Orderly assembly underpinning built-in asymmetry in the yeast centrosome duplication cycle requires cyclin-dependent kinase
Figures
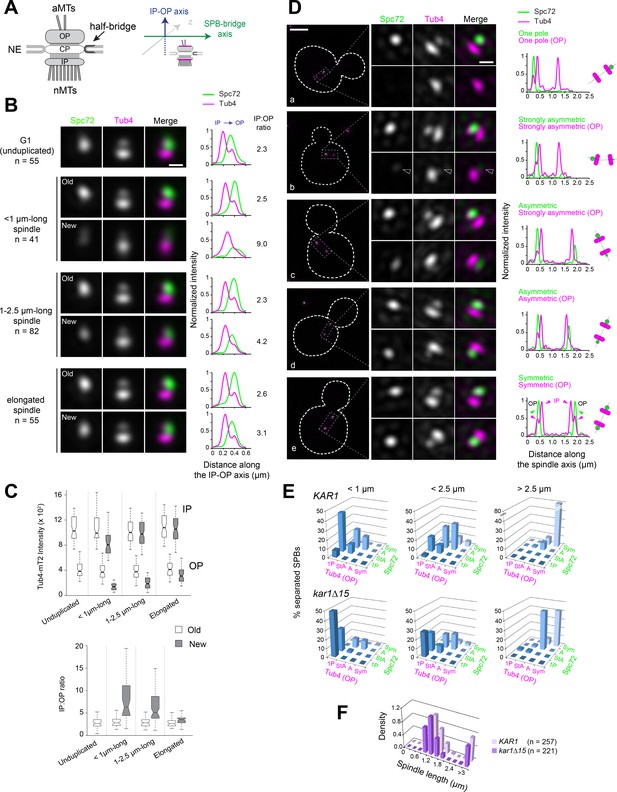
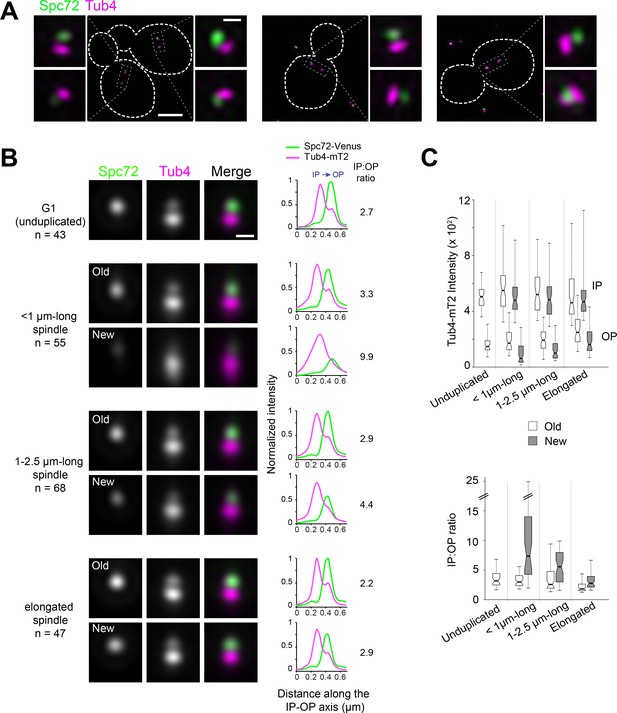
Tub4 accumulation at SPB outer plaques follows Spc72 asymmetric recruitment during spindle assembly.
(A) [Left] Simplified schematic of the SPB showing the inner (IP) and outer (OP) plaques that nucleate nuclear (nMTs) and cytoplasmic or astral microtubules (aMTs), respectively. The central plaque (CP) anchors the SPB in the nuclear envelope (NE). The half-bridge is a modified region of the nuclear envelope involved in SPB duplication; it is also able to nucleate microtubules during G1 phase of the cell cycle (Byers and Goetsch, 1975). [Right] To study the distribution of SPB components, SIM images of individual SPBs were aligned using Tub4-mT2 as a reference, as previous work showed that it is present at the inner and outer plaque in different amounts (see Burns et al., 2015 and Figure 1—figure supplement 2). SPBs were assigned to a cell cycle and spindle stage using bud morphology and the distances between SPBs, referred to as SPB inter-distances. (B) Average image from realigned SPBs in each class. Scale bar, 200 nm. The number of SPBs is indicated. Linescan analysis (5-px width) shows the intensity and localization of Spc72-Venus as well as the distribution of Tub4-mT2 at the IP and OP. Linescan intensities were normalized relative to maximal values at the old SPB of elongated spindles. The IP:OP ratio is based on the Tub4-mT2 signal. (C) In addition to averaging, the intensity of Tub4-mT2 at the inner and outer plaques was measured in individual images. Values are plotted by stage and SPB identity (top); IP:OP intensity ratios (bottom) for the same dataset are also shown. Boxplots depict the 5th, 25th, 50th, 75th and 95th centiles. Notches represent 95% CI of the median. (D) Representative SIM images showing the distribution of Spc72-Venus (green) and Tub4-mT2 (magenta) at SPBs of cells with short spindles. Merged images with cell outlines (scale bar, 2 µm) and single channels and merged cropped images for each SPB (scale bar, 200 nm) are shown paired with internally normalized linescan analysis (3-px width) to indicate fluorescence intensity along the spindle axis as represented in the cartoons: (a) Spc72 and Tub4 present at one outer plaque; (b–d) progressive decline of asymmetry for Spc72 followed by Tub4, in (b) a hollow arrowhead points to weak Spc72/Tub4 labels at the new SPB cytoplasmic face; (e) both components symmetric. Modes of label are classified as described in Materials and methods. (E) Quantitation of Spc72-Venus and Tub4-mT2 distribution by spindle stage in KAR1 and kar1∆15 strains. 1P=one pole; StA = strongly asymmetric; A = asymmetric; Sym = symmetric, as described in (D). Note that categories for Tub4 label are based on the relative intensity of the outer plaque foci only. Those correspond to the one (1P) or two (StA, A, Sym) outer peaks of the intensity profile (in magenta) along the spindle axis as indicated by arrowheads for the linescan in (e). By contrast the intensity profile for Spc72 is represented exclusively by one (1P) or two (StA, A, Sym) single peaks (in green). (F) Distribution of spindle lengths in the KAR1 and kar1∆15 asynchronous cell populations analyzed.
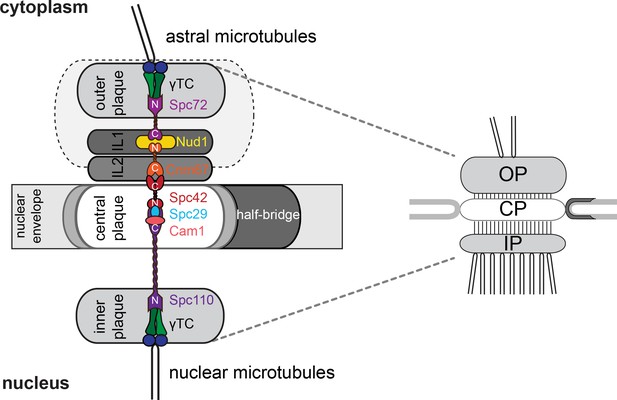
Simplified SPB structure.
Cartoon outlining key layers and sublayers making up the SPB (left) and their correspondence to the summary structure (right) used to describe landmark events in the SPB duplication pathway (Figure 2) and SPB 3-D realignment throughout.
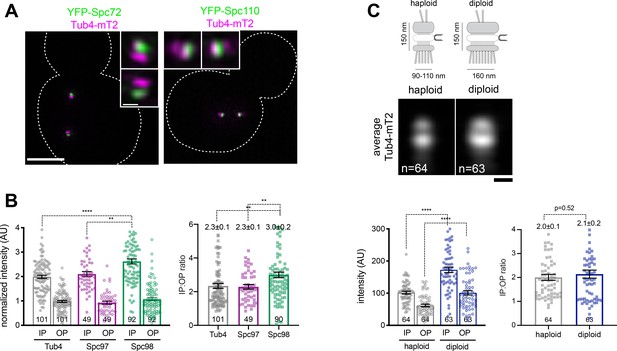
Quantitative detection of γTC by SIM shows SPB size scaling and roles for the outer plaque.
(A) SIM images of cells containing Tub4-mT2 (magenta) and YFP-Spc72 or YFP-Spc110 (green), which mark the SPB outer and inner plaque, respectively. Insets show SPB region. Bar, 2 µm and 200 nm (inset). (B) SIM images of strains containing Spc110-YFP and the indicated γTC component fused to mT2 were analyzed by fitting spots corresponding to the inner and outer plaques. Each individual value is plotted along with the mean, standard error and number of SPBs analyzed (n). Ratios were derived from intensity measurements. (C) Schematic showing the inner, central and outer plaques of the SPB along with the half-bridge. The average width and height based on EM measurements of haploid and diploid cells is shown (Adams and Kilmartin, 1999; Byers and Goetsch, 1974). Haploid and diploid cells containing Tub4-mT2 were imaged by SIM and analyzed as in (B). In addition, an average image was also created. n, number of cells. ****, p<0.0001; **, p<0.01. Note that in both (B) and (C), there is heterogeneity within individual SPBs at both the inner and outer plaque. Importantly, SIM was able to detect quantitative differences in inner and outer plaque intensity that scaled proportionally with SPB size, indicating that it can be used to assay changes at the SPB.
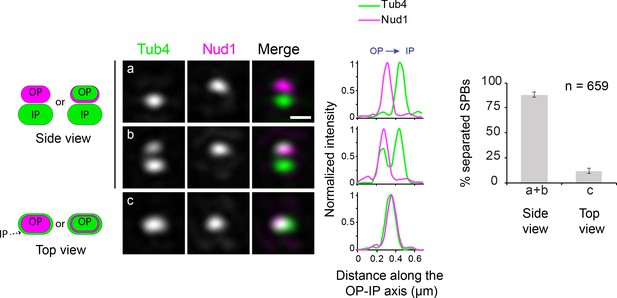
Discrimination between SPB side and top view in SIM images.
SIM images of wild type cells expressing Tub4-Venus and Nud1-mT2 were used to score the overall proportion of individual SPBs presented in side view (a-b) and top view (c). Note that the use of Nud1, a core component of the SPB outer plaque, further confirms the side view in SPBs lacking Tub4 outer plaque marking (b). Error bars, standard error of the proportion. Scale bar, 200 nm.
Spc72 and Tub4 asymmetric localization in the W303 yeast background.
A W303-derived yeast strain expressing Spc72-YFP (green) and Tub4-mT2 (magenta) was imaged by SIM. (A) Representative SIM images of cells with short spindles. Merged images with cell outlines (scale bar, 2 µm) and cropped merged images for each SPB (scale bar, 200 nm) are shown. (B–C) As in Figure 1, SIM images of individual SPBs were aligned using Tub4-mT2 as a reference and SPBs were assigned to a cell cycle and spindle stage using bud morphology and SPB inter-distances. (B) Averaged image from realigned SPBs in each class. Scale bar, 200 nm. The number of SPBs is indicated. Linescan analysis (5-px width; normalized relative to maximal intensity at the old SPB from elongated spindles) shows the intensity and localization of Spc72-YFP as well as the distribution of Tub4-mT2 at the IP and OP. The IP:OP ratio is based on the Tub4-mT2 signal. (C) In addition to averaging, the intensity of Tub4-mT2 at the inner and outer plaques was measured in individual images. Values are plotted by stage and SPB identity (top); IP:OP intensity ratios (bottom) for the same dataset are also shown.
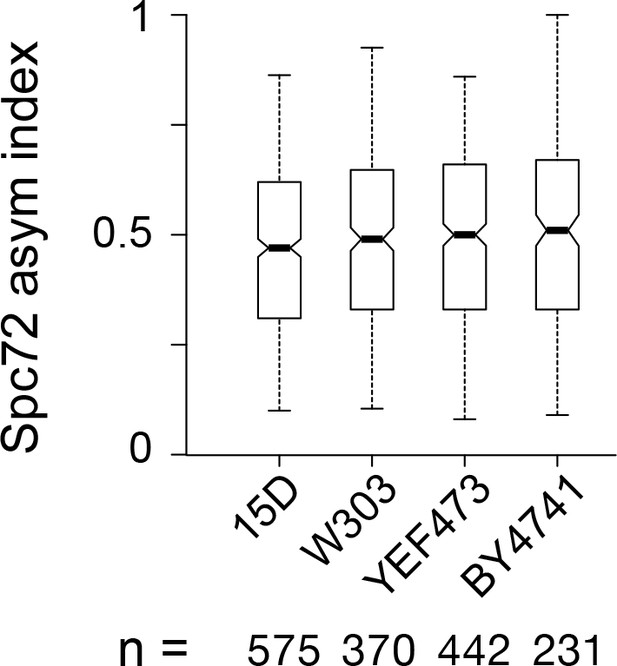
Spc72 asymmetry in multiple yeast genetic backgrounds.
Boxplot for the distribution of Spc72 asymmetry index in short spindles (<2.5 µm-long) for asynchronous cultures in the indicated wild type yeast backgrounds from wide-field images. Boxplots depict the 5th, 25th, 50th, 75th and 95th centiles. Notches represent 95% CI of the median.
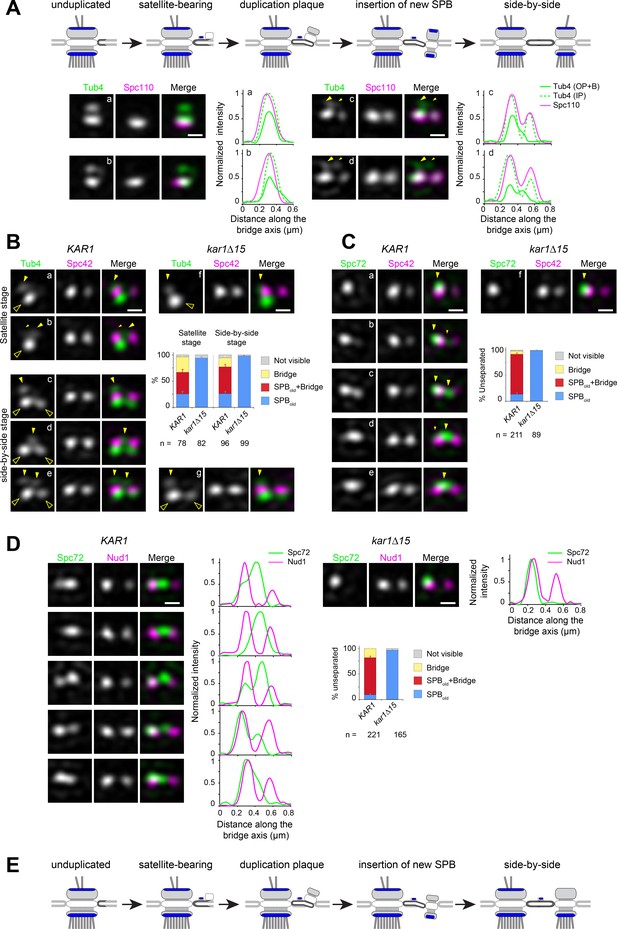
Spc72 and γTC are not added to the new SPB during its duplication.
(A–D) SIM image projections were obtained after 3D-Gaussian fit and realignment using the indicated reference label. Images were rotated to position the old SPB to the left. Scale bar, 200 nm. (A) (top) Model for landmark events along the SPB duplication pathway deduced by EM and SIM studies (after Adams and Kilmartin, 1999 and Burns et al., 2015). The presumed localization of γTC according to this model is suggested in blue. (bottom) Representative SIM images and corresponding linescan analysis along the inner or outer plaque (1px-width, parallel to the bridge axis, internally normalized) illustrating Tub4-Venus distribution (green) relative to mT2-Spc110 (magenta). Images are ordered along the presumed SPB duplication pathway according to structures seen in early or intermediate steps of SPB duplication leading to the completed side-by-side SPBs: single SPB showing either a single (a) or extended (b) cytoplasmic label; (c–d) side-by-side stage. Big and small yellow arrowheads point to Tub4 at the old SPB outer plaque and bridge, respectively, as described in the text (B–C) Using Spc42-mT2 intensity, SIM images were organized to represent stages of SPB duplication. (B) The distribution of Tub4-Venus (green) localization relative to Spc42-mT2 (magenta) during SPB duplication in KAR1 (a–e) versus kar1∆15 (f–g) cells and plot for distribution of modes of Tub4 localization at the cytoplasmic face of the SPB. Solid yellow arrowheads point to sites on the cytoplasmic face of the SPB hollow arrowheads indicate inner plaque(s) (C) The distribution of Spc72-Venus (green) localization relative to Spc42-mT2 (magenta) during SPB duplication in KAR1 (a–e) versus kar1∆15 (f) cells and plot for modes of Spc72-Venus distribution at unseparated SPBs. Solid arrowheads mark dual localization of Spc72 as described in the text. Error bars, standard error of the proportion. (D) Spc72-Venus (green) and Nud1-mT2 (magenta) were localized in KAR1 versus kar1∆15 cells by SIM. Representative images of cells undergoing SPB duplication, paired to linescan analysis (three px-width along the bridge axis; internally normalized) and plot for distribution of modes of Spc72 localization relative to Nud1. Error bars, standard error of the proportion. (E) New model depicting the temporality for completion of inner and outer plaque assembly relative to other landmark events in the SPB duplication cycle as emerging from the data presented here. In particular, the new SPB outer plaque is incompletely assembled at the side-by-side stage, and thus unable to recruit γTC.
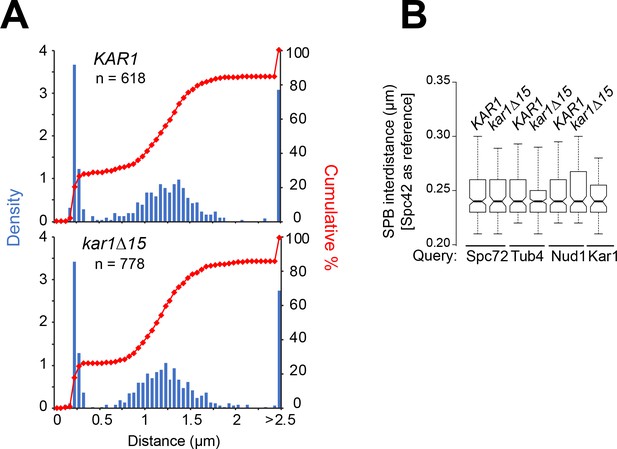
SPB inter-distance as basis for classification of SPBs by cell cycle stage.
(A) Distribution of SPB inter-distances using Spc42-mT2 as reference in asynchronous KAR1 or kar1∆15 cell populations analyzed by 3D-SIM. These values were used to classify SPBs into unseparated SPBs (<350 nm), SPBs in short spindles (<2.5 µm) and SPBs in elongated spindles (>2.5 µm). (B) Boxplot for distribution of SPB inter-distances in all unseparated SPBs selected for analysis from asynchronous cell populations in Figures 2 and 3 and Figure 2—figure supplement 2. In all cases, analysis was performed after Spc42-mT2 labels (reference channel) were subject to 3D-Gaussian fit and realignment, as described in the Materials and methods.
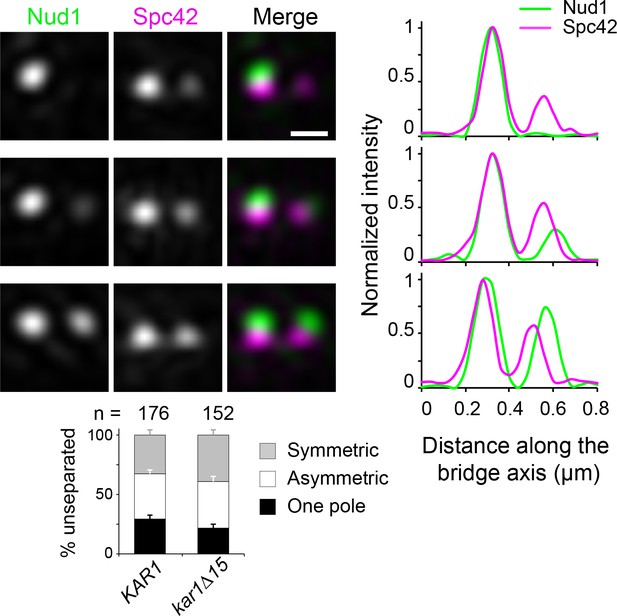
Modes of Nud1 localization in unseparated SPBs.
Nud1-Venus (green) and Spc42-mT2 (magenta) were localized in KAR1 versus kar1∆15 cells by SIM. Three representative images show modes of label in unseparated SPBs. Consistent with the previously shown order of satellite assembly (Burns et al., 2015), Nud1 did not mark the presumptive satellite in approximately 25% of cells containing two Spc42 signals, but otherwise significantly accumulated at the new SPB toward the side-by-side stage. This is quantitated below.
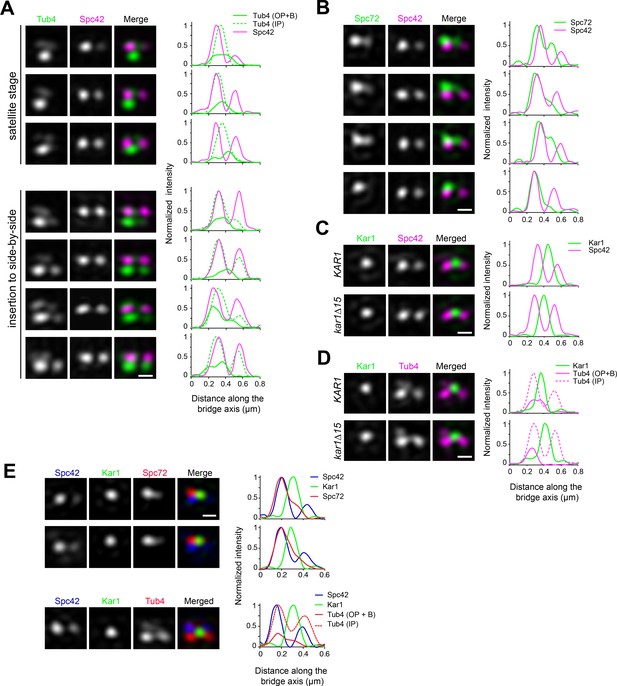
A subset of cytoplasmic nucleation sites coincides with Kar1 localization in unseparated SPBs.
(A–C) Comparative localization of Tub4-Venus, Spc72-Venus, Venus-Kar1 or Venus-Kar1∆15 (in green) relative to Spc42-mT2 (magenta). Representative SIM image projections were obtained after 3D-Gaussian fit and realignment using Spc42-mT2 as reference. The corresponding linescan for fluorescence intensity along the bridge axis is also shown. When indicated, separate profiles were obtained along inner and outer plaques (one pixel-width, parallel to the bridge axis). As quantified in Figure 2B–C, Tub4-Venus or Spc72-Venus exhibited dual localization at the cytoplasmic face of duplicated SPBs by distributing between the old SPB outer plaque and the bridge. By contrast, Venus-Kar1 or Venus-kar1∆15 was confined to the space between Spc42-mT2 foci, representing the exclusive localization to the bridge. (D) Partial overlap between Tub4-mT2 (magenta) and Venus-Kar1 (green) signals at the bridge was abolished when cells expressed instead Venus-Kar1∆15. Scale bar, 200 nm. (E) Three color SIM of unseparated SPBs containing YFP-Kar1 (green), Spc42-mCherry (blue) and Spc72-mT2 or Tub4-mT2 (red). Linescan analysis was carried out along inner and outer plaques (one pixel-width, parallel to the bridge axis). Scale bar, 200 nm.
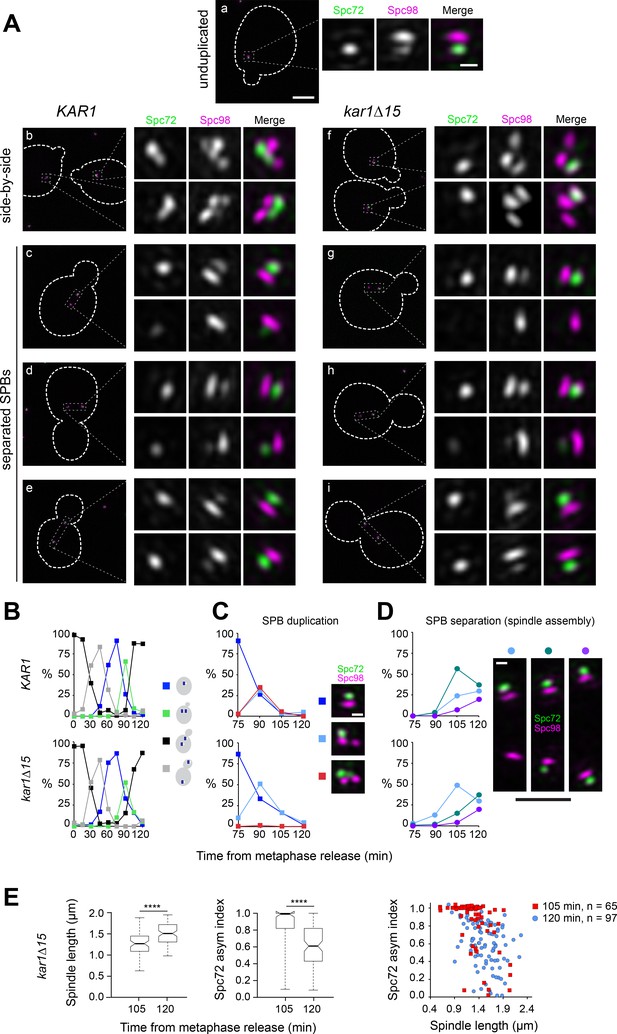
Spc72 accumulation at the new SPB outer plaque begins at the ~1 µm-long spindle stage.
(A–E) KAR1 or kar1∆15 cells co-expressing Spc72-Venus (green) and Spc98-mT2 (magenta) were synchronized in metaphase using PMET3-CDC20, then released into the cell cycle. (A) Representative SIM images showing the distribution of Spc72-Venus (green) and Spc98-mT2 (magenta). Merged images with cell outlines (scale bar, 2 µm) and cropped images of SPBs by channel or merged (scale bar, 200 nm) are shown: (a) unduplicated, (b, f) side-by-side (c–e and g–i), separated SPBs. (B) Cell cycle progression in terms of the spindle pathway. (C–D) The localization of Spc72-Venus and Spc98-mT2 was quantitated at the indicated times in cells with side-by-side SPBs (C; scale bar, 200 nm) or separated SPBs (D; scale bars: white, 200 nm and black, 1 µm) Total number of cells analyzed by time point was 70, 85, 107, 123, 70, 114, 150, 198 and 125 (KAR1); 96, 160, 103, 70, 128, 139, 320, 205 and 224 (kar1∆15). (E) Boxplots for distribution of spindle length or Spc72 asymmetry index (left) and scattered plot for Spc72 asymmetry index as a function of spindle length at the indicated time points were generated by 3-D SIM analysis. ****, p<0.0001 according to Mann Whitney two-tailed test.
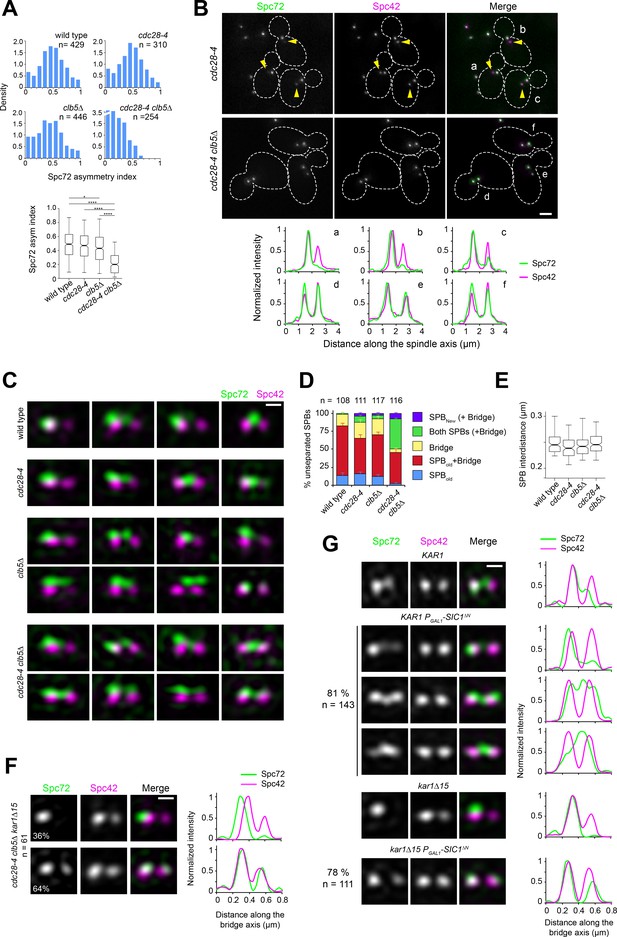
Spc72 asymmetric recruitment requires S-phase CDK.
(A–B) The distribution of Spc72-GFP (green) and Spc42-CFP (magenta) was compared in wild-type, clb5∆, cdc28-4 and cdc28-4 clb5∆ cells grown at 23°C. (A) Histograms show the distribution of Spc72 asymmetry index in cells with short spindles (<2.5 µm length) for each sample. Boxplots below depict the 5th, 25th, 50th, 75th and 95th centiles. Notches represent 95% CI of the median. *p=0.0189 and ****p<0.0001, according to Kruskal-Wallis and Dunn's multiple comparison tests. (B) Representative wide-field fluorescence images and corresponding linescan analysis along the spindle axis showing Spc72 asymmetry in cdc28-4, but not cdc28-4 clb5∆ cells. Yellow arrowheads point to new SPBs weakly labelled by Spc72-GFP, underscoring asymmetry. Scale bar, 2 µm. (C–D) SIM analysis of unseparated SPBs of the indicated strains. (C) Representative 3D-realigned images of unseparated SPBs labeled by Spc72-Venus (green) and Spc42-mT2 (magenta). Loss of asymmetry occurred in cdc28-4 clb5∆ cells in which Spc72 recruitment at the new SPB outer plaque began prior to SPB separation. Scale bar, 200 nm. (D) Quantitation of modes of Spc72 localization in unseparated SPBs of the indicated strains. Error bars, standard error of the proportion. (E) Boxplot showing the distance between Spc42 foci in all cells tallied. 5th, 25th, 50th, 75th and 95th centiles are shown. Notches represent 95% CI of the median. (F) Representative images for the two modes of Spc72 localization observed in unseparated SPBs of cdc28-4 clb5∆ kar1∆15 asynchronous cells. Images were subject to realignment based on the Spc42-mT2 label. The corresponding linescans for fluorescence intensity along the bridge axis (3-px width; internally normalized) are also shown. (G) KAR1 or kar1∆15 PGAL1-SIC1∆N cells (or control cells carrying a vector instead of pLD1) containing Spc72-Venus (green) and Spc42-mT2 were grown in synthetic-raffinose medium to early log phase at 25ºC and induced at the same temperature by addition of 3% galactose. After 1.5 hr induction, SIM images were acquired and the SPBs in small-budded cells analyzed. Uniform cell cycle arrest was observed after 3.5 hr. Representative SIM images of Spc72-Venus (green) and Spc42-mT2 (magenta) are shown with corresponding linescans (3-px width; internally normalized). Scale bar, 200 nm.
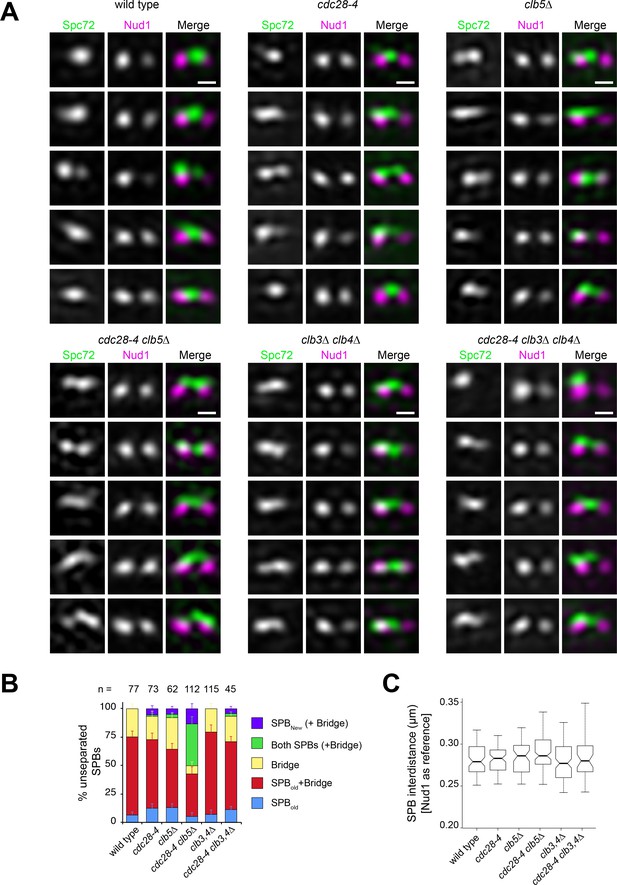
S phase CDK inactivation anticipates Spc72 recruitment at the new outer plaque in unseparated SPBs.
Wild-type and cdc28-4 and cyclin mutant strains containing Spc72-Venus (green) and Nud1-mT2 (magenta) were imaged by SIM and subject to 3-D Gaussian fit and realignment, in reference to Nud1, to determine the distribution of Spc72 relative to Nud1. (A) Representative SIM images showing unseparated SPBs. Scale bar, 200 nm. (B) Quantitation of Spc72 distribution in all strains analyzed. Bars, standard error of the proportion. (C) Boxplot for distance between Nud1 unseparated foci for all cells analyzed. Boxplots depict the 5th, 25th, 50th, 75th and 95th centiles. Notches represent 95% CI of the median.
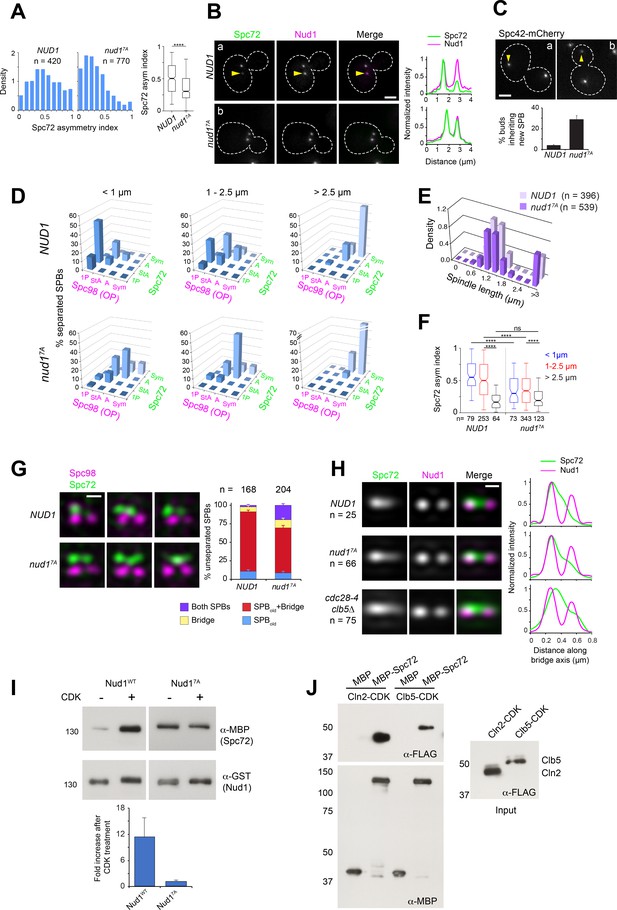
Substitutions cancelling CDK phosphorylation in Nud1 result in loss of Spc72 asymmetry.
(A–B) Cells expressing wild type Nud1-CFP or Nud17A-CFP along with Spc72-GFP were analyzed by wide-field fluorescence microscopy. (A) Histogram showing the distribution of Spc72 asymmetry index in each sample. ****p<0.0001 according to the Mann Whitney two-tailed test. (B) Representative fluorescence images and corresponding linescan analysis along the spindle axis. Yellow arrowheads highlight asymmetric localization pointing to weaker label at the new SPB. Scale bar, 2 µm. (C) Proportion of NUD1 versus nud17A cells in which the new SPB was inherited by the bud. Mean of three counts of 200 cells is shown, error bars SD. (D) Distribution of Spc98-mT2 and Spc72-Venus by spindle stage was quantitated as described in Figure 1. (E) The spindle length distribution within the cell populations is shown. (F) Boxplot analysis showing Spc72 asymmetry index by spindle stage. 5th, 25th, 50th, 75th and 95th centiles are shown. Notches represent 95% CI of the median. ****, p<0.0001 according to Kruskal-Wallis and Dunn's multiple comparison tests. (G) (Left) Representative SIM images from cells with side-by-side SPBs to compare the distribution of Spc72 in NUD1 versus nud17A cells. In nud17A cells, Spc72-Venus (green) was incorporated into the SPB outer plaque in a fraction of cells with side-by-side SPBs. Bar, 200 nm. (Right) Quantification of modes of Spc72 recruitment in unseparated SPBs of NUD1 versus nud17A cells. Error bars, standard error of the proportion. (H) Comparison of the extent of disruption of Spc72 asymmetry in nud17A versus cdc28-4 clb5∆ cells assessed in averaged images of unseparated SPBs from asynchronous cell populations, compiled after 3D-realignment in reference to Nud1 label. The corresponding linescans for fluorescence intensity along the bridge axis (3-px width; internally normalized) are shown. (I) Effect of CDK phosphorylation of Nud1 vs Nud17A on Spc72 binding in vitro. (Top) A mixture of Cln2 and Clb5-CDK was used to phosphorylate GST-Nud1 or GST-Nud17A bound to beads. After extensive washing to terminate the kinase reaction, beads were incubated with identical amounts of purified MBP-Spc72 for 2 hr followed by repeated washes. Eluted fractions were resolved in SDS-PAGE and the presence of Nud1 and Spc72 determined by western blot analysis. (Bottom) Plot representing average fold increase in Nud1-Spc72 binding upon CDK treatment from three independent experiments. Error bar, SD. (J) Ability of MBP-Spc72 to pull down Cln2 or Clb5 as part of CDK complexes in vitro. An equivalent amount of input (right, 5 s exposure) was analyzed under identical conditions to the pull-down (left, 20 s exposure).
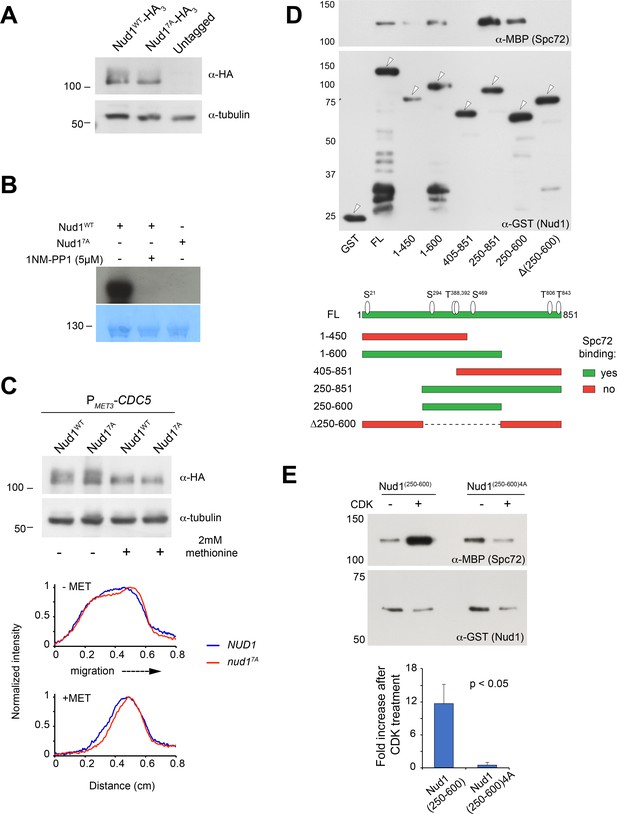
Nud1 and Nud17A characterization in vivo and in vitro.
(A) Nud1WT-HA3 and Nud17A-HA3 levels assessed by western blot analysis of cell extracts prepared from asynchronous cultures of the parent strains analyzed in Figure 5. (B) Kinase assay using 4 µg of purified MBP-Nud1WT or MBP-Nud17A and 19 ng of purified Twin-Strep-Clb5∆db/Cdc28-as/Cks1 was carried out as described under Materials and methods. 5 µM 1NM-PP1 was added to inhibit Cdc28-as (analog-sensitive CDK). Reactions were resolved by SDS-PAGE and blotted onto a PVDF membrane (bottom) that was exposed to film overnight (top) (C) CDK sites in Nud1 contribute to bulk phosphorylation in vivo. Western blot analysis of whole cell extracts from cells expressing Nud1WT-HA3 or Nud17A-HA3 before or after depletion of Cdc5 under the control of the repressible MET3 promoter was carried out as described under Materials and methods. Cdc5 also contributes to Nud1 mobility shift by phosphorylation. A further decrease in mobility shift was observed in cells expressing Nud17A versus wild type Nud1 following Cdc5 depletion. Linescans along the direction of electrophoretic migration show the relative down-shift of Nud17A-HA3 (in red). (D) A Nud1 domain between amino acid positions 250–600 was necessary and sufficient for Spc72 binding in vitro. Previous yeast two-hybrid studies suggested that the Nud1 C-terminus (residues 405–852) associated with Spc72 (Gruneberg, 2000). (E) Spc72 pull down by Nud1(250-600) showing enhanced binding following CDK phosphorylation as described for full-length Nud1 in Figure 5J.
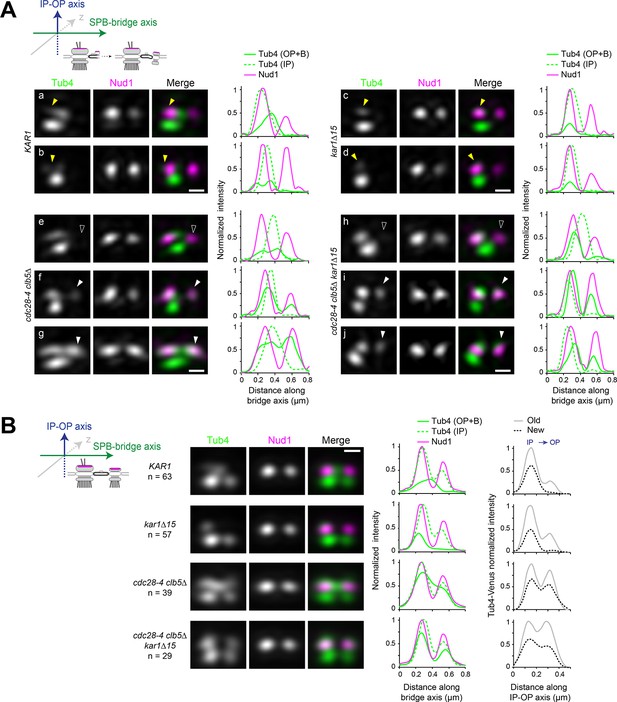
Tub4 IP:OP ratio and the sequence for new SPB assembly are perturbed in cdc28-4 clb5∆ cells.
(A–B) SIM analysis of the indicated strains expressing Tub4-Venus and Nud1-mT2 as a reference for the outer plaque. Cartoons indicate the stage in each case and label used for 3D-realignment. Scale bar, 200 nm. (A) Individual SIM images and linescan analysis (1-px width; internally normalized) for the indicated strains corresponding to unseparated SPBs prior to nucleation establishment at the new SPB inner plaque, realigned with respect to Nud1-mT2. Tub4 colocalized partly with Nud1 at the old SPB in wild type cells (a-b; yellow arrowhead) or exclusively in kar1∆15 cells (c-d; yellow arrowhead). By contrast, in cdc28-4 clb5∆ and cdc28-4 clb5∆ kar1∆15 cells at this stage, Tub4 additionally colocalized with the new Nud1 focus (compare (e) with (f-g) or (h) with (i-j); hollow versus solid white arrowheads). (B) Averaged images compiled after 3D-realignment with respect to Nud1-mT2 of side-by-side SPBs. Linescans for fluorescence intensity along the bridge and IP-OP axis are also shown (1-px width; internally normalized).
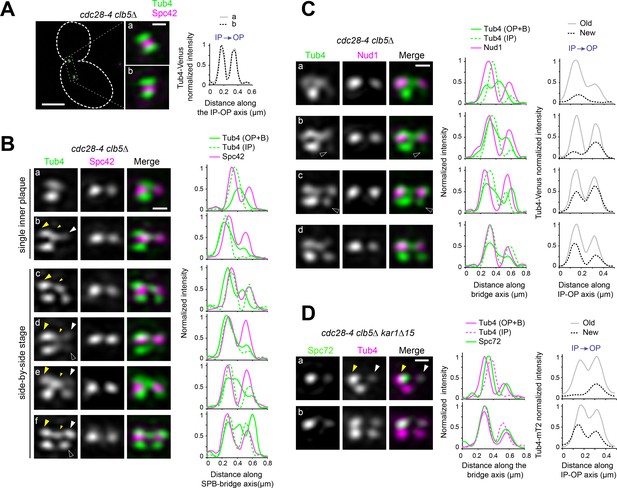
Tub4 IP:OP ratio and sequence of Tub4 recruitment during SPB duplication in cdc28-4 clb5∆ cells.
(A–B) Representative SIM images for Tub4-Venus localization relative to Spc42-mT2 in cdc28-4 clb5∆ cells showing a marked decrease in the IP:OP ratio. (A) Merged image with cell outline (scale bar, 2 µm) and cropped merged images of SPBs (scale bar, 200 nm) corresponding to a cell with a short spindle. Linescan analysis (3-px width) along the IP-OP axis for each SPB is also shown. (B) Realigned SIM images of unseparated SPBs in reference to Spc42-mT2. Tub4 distributed between the old SPB outer plaque and the bridge during duplication (big and small yellow arrowheads, respectively) but was also recruited to the new SPB outer plaque. Early in duplication, some cells appeared to incorporate Tub4 at the new outer plaque before the inner plaque was complete (b, white arrowhead) but others preserved the correct order (c). Hollow white arrowheads point to weaker IP signal relative to OP at the new SPB, also suggestive of perturbed order. Linescan analysis along inner or outer plaque (1px-width) is also shown. (C) Selected realigned images of unseparated SPBs in cdc28-4 clb5∆ cells expressing Tub4-Venus and Nud1-mT2 from the dataset used to generate the average image shown in Figure 6D alongside linescan analysis. Cells began to add Tub4 to the new inner plaque before (a) or after (b) incorporation of Tub4 to the outer plaque. Accordingly, cells at the side-by side-stage could exhibit a weaker Tub4 signal at the new inner plaque relative to the new outer plaque (c, hollow arrowhead). (D) Spc72 recruitment at the new SPB outer plaque may be detected in SIM images of cdc28-4 clb5∆ kar1∆15 expressing Spc72-Venus and Tub4-mT2. During duplication, Spc72-Venus was present at the old SPB outer plaque (yellow arrowhead) but additionally marked the presumptive new SPB outer plaque (white arrowhead) in 5 out of 15 cells showing a single inner plaque focus of Tub4-mT2 (a). Aberrant IP:OP ratio also trailed the premature incorporation of Spc72 at the side-by-side stage (b).
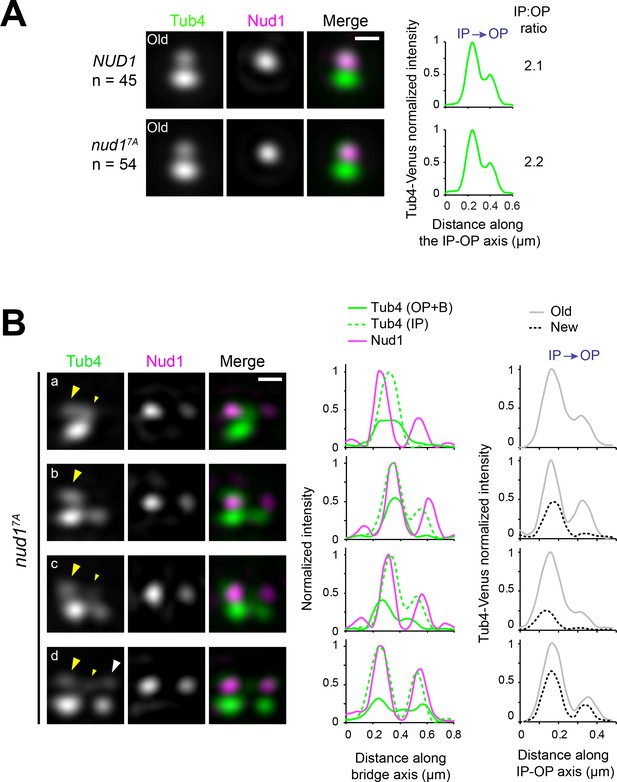
Nud17A anticipates Spc72 recruitment without otherwise affecting the Tub4 IP:OP ratio.
(A) Tub4 IP:OP ratio in SPBs of NUD1 versus nud17A cells carrying short spindles (<2.5 µm-long). SIM averaged images of old SPBs obtained after 3D-Gaussian fitting and realignment using Tub4-Venus as reference. Scale bar, 200 nm. The number of SPBs is indicated. Linescan analysis for Tub4-Venus intensity along the IP-OP axis is shown. (B) Representative images of unseparated SPBs in asynchronous nud17A cells following 3D-Gaussian fitting and realignment using Nud17A-CFP as reference. Tub4-Venus distributed between the old SPB and bridge during duplication (a-c; big and small yellow arrowheads, respectively) but was prematurely added to the new SPB outer plaque at the side-by-side stage (after a Tub4 focus was established at the new inner plaque) without otherwise perturbing the IP:OP ratio (d).
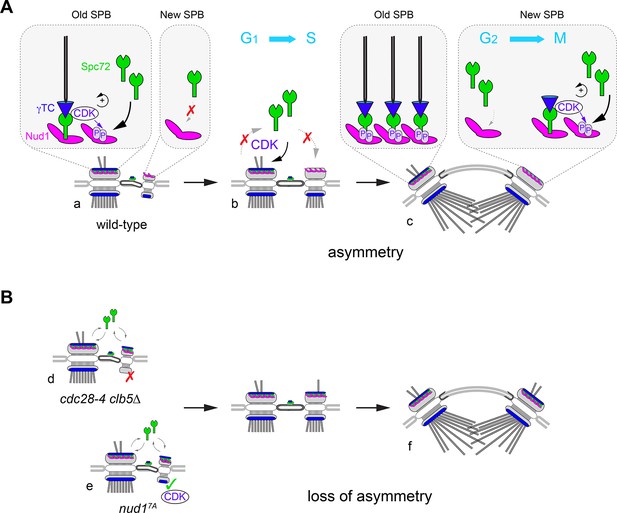
A model for CDK control of intrinsic SPB asymmetry.
Our data suggest that CDK phosphosites in Nud1 increase affinity toward Spc72. (A) Because the old SPB inherits Spc72 and Nud1 from the preceding cell cycle (a), CDK recruitment to the old SPB via Spc72 promotes phosphorylation of Nud1 that may reinforce Spc72 retention in wild type cells. De novo Nud1 added to the new SPB (from the satellite state) is presumably not phosphorylated and would have low affinity for Spc72. As a result (b), S-phase CDK enforces structural asymmetry during the side-by-side stage. By onset of spindle assembly (c), low-affinity binding of Spc72 at the new SPB outer plaque may redirect CDK to phosphorylate Nud1 and trigger the phospho-dependent increase in affinity toward Spc72 that accelerates completion of the new SPB outer plaque. Thus, CDK recruitment via Spc72 translates phospho-dependent affinity between Spc72 and Nud1 into intrinsic structural asymmetry. This built-in delay in new outer plaque maturation manifests in temporal asymmetry in γTC recruitment and aMT organization. (B) In the absence of Nud1 phospho-dependent control, Spc72 is no longer restricted to the old SPB during the side-by-side stage with concomitant disruption of intrinsic asymmetry. The more severe phenotype observed upon loss of S-phase CDK implicates further substrates (see text). In cdc28-4 clb5∆ cells (d), Spc72 symmetry is compounded by mis-regulation of the inner plaque allowing for the incorporation of γTC to the new SPB in the reverse order - first at the outer plaque and then at of the inner plaque. By contrast (e), loss of Nud1 phosphosites in nud17A cells, decreases Spc72 asymmetry without otherwise perturbing inner plaque function. Order of γTC addition is retained but acquisition by the outer plaque is advanced as it precedes SPB separation. In both mutants, short spindles display significant loss of outer plaque structural asymmetry (f).
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Saccharomyces cerevisiae) | NUD1 | Saccharomyces Genome Database | SGD:S000005900 | |
| Gene (Saccharomyces cerevisiae) | SPC72 | Saccharomyces Genome Database | SGD:S000000045 | |
| Strain, strain background (S. cerevisiae) | 15DaubA | Richardson et al., 1989. doi:0092-8674(89)90768-X [pii] | ||
| Strain, strain background (S. cerevisiae) | W303 | Saccharomyces Genome Database | ||
| Strain, strain background (S. cerevisiae) | BY4741 | Saccharomyces Genome Database | ||
| Strain, strain background (S. cerevisiae) | YEF473 | Bi and Pringle, 1996. doi:10.1128/mcb.16.10.5264 | ||
| Strain, strain background (E. coli) | Rosetta 2 cells | Novagen (Merck) | # 71400 | |
| Strain, strain background (S. cerevisiae) | MGY70 | Geymonat et al., 2009. doi: 10.1007/978-1-60327-993-2_4. | ||
| Strain, strain background (S. cerevisiae) | MGY139 | Geymonat et al., 2009. doi: 10.1007/978-1-60327-993-2_4. | ||
| Antibody | anti-HA 12CA5 (Mouse monoclonal) | Roche | #11583816001 RRID:AB_514505 | WB (1:1000) |
| Antibody | anti-α tubulin B-5-1-2 (Mouse monoclonal) | Sigma (Merck) | T5168 RRID:AB_477579 | WB (1:1000) |
| Antibody | anti-GST 3G10/1B3 (Mouse monoclonal) | Abcam | ab92 RRID:AB_307067 | WB (1:1000) |
| Antibody | anti-MBP (Mouse monoclonal) | New England Biolabs | E8032 RRID:AB_1559730 | WB (1:1000) |
| Chemical compound, drug | paraformaldehyde EM Grade | Ted Pella | # 18501 | |
| Software, algorithm | FIJI | https://fiji.sc/ | RRID:SCR_002285 | |
| Software, algorithm | Plugins for FIJI | http://research.stowers.org/imagejplugins | ||
| Other | Dako mounting media | Agilent Technologies | # S3023 |
Additional files
-
Supplementary file 1
Yeast strains.
- https://cdn.elifesciences.org/articles/59222/elife-59222-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59222/elife-59222-transrepform-v2.pdf





