Conservation of peripheral nervous system formation mechanisms in divergent ascidian embryos
Figures
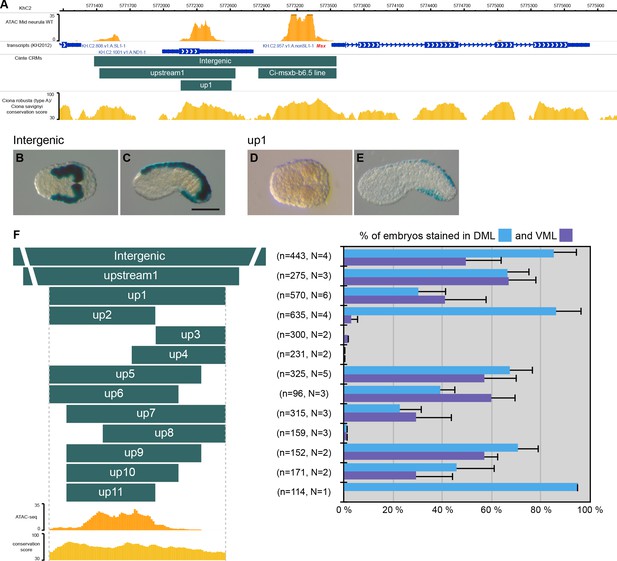
CRMs controlling Ciinte.Msx expression in VDML.
(A) Snapshot of the Ciinte.Msx locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between C. robusta and C. savignyi (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). (B–E) Representative examples of X-gal stained embryos at late gastrula stages (B, D) and early tailbud stages (C, E) following C. intestinalis embryos electroporation of Ciinte.Msx-Intergenic (B, C) and Ciinte.Msx-up1 (D, E). Embryos are shown in dorsal view (B, D) and in lateral view with dorsal to the top (C, E), and anterior to the left. Scale bar: 100 μm. (F) Schematic representation of the various constructs and their activity at early tailbud stages in DML (blue) and VML (purple) (n indicates the total number of embryos examined; N indicates the number of independent experiments).
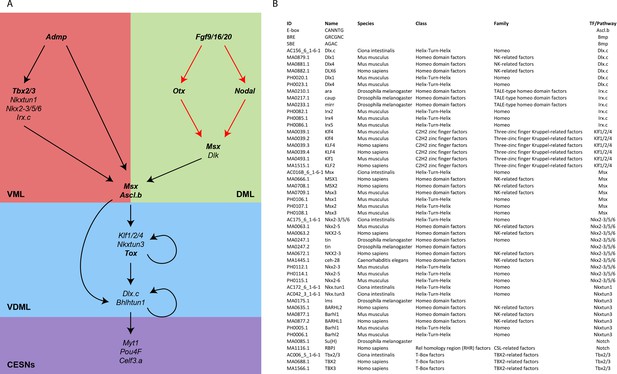
Developmental regulators of caudal PNS in C. intestinalis.
(A) Provisional GRN for caudal PNS specification in C. intestinalis. The genes depicted and their interactions are based on previous publications (Roure et al., 2014; Pasini et al., 2006; Roure and Darras, 2016; Waki et al., 2015; Joyce Tang et al., 2013; Bertrand et al., 2003) and hypotheses described in the Materials and methods section. Genes whose requirement for caudal PNS formation has been shown by loss-of-function are in bold. Demonstrated direct regulations are shown in red. (B) List of matrices and consensus motifs used to identify TFBS in CRMs.
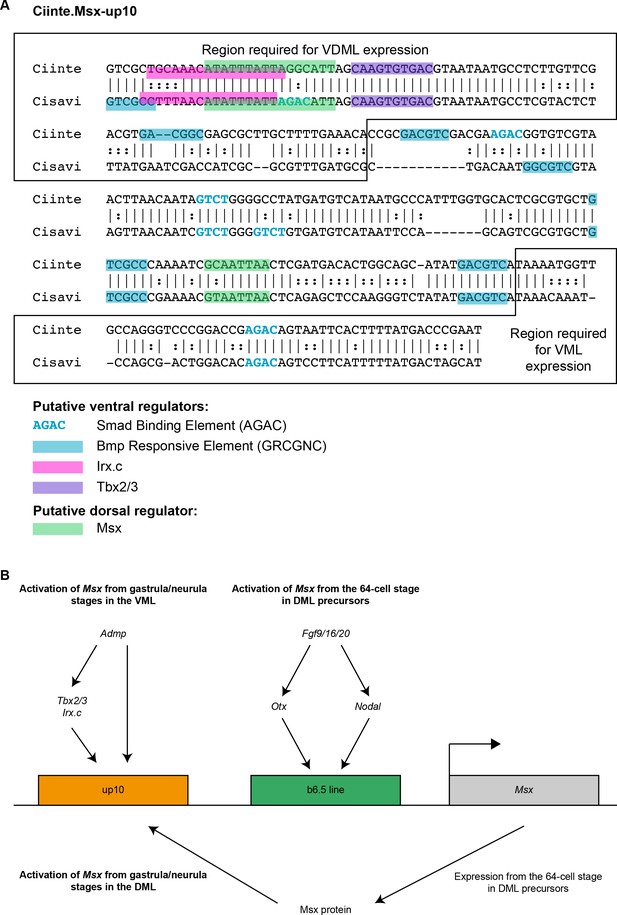
Model for Ciinte.Msx regulation.
(A) Identification of putative TFBS for candidate upstream factors in Ciinte. Msx-up10 aligned with its counterpart from C. savignyi. All putative sites for ventral factors (SBE, BRE, Tbx2/3, Nkxtun1, Nkx2-3/5/6 and Irx.c) and dorsal factors (Msx and Su(H)/Rbpj) have been mapped, but only conserved sites are shown: 4 BRE, 3 SBE, 1 Irx.c, 1 Tbx2/3 and 2 Msx. The region deleted in up2 and up11 abuts a BRE and contains a SBE; the loss of these sites might thus be responsible of the absence of activity in the VML. The region deleted in up8 and required for VDML activity contains 1 BRE, 1 Irx.c, 1 Tbx2/3 and 1 Msx sites. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used. (B) Working model for Msx transcriptional regulation in the VDML. Msx expression is initiated in DML precursors at the 64-cell stage (Roure et al., 2014) through the proximal/early CRM (denoted b6.5 line, green) activated by Otx and Nodal (via Smad2/3). The Msx protein produced via this regulation would activate Msx transcritption in the DML at gastrula/neurula stages via the distal/late CRM (denoted up10, orange). The same distal CRM would activate Msx expression in the VML directly by Bmp signaling activated by the Admp ligand (via the SBEs and BREs) or Admp targets (Irx.c and Tbx2/3).
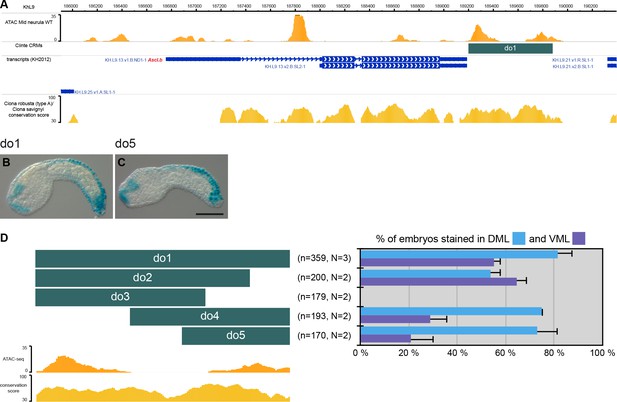
CRMs controlling Ciinte.Ascl.b expression in VDML.
(A) Snapshot of the Ciinte.Ascl.b locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between C. robusta and C. savignyi (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). (B–C) Representative examples of X-gal stained embryos at early tailbud stages following C. intestinalis embryos electroporation of Ciinte.Ascl.b-do1 (B) And Ciinte.Ascl.b-do5 (C). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 100 μm. (D) Schematic representation of the various constructs and their activity at early tailbud stages in DML (blue) and VML (purple) (n indicates the total number of embryos examined; N indicates the number of independent experiments).

Putative TFBS.
Identification of putative TFBS for candidate upstream factors in Ciinte.Ascl.b-do2 aligned with its counterpart from C. savignyi. All putative sites for ventral factors (SBE, BRE, Tbx2/3, Nkxtun1, Nkx2-3/5/6 and Irx.c) and dorsal factors (Msx and Su(H)/Rbpj) have been mapped, but only conserved sites are shown: 2 SBE, 3 Tbx2/3, 2 Nkx2-3/5/6 and 2 Msx sites. The region important for VML expression (deleted in do4) contains 2 Tbx2/3 and 1 Nkx2-3/5/6 sites. Since Ascl.b is expressed after Msx (Roure and Darras, 2016), it could be directly regulated by Msx in both VML and DML. However, the region required for VDML activity (deleted in do3) contains a Tbx2/3 site. This suggest that Ascl.b expression relies on additional unidentified factors. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used.
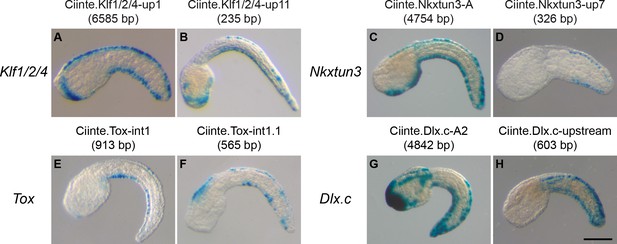
CRMs with activity in the tail epidermis midlines for Ciinte.Klf1/2/4, Ciinte.Nkxtun3, Ciinte.Tox, and Ciinte.Dlx.c.
Representatives examples of X-gal stained embryos at tailbud stages following C. intestinalis embryos electroporation of C. intestinalis genomic regions for Klf1/2/4 (A, B), Nkxtun3 (C, D), Tox (E, F) and Dlx.c (G, H). For each gene, an example for the largest and the smallest regions with robust VDML activity are shown (the size of the region is shown between parentheses after the CRM's name). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 100 μm.
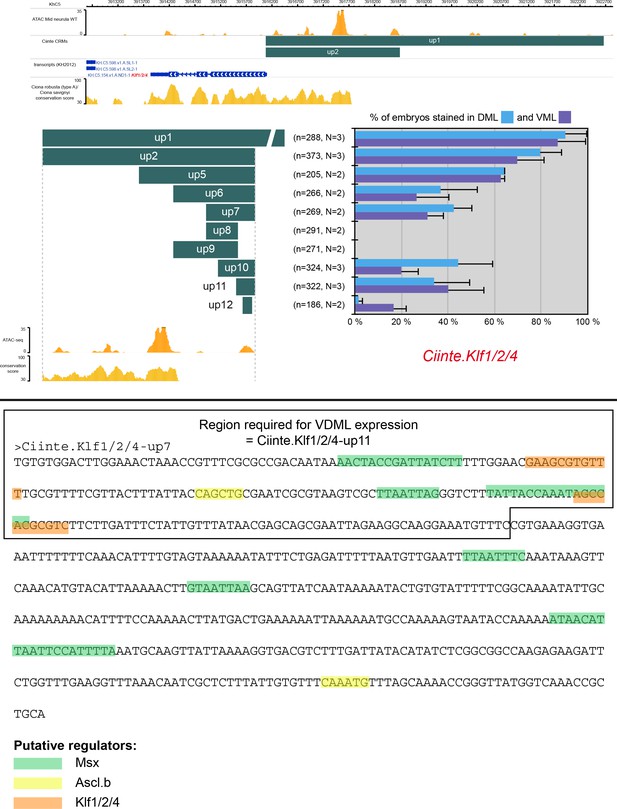
CRMs controlling Ciinte.Klf1/2/4 expression in VDML.
(Top) Snapshot of the Ciinte.Klf1/2/4 locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between C. robusta and C. savignyi (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Schematic representation of the various constructs and their activity at tailbud stages in DML (blue) and VML (purple) (n indicates the total number of embryos examined, N indicates the number of independent experiments). (Bottom) Identification of putative TFBS for candidate upstream factors in Ciinte.Klf1/2/4-up7 (604 bp). Note that all predicted sites for Msx (6 sites) (Schwartz et al., 2003), Ascl.b (2 sites) (Davidson and Erwin, 2006), and Klf1/2/4 (2 sites) are depicted since this region does not align with the C. savignyi genome (see top panel). The abundance of sites for Msx makes it a likely activator. Up11 is a smaller derivative of up7 (235 bp fragment long) that behaves as a minimal VDML enhancer (Figure 3B) and is deleted in the up8 region (inactive). It is located 2.4 kb upstream of Klf1/2/4, corresponds to an ATAC-seq enrichment detected from late gastrula stages (Supplementary file 2) and contains the 2 Klf1/2/4 sites, suggesting it might be involved in autoregulation/maintenance. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used.
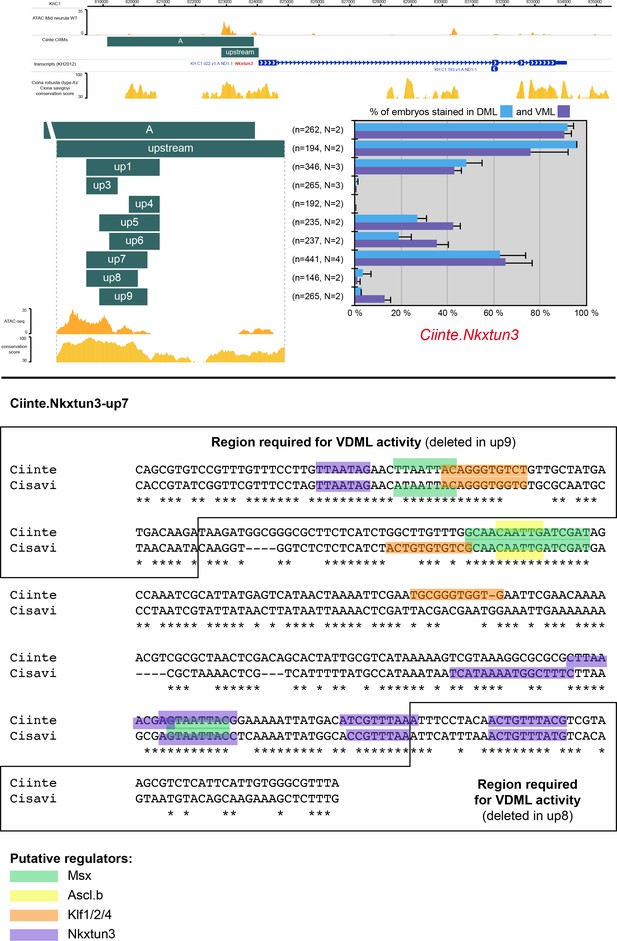
CRMs controlling Ciinte.Nkxtun3 expression in VDML.
(Top) Snapshot of the Ciinte.Nkxtun3 locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between C. robusta and C. savignyi (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Schematic representation of the various constructs and their activity at tailbud stages in DML (blue) and VML (purple) (n indicates the total number of embryos examined, N indicates the number of independent experiments). (Bottom) Identification of putative TFBS for candidate upstream factors in Ciinte.Nkxtun3-up7 aligned with its counterpart from C. savignyi. All putative sites for Msx, Ascl.b, Klf1/2/4 and Nkxtun3 have been mapped, but only conserved sites are shown: 3 Msx, 1 Ascl.b, 2 Klf1/2/4, and 5 Nkxtun3 sites. Deletions on either end of up7 that abolish activity remove 1 Nkxtun3 site, and 1 Msx, 1 Klf1/2/4, and 1 Nkxtun3 sites, respectively. This is suggestive of activation by Msx and Klf1/2/4, and autoregulation/maintenance by Nkxtun3. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used.
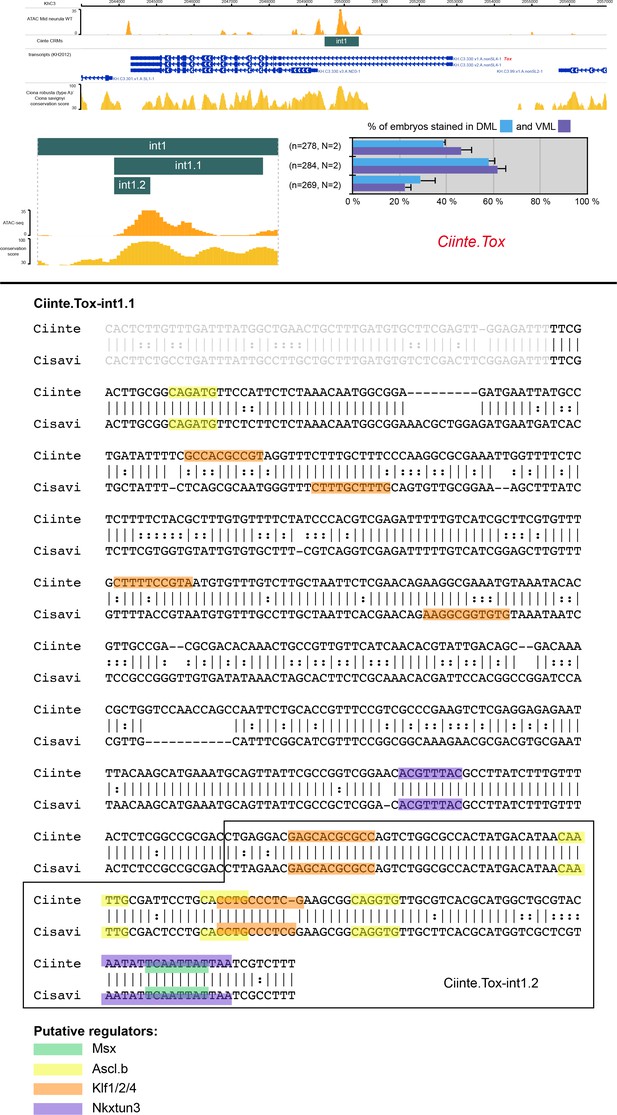
CRMs controlling Ciinte.Tox expression in VDML.
(Top) Snapshot of the Ciinte.Tox locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between C. robusta and C. savignyi (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Schematic representation of the various constructs and their activity at tailbud stages in DML (blue) and VML (purple) (n indicates the total number of embryos examined, N indicates the number of independent experiments). (Bottom) Identification of putative TFBS for candidate upstream factors in Ciinte.Tox-int1.1 aligned with its counterpart from C. savignyi. All putative sites for Msx, Ascl.b, Klf1/2/4 and Nkxtun3 have been mapped, but only conserved sites are shown: 1 Msx, 4 Ascl.b, 4 Klf1/2/4, and 3 Nkxtun3 sites. The smaller region Ciinte.Tox-int1.2 contains 3 Ascl.b and 2 Klf1/2/4 suggesting activation by Ascl.b and Klf1/2/4. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used.
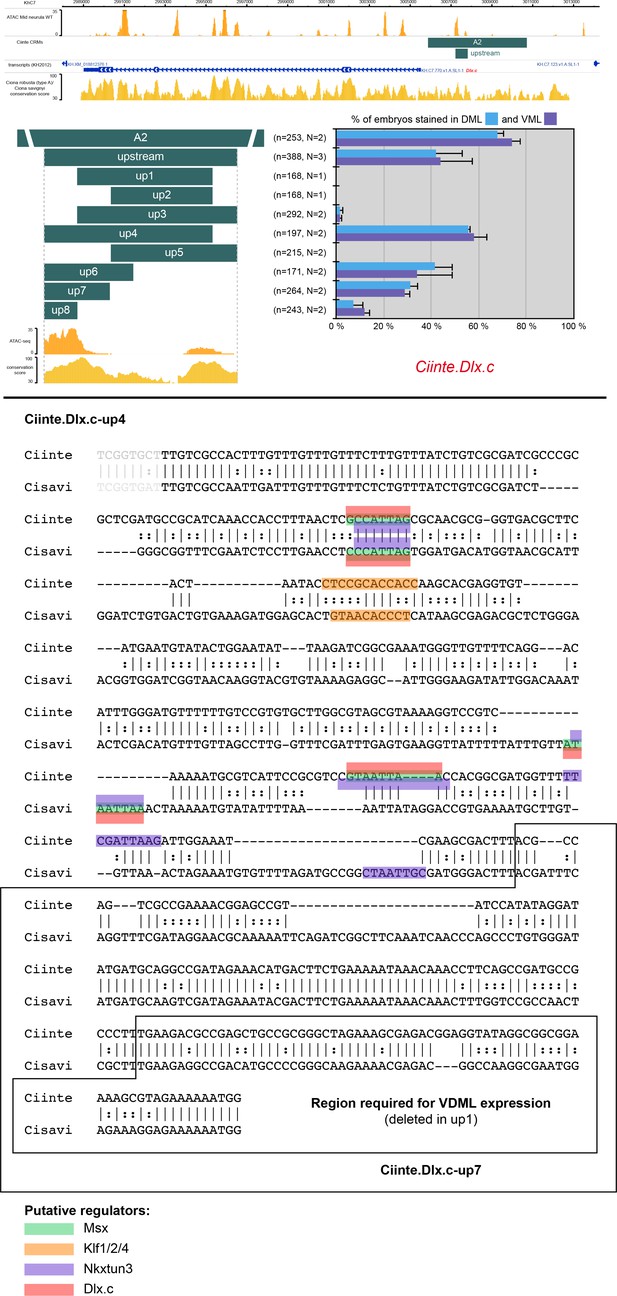
CRMs controlling Ciinte.Dlx.c expression in VDML.
(Top) Snapshot of the Ciinte.Dlx.c locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between C. robusta and C. savignyi (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Schematic representation of the various constructs and their activity at tailbud stages in DML (blue) and VML (purple) (n indicates the total number of embryos examined, N indicates the number of independent experiments). (Bottom) Identification of putative TFBS for candidate upstream factors in Ciinte.Dlx.c-up4 aligned with its counterpart from C. savignyi. All putative sites for Msx, Ascl.b, Klf1/2/4, Nkxtun3, and Dlx.c have been mapped, but only conserved sites are shown: 2 Msx, 1 Klf1/2/4, 3 Nkxtun3, and 2 Dlx.c sites. However, these sites are absent from the essential region deleted in up1. Consequently, unidentified factors are likely to regulate Ciinte.Dlx.c-up4 activity and Dlx.c expression. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used.
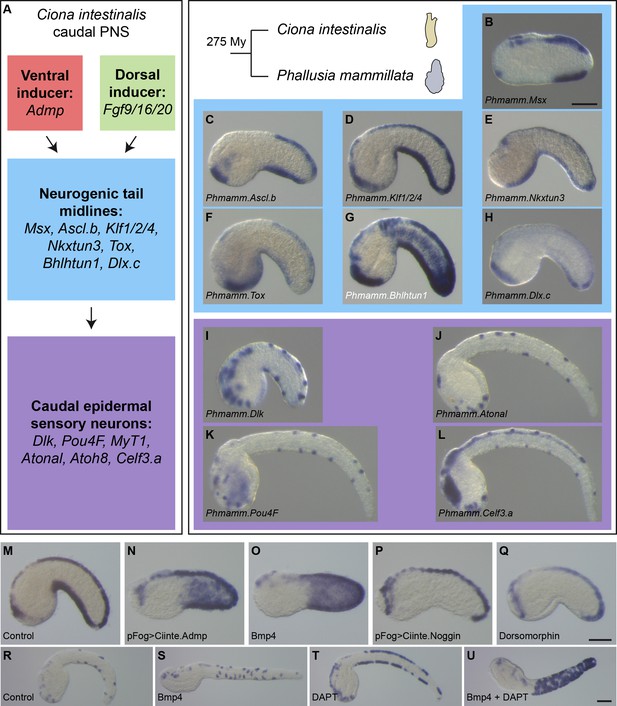
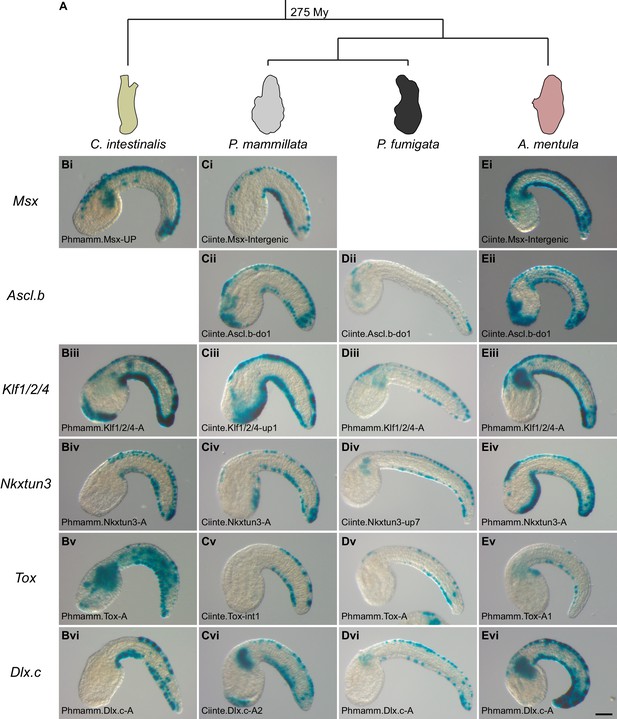
Expression and regulation of caudal PNS genes in Phallusia mammillata.
(A) Schematic summary of caudal PNS specification and molecular regulators in Ciona intestinalis (adapted from Roure et al., 2014; Pasini et al., 2006; Roure and Darras, 2016; Waki et al., 2015; Joyce Tang et al., 2013, more details can be found in Figure 1—figure supplement 1A). (B–L) Expression of the P. mammillata orthologs of the C. intestinalis caudal PNS genes. In situ hybridization of a late neurula for Msx (B), at early tailbud stages for Ascl.b (C), Klf1/2/4 (D), Nkxtun3 (E), Tox (F), Bhlhtun1 (G), Dlx.c (H) and Dlk (I); and at mid/late tailbud stages for Atonal (J), Pou4F (K), and Celf3.a (L). (M–Q) Expression of Phmamm-Klf1/2/4 at early tailbud stages in control embryos (M), following electroporation of pFog >Ciinte.Admp (N) or pFog >Ciinte.Noggin (P); or following treatment with Bmp4 protein (O) or Dorsomorphin (Q). (R–U) Expression of Phmamm.Pou4F at late mid/late tailbud stages in control embryos (R), following treatment with Bmp4 protein (S), DAPT (T) or a combination of Bmp4 and DAPT (U). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bars: 50 μm.
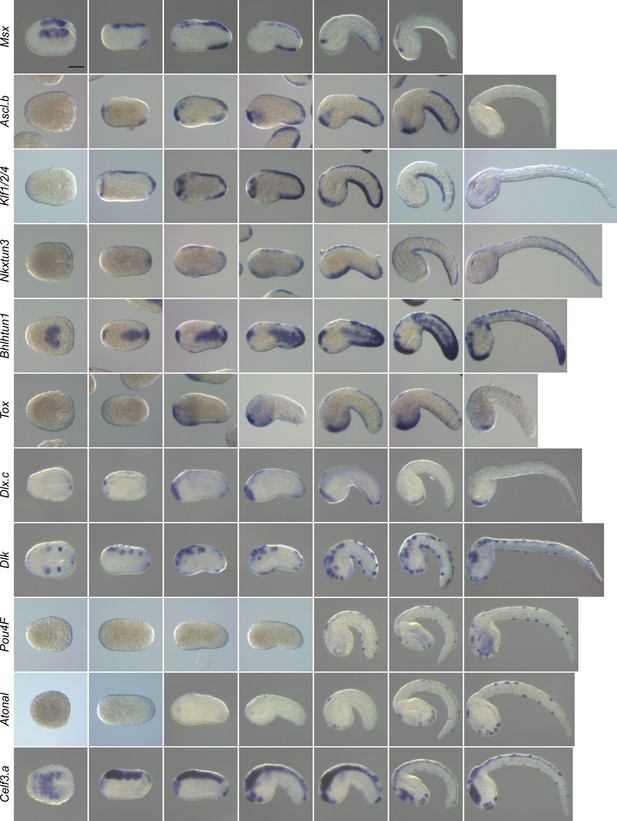
Expression pattern of caudal PNS genes in Phallusia mammillata.
In situ hybridization results at several developmental stages for the P. mammillata orthologs of the C. intestinalis caudal PNS genes: Msx, Ascl.b, Klf1/2/4, Nkxtun3, Bhlhtun1, Tox, Dlx.c, Dlk, Pou4F, Atonal, and Celf3.a. Scale bar: 50 μm.
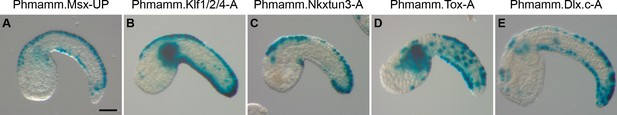
CRMs with activity in the tail epidermis midlines for Phmamm.Msx, Phmamm.Klf1/2/4, Phmamm.Nkxtun3, Phmamm.Tox, and Phmamm.Dlx.c.
Representatives examples of X-gal staining at tailbud stages following P. mammillata embryos electroporation of P. mammillata genomic regions for Msx (A), Klf1/2/4 (B), Nkxtun3 (C), Tox (D), and Dlx.c (E). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 50 μm.

CRMs controlling Phmamm.Msx expression in VDML.
(Top) Snapshot of the Phmamm.Msx locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between P. mammillata and P. fumigata (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Representative examples of X-gal staining at tailbud stages following P. mammillata embryos electroporation of P. mammillata genomic regions for Msx (Phmamm.Msx-UP (same picture as Figure 5A) and Phmamm.Msx-LED). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 50 μm. Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments). (Bottom) Identification of putative TFBS for candidate upstream factors in Phmamm.Msx-LED aligned with its counterpart from P. fumigata. All putative sites for ventral factors (SBE, BRE, Tbx2/3, Nkxtun1, Nkx2-3/5/6 and Irx.c) and dorsal factors (Msx and Su(H)/Rbpj) have been mapped, but only conserved sites are shown: 5 SBE, 2 BRE, 1 Nkxtun1, 1 Nkx2-3/5/6, and 1 Msx sites. When comparing with Ciinte.Msx-up10 (Figure 1—figure supplement 2), we found shared sites for direct activation by Bmp signaling (SBE and BRE), and for activation by Msx itself. However, sites for ventral TFs were different: Nkxtun1 and Nkx2-3/5/6 in Phmamm.Msx-LED and Irx.c and Tbx2/3 in Ciinte.Msx-up10. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used.
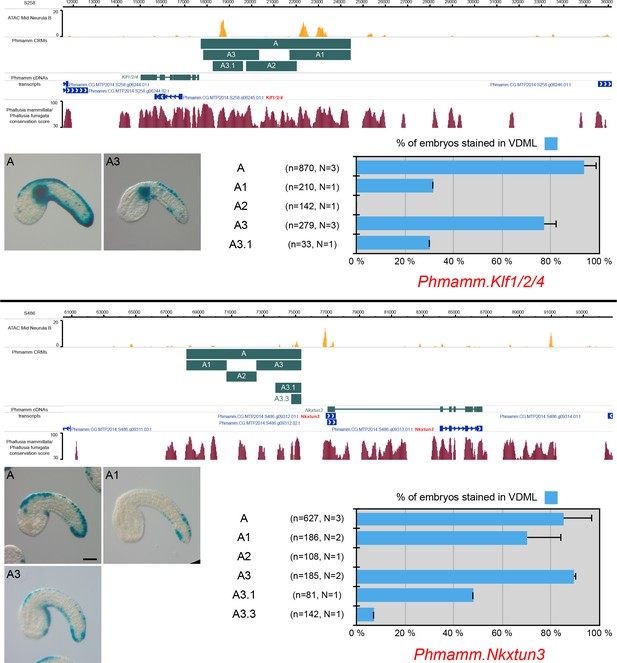
CRMs controlling Phmamm.Klf1/2/4 and Phmamm.Nkxtun3 expression in VDML.
Snapshots of the Phmamm.Klf1/2/4 and Phmamm.Nkxtun3 loci depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between P. mammillata and P. fumigata (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Representative examples of X-gal staining at tailbud stages following P. mammillata embryos electroporation of P. mammillata genomic regions for Klf1/2/4 (Phmamm.Klf1/2/4-A (same picture as Figure 5B) and Phmamm.Klf1/2/4-A3) and Nkxtun3 (Phmamm.Nkxtun3-A (same picture as Figure 5C), Phmamm.Nkxtun3-A1 and Phmamm.Nkxtun3-A3). Note that two separate VDML CRMs have been identified for Nkxtun3: a distal one, Phmamm.Nkxtun3-A1, and a proximal one, Phmamm.Nkxtun3-A3. This could be a case of redundant or shadow enhancers, but the distal CRM was preferentially active in posterior VDML while the proximal one did not show differential activity along the antero-posterior axis. Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 50 μm. Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments).
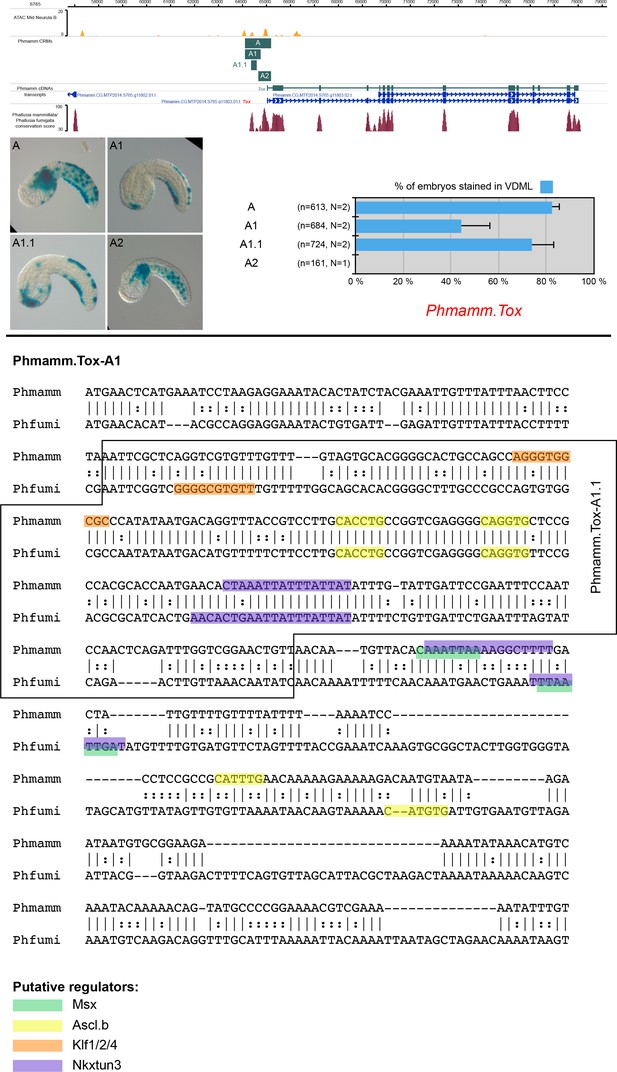
CRMs controlling Phmamm.Tox expression in VDML.
(Top) Snapshot of the Phmamm.Tox locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between P. mammillata and P. fumigata (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Representative examples of X-gal staining at tailbud stages following P. mammillata embryos electroporation of P. mammillata genomic regions for Tox (Phmamm.Tox-A (same picture as Figure 5D), Phmamm.Tox-A1, Phmamm.Tox-A1.1 and Phmamm.Tox-A2). Phmamm.Tox-A (1093 bp) was the longest region tested for Phmamm-Tox, and it was found active in tail muscle in addition to VDML. Both activities could be separated into two sub-domains of Phmamm.Tox-A: Phmamm.Tox-A1 (VDML) and Phmamm.Tox-A2 (muscle). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 50 μm. Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments). (Bottom) Identification of putative TFBS for candidate upstream factors in Phmamm.Tox-A1 aligned with its counterpart from P. fumigata. All putative sites for Msx, Ascl.b, Klf1/2/4, and Nkxtun3 have been mapped, but only conserved sites are shown: 1 Msx, 3 Ascl.b, 1 Klf1/2/4, and 2 Nkxtun3 sites. Comparison with Ciinte.Tox-int1.2 (Figure 3—figure supplement 3) was suggestive of a shared regulation by Ascl.b. Note that the size of the highlighted site for of a given TF may vary depending on the matrix used.
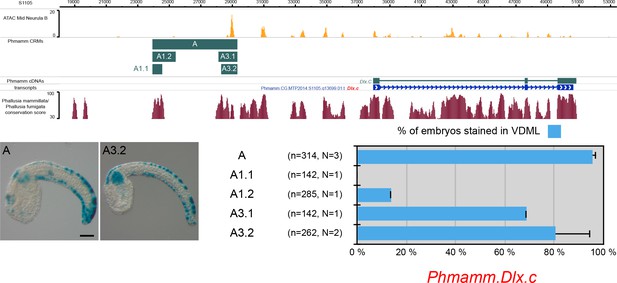
CRMs controlling Phmamm.Dlx.c expression in VDML.
Snapshot of the Phmamm.Dlx.c locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between P. mammillata and P. fumigata (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Representative examples of X-gal staining at tailbud stages following P. mammillata embryos electroporation of P. mammillata genomic regions for Dlx.C (Phmamm.Dlx.c-A (same picture as Figure 5E), and Phmamm.Dlx.c-A3.2). Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 50 μm. Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments).
Conserved VDML activity during enhancer swaps in Phlebobranchia (A).
Representative examples of X-gal stained embryos at tailbud stages following C. intestinalis (B), P. mammillata (C), P. fumigata (D), or A. mentula (E) embryo electroporation with C. intestinalis or P. mammillata CRMs. All shown CRMs for the genes Msx (i), Ascl.b (ii), Klf1/2/4 (iii), Nkxtun3 (iv), Tox (v), and Dlx.c (vi) are active in VDML in their species of origin. The name of the electroporated CRM is indicated on each picture. Details for each experiment can be found in Figure 6—figure supplement 1 and Figure 6—figure supplement 2. Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 50 μm.
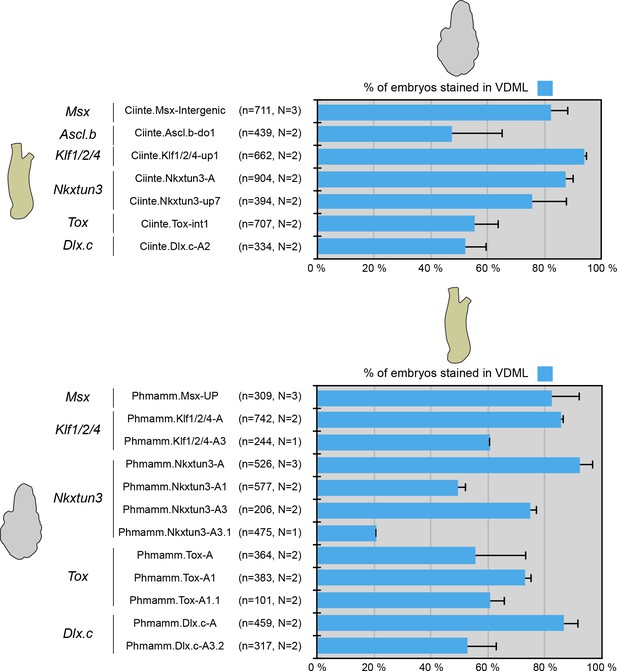
Conserved VDML activity during enhancer swaps between C. intestinalis and P. mammillata.
Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments).
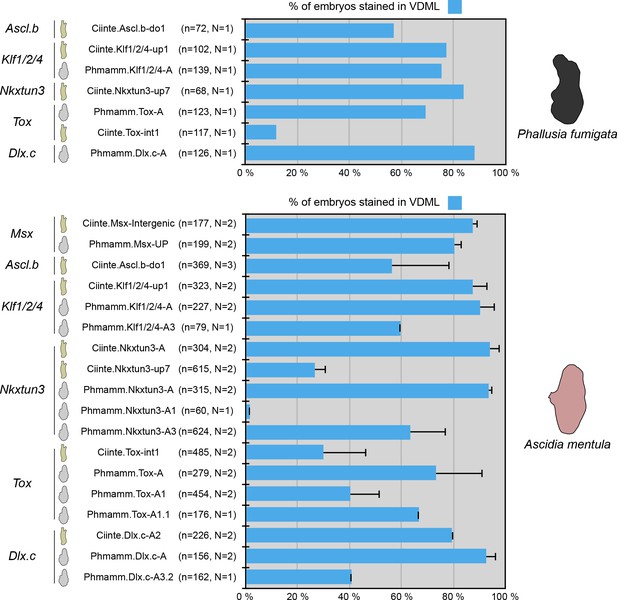
C. intestinalis and P. mammillata midline CRMs are active in VDML of P. fumigata and A. mentula embryos.
Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments).
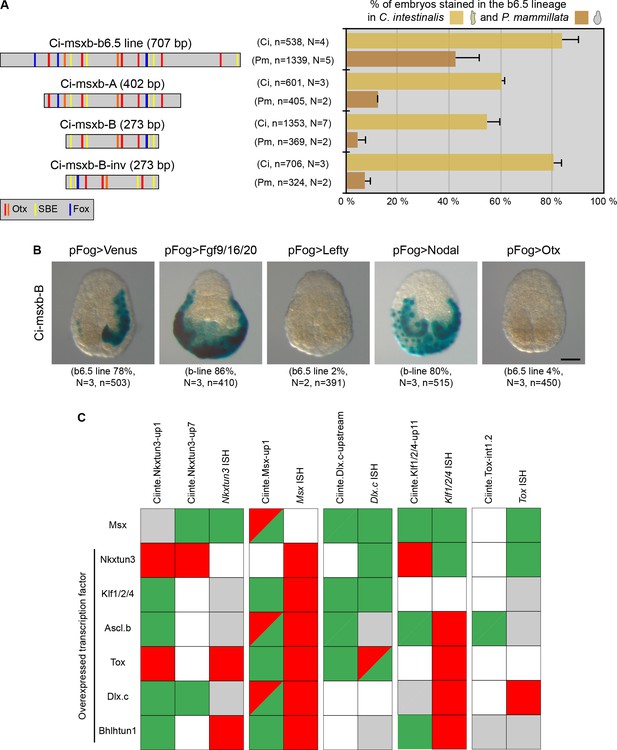
Minimal CRMs are not robust to phylogenetic and genetic challenges.
(A) Structure of the early DML CRM 'Ci-msxb-b6.5 line' and its derivatives (adapted from Roure et al., 2014), and in vivo transcriptional activity in the embryos of C. intestinalis and P. mammillata (n indicates the total number of embryos examined, N indicates the number of independent experiments). (B) Effects of various overexpressions on Ci-msxb-B activity. The indicated factors (Fgf9/16/20, Lefty, Nodal and Otx) were expressed under the control of the early ectodermal driver pFog. Embryos at gastrula/neurula stages are shown in neural plate view with anterior to the top (n indicates the total number of embryos examined, N indicates the number of independent experiments). Scale bar: 50 μm. (C) Consequences of caudal midline TF overexpression on the activity of VDML CRMs. The effects were summarized as: activation (green), repression (red), no effect (gray), or not done (white). They were compared with results of endogenous gene expression from Roure and Darras, 2016.
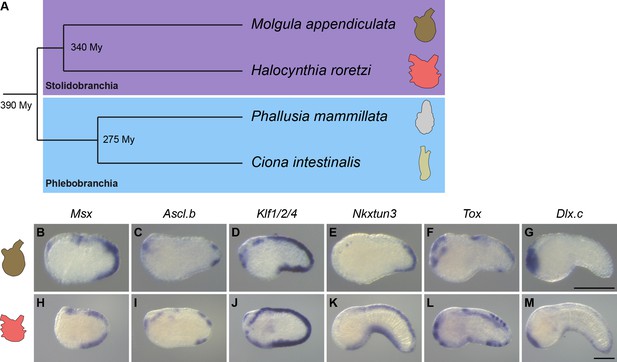
Overall conservation of midline gene expression in Stolidobranchia ascidians.
(A) Phylogenetic relationships between C. intestinalis, P. mammillata, M. appendiculata and H. roretzi (estimated divergence times come from Delsuc et al., 2018). (B–M) In situ hybridization for Msx (B, H) at neurula stages, for Ascl.b (C, I), Klf1/2/4 (D, J), Nkxtun3 (E, K), Tox (F, L), and Dlx.c (G, M) at tailbud stages in embryos of M. appendiculata (B–G) and H. roretzi (H–M). Note that all genes except Moappe.Dlx.c are expressed in tail midlines. Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bars: 100 μm.
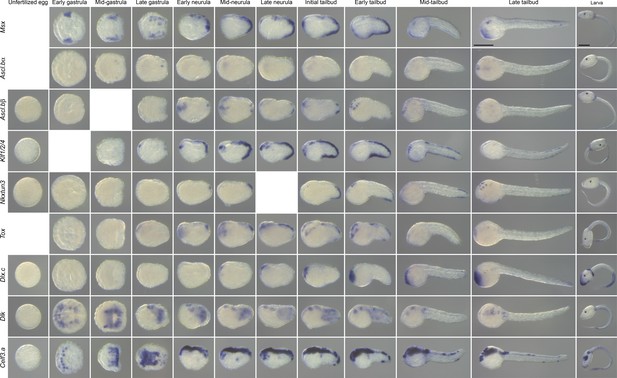
Expression pattern of caudal PNS genes in M. appendiculata.
In situ hybridization results at several developmental stages for the M. appendiculata orthologs of the C. intestinalis caudal PNS genes: Msx, Ascl.b (Ascl.bα and Ascl.bβ), Klf1/2/4, Nkxtun3, Tox, Dlx.c, Dlk, and Celf3.a. Scale bars: 100 μm.
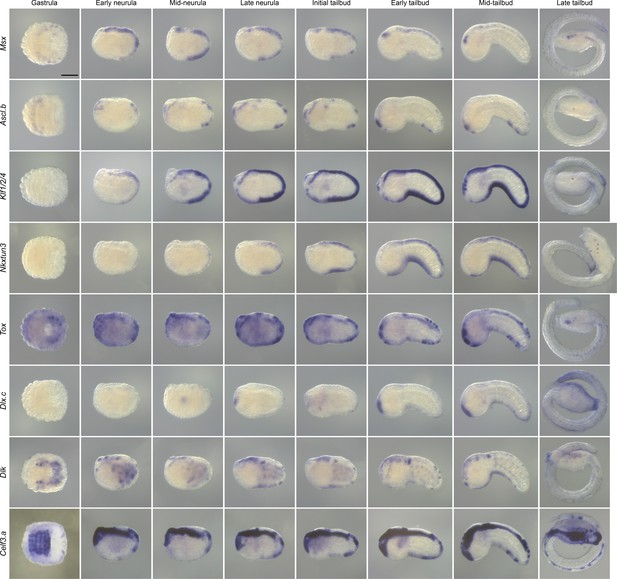
Expression pattern of caudal PNS genes in H. roretzi.
In situ hybridization results at several developmental stages for the H. roretzi orthologs of the C. intestinalis caudal PNS genes: Msx, Ascl.b, Klf1/2/4, Nkxtun3, Tox, Dlx.c, Dlk, and Celf3.a. Scale bar: 100 μm.
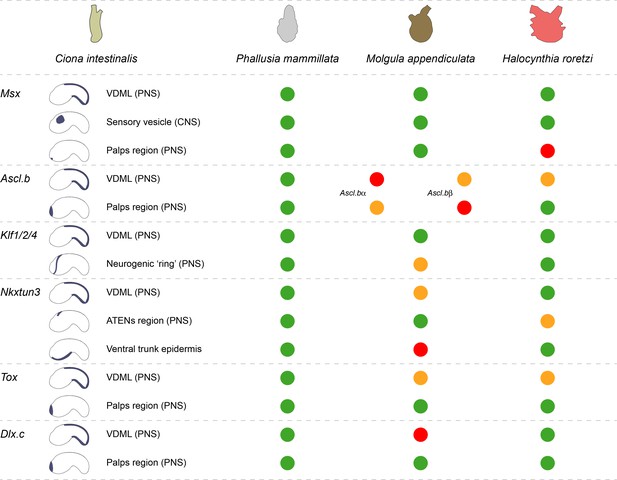
Comparison of midline TFs expression in C. intestinalis, P. mammillata, M. appendiculata, and H. roretzi.
For each gene, the main sites of expression from late neurula stages in C. intestinalis (Roure and Darras, 2016; Imai et al., 2004) are schematized on the left. For the other species, identical pattern is represented as a green circle, expression in part of the tissue as an orange circle, and absence of expression as a red circle. For simplicity, developmental timing has not been described, but it has been taken into account for comparisons. For example, Ciinte.Ascl.b is expressed in the palps forming region from initial tailbud to late tailbud stages, while Moappe.Ascl.bα was detected in the palps only at late tailbud stages. This difference was depicted by an orange circle.
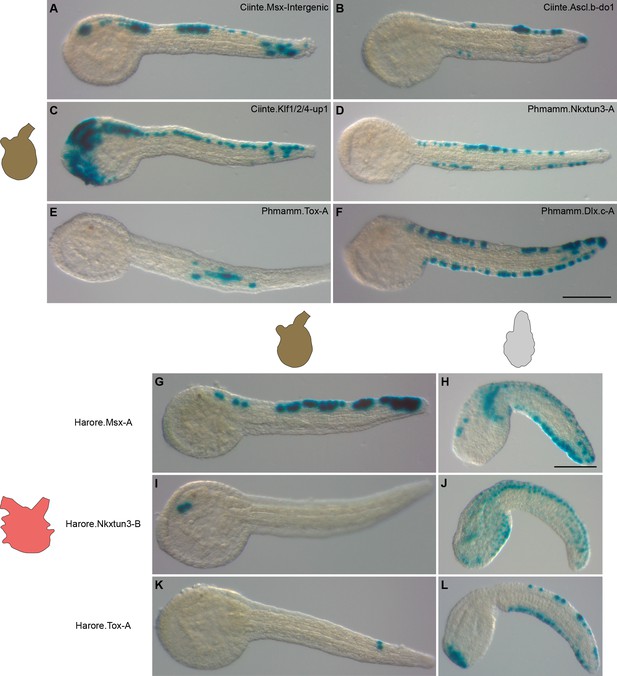
Divergence in gene regulation in Stolidobranchia ascidians.
(A–F) Activity of various CRMs from C. intestinalis and P. mammillata in the embryos of M. appendiculata. The name of the electroporated CRM is indicated on each picture. Details for each experiment can be found in Figure 9—figure supplement 1. (G–L) Activity of various genomic regions from H. roretzi in the embryos of M. appendiculata (G, I and K) and P. mammillata (H, J and L). Details for each experiment can be found in Figure 9—figure supplement 2. Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bars: 100 μm.
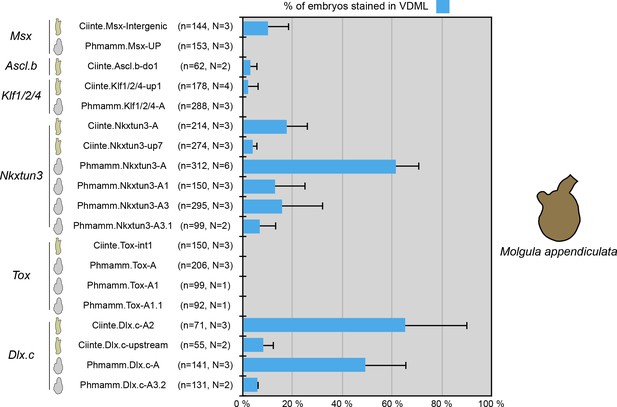
Activity of VDML CRMs from C. intestinalis and P. mammillata in the embryos of M. appendiculata.
Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments).
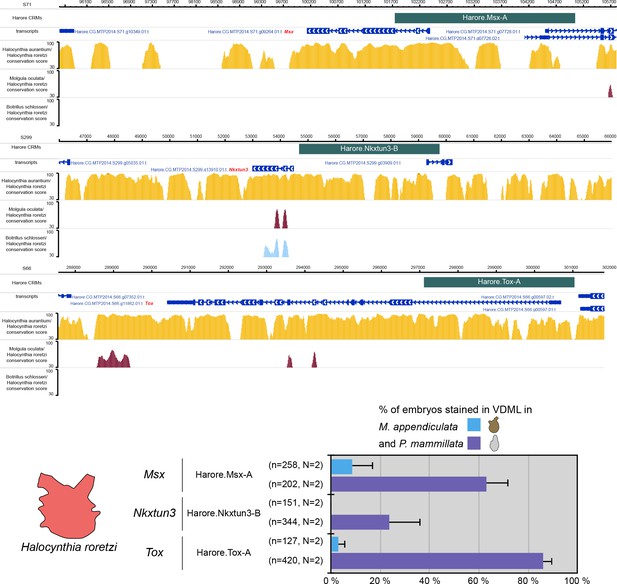
Activity of genomic regions from H. roretzi in the embryos of P. mammillata and M. appendiculata.
Snapshots of the Harore.Msx, Harore.Nkxtun3 and Harore.Tox loci depicting tested genomic regions, transcript models and conservation between H. roretzi and H. aurantium, M. oculata and Botryllus schlosseri (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments).
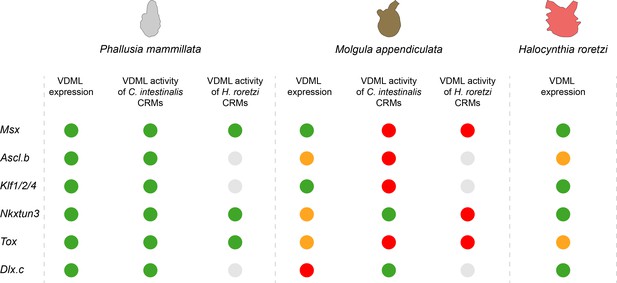
Summary of comparative results obtained in the present study.
For each gene, results of in situ hybridization and enhancer swaps are summarized. VDML expression (in situ hybridization) or activity (transcriptional assay) is represented as a green circle, expression in part of the VDML as an orange circle, and lack of expression or activity as a red circle. Gray circle: not done.
Additional files
-
Supplementary file 1
List of all genomic regions tested in the present study.
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp1-v2.xlsx
-
Supplementary file 2
Genome browser view for each locus of the seven caudal PNS neurogenic TFs in Ciona robusta.
Tested CRMs were added to the data extracted from the Aniseed website (https://www.aniseed.cnrs.fr/; Dardaillon et al., 2020).
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp2-v2.pdf
-
Supplementary file 3
Activity of various genomic regions for the gene Ciinte.Bhlhtun1.
(Top panel) Snapshot of the Ciinte.Bhlhtun1 locus depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between C. robusta and C. savignyi (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). (Middle panel) Representative examples of X-gal-stained embryos at tailbud stages following electroporation of Ciinte.Bhlhtun1-upstream, Ciinte.Bhlhtun1-up1 and Ciinte.Bhlhtun1-down1. Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 100 μm. (Bottom panel) Schematic representation of the various constructs and their activity at tailbud stages in DML (blue) and VML (purple) (n indicates the total number of embryos examined, N indicates the number of independent experiments). Note that while VDML activity is rare, activity can be detected at other sites of endogenous Ciinte.Bhlhtun1 expression: anterior epidermis around the palps for Ciinte.Bhlhtun1-up1, and notochord and stomodeum for Ciinte.Bhlhtun1-down1.
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp3-v2.pdf
-
Supplementary file 4
Activity of various genomic regions for the genes Phmamm.Ascl.b and Phmamm.Bhlhtun1.
Snapshots of the Phmamm.Ascl.b and Phmamm.Bhlhtun1 loci depicting ATAC-seq profile at mid-neurula stages, tested genomic regions, transcript models and conservation between P. mammillata and P. fumigata (from https://www.aniseed.cnrs.fr/ and Dardaillon et al., 2020; Madgwick et al., 2019). Representative examples of X-gal stained embryos at tailbud stages following electroporation of Phmamm.Ascl.b-A (no activity), Phmamm.Ascl.b-B (activity in palps and anterior nervous system), and Phmamm.Bhlhtun1-A (activity in notochord, endodermal strand and tail tip) into P. mammillata embryos. Embryos are shown in lateral view with dorsal to the top and anterior to the left. Scale bar: 50 μm. Schematic representation of the various constructs and their activity at tailbud stages in VDML (blue) (n indicates the total number of embryos examined, N indicates the number of independent experiments). Note that while VDML activity is rather robust for Phmamm.Bhlhtun1-A, it was not considered further since this activity was restricted to the very posterior cells of the midlines at the tail tip.
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp4-v2.pdf
-
Supplementary file 5
Genome browser view for each locus of the seven caudal PNS neurogenic TFs in Phallusia mammillata.
Tested CRMs and predicted cDNAs were added to the data extracted from the Aniseed website (https://www.aniseed.cnrs.fr/; Dardaillon et al., 2020).
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp5-v2.pdf
-
Supplementary file 6
Identification of VDML CRMs for Phfumi.Msx and Asment.Msx genes.
(Top) Snapshot of the Phfumi.Msx locus. (Middle) Activity of Phfumi.Msx and Asment.Msx CRMs at tailbud stages in VDML (blue) of C. intestinalis and P. mammillata embryos (n indicates the total number of embryos examined, N indicates the number of independent experiments). (Bottom) Representative examples of X-gal staining at tailbud stages (embryos in lateral view with dorsal to the top and anterior to the left, scale bar: 50 μm).
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp6-v2.pdf
-
Supplementary file 7
Genome browser view for each locus for three caudal PNS neurogenic TFs in Halocynthia roretzi.
Tested CRMs were added to the data extracted from the Aniseed website (https://www.aniseed.cnrs.fr/; Dardaillon et al., 2020).
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp7-v2.pdf
-
Supplementary file 8
List of DNA clones used for in situ hybridization.
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp8-v2.xlsx
-
Supplementary file 9
Various sequences.
Predicted cDNA sequences for genes in C. intestinalis, P. mammillata and M. appendiculata: transcripts models from RNA-seq data and ESTs sequences were used to build the cDNA sequences. Open reading frame is highlighted in bold. Sequence of the genomic region Asment.Msx-up isolated from A. mentula.
- https://cdn.elifesciences.org/articles/59157/elife-59157-supp9-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59157/elife-59157-transrepform-v2.pdf





