the pre-estradiol value for each sample. (C) Replicate experiment to that described in Figure 3C, using an isogenic strain (JRY12170).
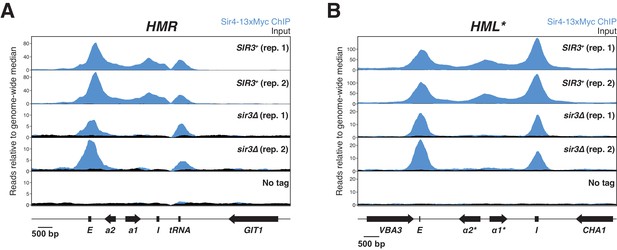
ChIP-seq with sonicated chromatin.
All panels show Sir4-13xMyc in blue and input in black. Read counts were normalized to the non-heterochromatin genome-wide median. IP and input values are plotted on the same scale. Cells with SIR3 (JRY12171, JRY12172), sir3∆ (JRY12167, JRY12168), and a no-tag control (JRY12169) were grown to mid-log phase and fixed for 15 min in formaldehyde. (A) ChIP-seq for Sir4-13xMyc at HMR. (B) ChIP-seq for Sir4-13xMyc at HML*.
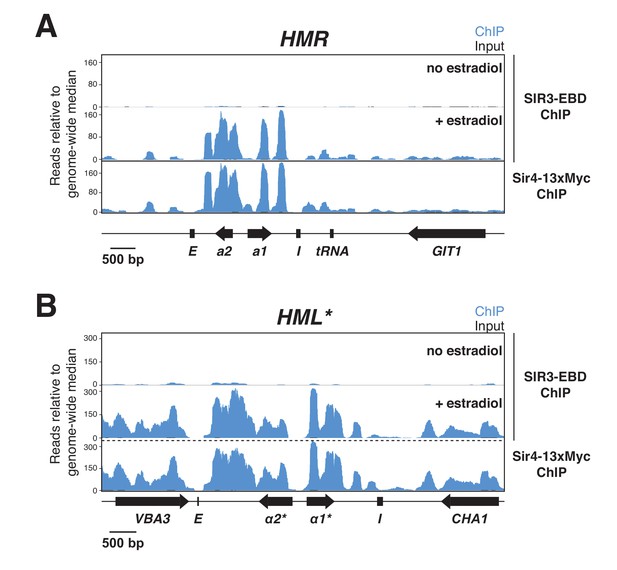
ChIP-seq for Sir3-EBD.
All panels show ChIP-seq signal in blue and input in black. Read counts were normalized to the non-heterochromatin genome-wide median. IP and input values are plotted on the same scale. Cells with SIR3-EBD (JRY12170) were grown to mid-log phase with or without estradiol, then fixed for 15 min in formaldehyde. Also shown is Sir4-13xMyc ChIP signal from Figure 3—figure supplement 1A. (A) Sir3-EBD and Sir4-13xMyc ChIP-seq at HMR. (B) Sir3-EBD and Sir4-13xMyc ChIP-seq at HML*.
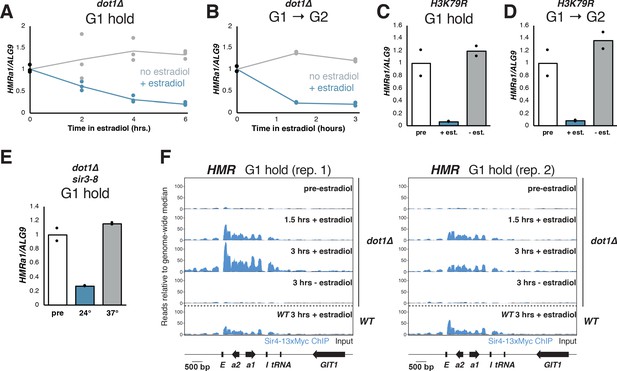
Cells without H3K79 methylation established silencing without cell-cycle progression.
(A) Cultures of dot1∆ cells (JRY12443, JRY12445) were arrested in G1 with α factor, then split, with half receiving ethanol and the other half receiving estradiol. Silencing was monitored by RT-qPCR over time after estradiol addition. (B) dot1∆ mutants were arrested in G1 with α factor, then released to G2/M by addition of protease and nocodazole, and either ethanol or estradiol. Silencing was monitored by RT-qPCR over time after estradiol addition. (C) Cultures of cells in which lysine 79 was mutated to arginine in both HHT1 and HHT2 in two isogenic strains (H3K79R; JRY12851, JRY12852) were arrested in G1 with α factor, then split, with one sub-culture receiving ethanol and the other receiving estradiol. Silencing was assayed by RT-qPCR after 6 hr in ethanol. (D) H3K79R cells were arrested in G1 with α factor, then released to nocodazole with protease and nocodazole and either estradiol or ethanol. Silencing was assayed by RT-qPCR after 3 hr in estradiol. The pre-estradiol sample for this experiment was the same culture used in (C). (E) Cultures of dot1∆ sir3-8 cells (JRY12859, JRY12890) were grown at the non-permissive temperature for sir3-8 (37°C) and arrested in G1 with α factor, then split, with half shifted to the permissive temperature (24°C) and other half staying at the non-permissive temperature. Silencing was assayed by RT-qPCR after 6 hr. (F) Cultures of dot1∆ cells (JRY12443, JRY12444) were arrested in G1 with α factor (‘pre-estradiol’), then split, with half the culture receiving ethanol, and the other half receiving estradiol. After 1.5 hr and after 3 hr, samples were fixed for 15 min in formaldehyde and collected for ChIP. Sir4-13xMyc ChIP-seq signal is in blue and input in black, each normalized to the non-heterochromatin genome-wide median and plotted on the same scale. Also displayed are two replicates of wild-type G1 cells after 3 hr in estradiol from Figure 3C and Figure 3—figure supplement 1C.
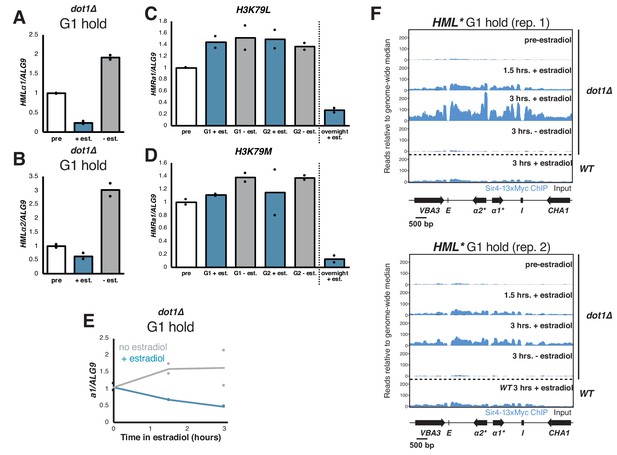
Silencing establishment in dot1∆ cells.
(A) dot1∆ cells (JRY12443) were arrested in G1 with α factor, then split, with one sub-culture receiving estradiol and the other receiving ethanol. Cells were collected after 6 hr in estradiol and RT-qPCR was performed for hmlα1*. The data in this plot are also shown in Figure 5B for comparison with other mutants. (B) RT-qPCR for hmlα2* in the cells described in (A). (C) Cells with lysine 79 of H3 mutated to leucine in both HHT1 and HHT2 in two isogenic strains (H3K79L; JRY12854, JRY12855) were arrested in G1 with α factor, then split four ways. Two cultures were kept in G1, with one receiving estradiol and the other ethanol. The other two cultures were released to G2/M by addition of protease and nocodazole, and either estradiol or ethanol. Cells were collected after 3 hr for the G2/M samples and after 6 hr for the G1 samples, and RT-qPCR was performed for HMRa1. Also shown is a sample grown overnight in medium with estradiol. (D) Cells with lysine 79 of H3 mutated to methionine in both HHT1 and HHT2 in two isogenic strains (H3K79M; JRY12857, JRY12858) were subjected to the same experiment described in (C). (E) RT-qPCR analysis of silencing establishment at HMRa1 from samples that were used for ChIP-seq shown in F and Figure 4F. (F) Sir4-13xMyc binding at HML* from the same samples displayed in Figure 4F. Sir4-13xMyc is in blue and input in black, plotted on the same scale. Read counts were normalized to the non-heterochromatin genome-wide median.
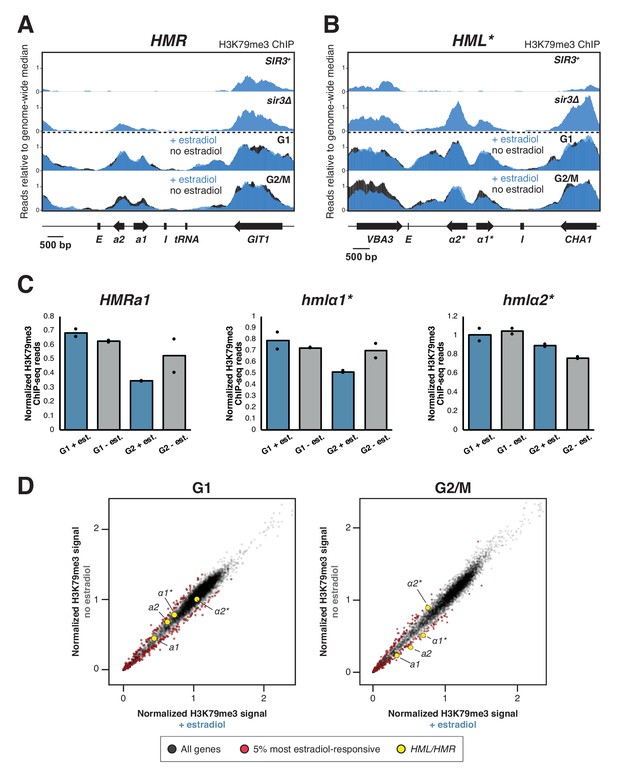
H3K79 trimethylation dynamics during silencing establishment.
SIR3-EBD cultures (JRY12169, JRY12170) were arrested in G1 with α factor, then split four ways. Two cultures were kept in G1, with one receiving estradiol (blue) and the other ethanol (black). The other two cultures were released to G2/M by addition of protease and nocodazole, and either estradiol (blue) or ethanol (black). Cells were collected after 3 hr and subjected to ChIP-seq for H3K79me3. (A) ChIP-seq for H3K79me3 at HMR. For the top two panels, cultures of SIR3(JRY12171) and sir3∆ (JRY12167) cells were grown to mid-log phase and subjected to ChIP-seq for H3K79me3. All data are plotted on the same scales. For the G1 and G2/M samples, the plotted value is the average of two biological replicates. (B) ChIP-seq for H3K79me3 at HML*. (C) Total ChIP-seq coverage was calculated for each gene and scaled to the length of the gene and the genome-wide mean coverage. Displayed is the normalized H3K79me3 signal for HMRa1, hmlα1*, and hmlα2*. (D) Normalized H3K79me3 signal for all genes after estradiol or ethanol addition, in G1 (left) and in G2/M (right). The plotted values represent the average of two biological replicates. The 5% most estradiol-responsive genes are shaded in red and the genes at HML and HMR are enlarged and shaded in yellow.
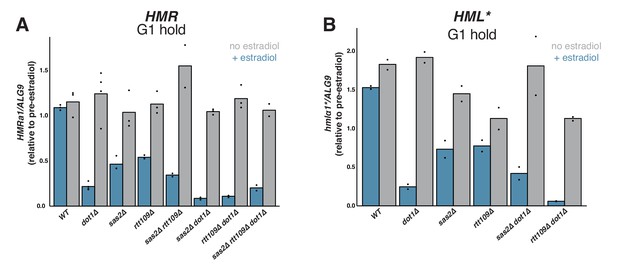
Effects of SAS2 and RTT109 on silencing establishment in G1.
For all strains, cultures were arrested in G1 with α factor, then split, with one sub-culture receiving estradiol and the other receiving ethanol. Silencing was assayed by RT-qPCR 6 hr after additions. Each sample was normalized to its own pre-estradiol value. The following strains were used. WT: JRY12169; dot1∆: JRY12443, JRY12445; sas2∆: JRY12615, JRY12616; rtt109∆: JRY12689, JRY12690; sas2∆ rtt109∆: JRY12765, JRY12766; sas2∆ dot1∆: JRY12618, JRY12619; rtt109∆ dot1∆: JRY12691, JRY12692; sas2∆ rtt109∆ dot1∆: JRY12767, JRY12768. (A) Silencing establishment of HMRa1 by RT-qPCR. The level of repression observed in each mutant was significantly greater than in wild type (Two-tailed T-test; p<0.005 for each pair-wise comparison). The level of repression observed in sas2∆ dot1∆ and rtt109∆ dot1∆ double mutants was significantly greater than in the dot1∆ single mutant (p<0.01 for each pair-wise comparison), but there was no significant difference between the values from the dot1∆ single mutant and the triple mutant sas2∆ rtt109∆ dot1∆ (p=0.70). (B) Silencing establishment of hmlα1* by RT-qPCR in a subset of mutant strains. The level of repression for each mutant was significantly greater than in wild type (p<0.05 for each pair-wise comparison). The level of repression observed in the rtt109∆ dot1∆ double mutant was significantly greater than in the dot1∆ single mutant (p=0.028), but there was no significant difference between the values from sas2∆ dot1∆ double mutant and the dot1∆ single mutant (p=0.19).
-
Figure 5—source data 1
P-values for comparisons displayed in Figure 5.
- https://cdn.elifesciences.org/articles/58910/elife-58910-fig5-data1-v2.xlsx
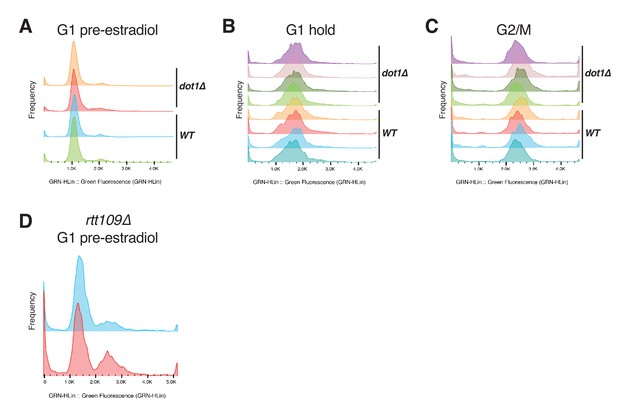
Representative flow cytometry profiles.
(A) Wild-type (JRY12169) and dot1∆ (JRY12443) cells arrested in G1 with α factor for ~2 hr. (B) Wild-type and dot1∆ cells kept in G1 for 6 hr, as in, for example Figure 1D. (C) Wild-type and dot1∆ cells after 3 hr in G2/M, as in, for example Figure 1E. (D) Cells lacking RTT109 (JRY12689, JRY12691) arrested in G1 with α factor for ~3 hr did not arrest uniformly.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Saccharomyces cerevisiae) | Various | This paper | NCBITaxon:4932 | SeeSupplementary file 1a |
| Antibody | Anti-c-myc beads (mouse monoclonal) | Thermo Fisher Scientific | Cat # 88842 | 50 µL per IP |
| Antibody | Anti-H3K79me3 (rabbit polyclonal) | Diagenode | Cat # C15410068 | 5 µL per IP |
| Antibody | Anti-ERα (rabbit polyclonal) | Santa Cruz Biotechnology | Cat # sc-8002, RRID:AB_627558 | 25 µL per IP |
| Antibody | Anti-Hexokinase (Rabbit polyclonal) | Rockland | Cat # 100–4159, RRID:AB_219918 | (1:20,000) |
| Antibody | Anti-V5 (Mouse monoclonal) | Thermo Fisher Scientific | Cat # R960-25, RRID:AB_2556564 | (1:2,500) |
| Antibody | IRDye 800CW anti-mouse (Goat polyclonal) | Li-Cor | Cat # 926–32210, RRID:AB_621842 | (1:20,000) |
| Antibody | IRDye 680RD anti-rabbit (Goat polyclonal) | Li-Cor | Cat # 926–68070, RRID:AB_10956588 | (1:20,000) |
| Recombinant DNA reagent | HML* | This paper | Mutated allele of HML | |
| Recombinant DNA reagent | SIR3-EBD | This paper | Fusion protein of Sir3 and EBD of mammalian ERα | |
| Sequence-based reagent | Various oligonucleotides | This paper | qPCR primers | SeeSupplementary file 1b |
| Sequence-based reagent | smRNA-FISH probes | Biosearch Technologies | SeeSupplementary file 1c | |
| Commercial assay or kit | RNEasy Mini Kit | Qiagen | Cat # 74104 | |
| Commercial assay or kit | Qiaquick PCR purification kit | Qiagen | Cat # 28104 | |
| Commercial assay or kit | NEBNext Ultra II Library prep kit | NEB | Cat # 37645L | |
| Commercial assay or kit | Superscript III reverse transcriptase kit | Thermo Fisher Scientific | Cat # 18080044 | |
| Commercial assay or kit | DyNamo HS SYBR Green qPCR kit | Thermo Fisher Scientific | Cat # F410L | |
| Peptide, recombinant protein | Catalase | Sigma-Aldrich | Cat # C3515 | |
| Peptide, recombinant protein | Proteinase K | NEB | Cat # P8107S | |
| Peptide, recombinant protein | Glucose oxidase | Sigma-Aldrich | Cat # G2133 | |
| Peptide, recombinant protein | Zymolyase-100T | VWR | Cat # IC320932 | |
| Peptide, recombinant protein | Protein A beads | Thermo Fisher Scientific | Cat # 10002D | |
| Peptide, recombinant protein | α-factor peptide | Elim Bio | ||
| Peptide, recombinant protein | Uracil-DNA Glycosylase | Thermo Fisher Scientific | Cat # EN0362 | |
| Peptide, recombinant protein | Micrococcal nuclease | Worthington Biochemical | Cat # LS004798 | |
| Commercial assay or kit | RNase-Free DNase Set | Qiagen | Cat # 79254 | |
| Chemical compound, drug | Nocodazole | Sigma-Aldrich | Cat # M1404 | |
| Chemical compound, drug | β-estradiol | Sigma-Aldrich | Cat # E8875 | |
| Chemical compound, drug | cOmplete EDTA-free protease inhibitor | Simga-Aldrich | Cat # 11873580001 | |
| Chemical compound, drug | 3-indoleacetic acid | Sigma-Aldrich | Cat # I2886 | |
| Software, algorithm | SAMtools | doi:10.1093/bioinformatics/btp352 | RRID:SCR_002105 | |
| Software, algorithm | Ggplot2 | doi:10.1007/978-0-387-98141-3 | RRID:SCR_014601 | |
| Software, algorithm | Bowtie2 | doi:10.1038/nmeth.1923 | RRID:SCR_005476 | |
| Software, algorithm | FISH-quant | doi:10.1038/nmeth.2406 | ||
| Software, algorithm | FlowJo | BD Life Sciences | RRID:SCR_008520 |
Additional files
-
Source code 1
Python script for bedgraph generation.
- https://cdn.elifesciences.org/articles/58910/elife-58910-code1-v2.zip
-
Source code 2
Python script for bedgraph normalization.
- https://cdn.elifesciences.org/articles/58910/elife-58910-code2-v2.zip
-
Supplementary file 1
Supplementary File 1a: Yeast strains used in this study. All strains listed were generated for this study and derived from the W303 background. Supplementary File 1b: Oligonucleotides used for RT-qPCR Supplementary File 1c: Probes used for smRNA-FISH.
- https://cdn.elifesciences.org/articles/58910/elife-58910-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/58910/elife-58910-transrepform-v2.pdf





