Comparison of induced neurons reveals slower structural and functional maturation in humans than in apes
Figures
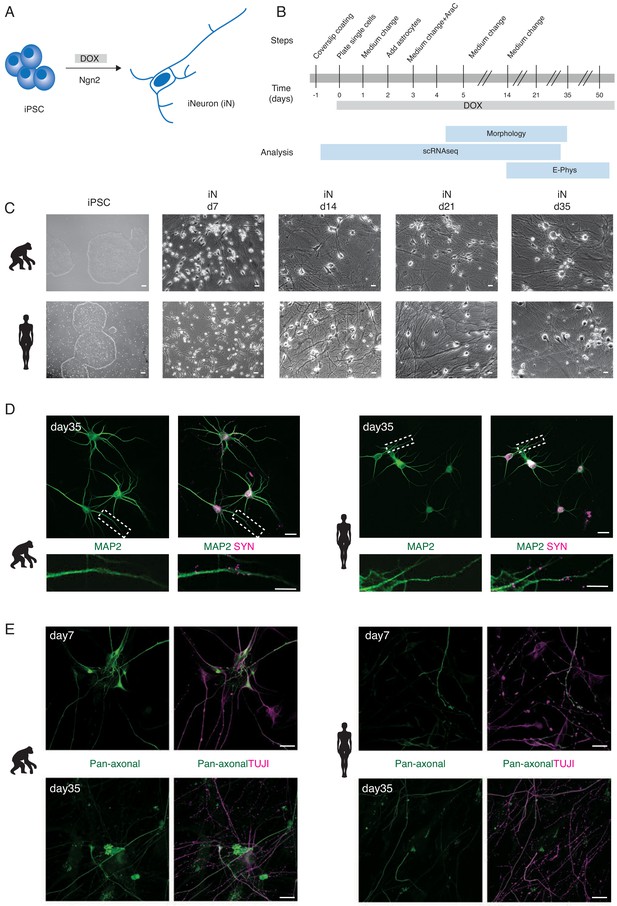
Generation of human and ape iNs.
NGN2-induced iNs generated from human and ape pluripotent stem cells (A, B) differentiate into mature neurons and express neuronal markers (C, D, E). (A) iNs are generated from iPSCs/ESCs upon DOX-induced overexpression of mouse NGN2. (B) Schematic of the experimental pipeline for iNs structural and functional analysis at different time points. The expression of NGN2 was induced at d0 by adding DOX to the culture medium. iNs were either collected for single-cell RNA sequencing (scRNAseq), used for electrophysiological recordings (E-phys), or fixed for morphological analysis (morphology). Single-cell RNA sequencing (scRNAseq) was performed for one chimpanzee (SandraA) and three human (409B2, SC102A1, and H9) cell lines, electrophysiology was performed using all eight cell lines and morphological analyses were done with two chimpanzee (SandraA and JoC), the bonobo (BmRNA), and three human (409B2, SC102A1, and H9) cell lines. (C) Phase-contrast images of iPSCs and iNs maturation for chimpanzee (SandraA) and human (409B2) lines. Scale bars are 10 µm. (D) Mature chimpanzee (SandraA) and human (409B2) iNs express the cytoskeletal marker MAP2 (green) and the pre-synaptic marker SYN1 (magenta). Top: low-magnification view. Scale bars are 20 µm. Bottom: high-magnification view, showing SYN1 puncta in juxtaposition with a MAP2-positive neurite. Scale bars are 10 µm. (E) Distribution of TUJI (TUJI, magenta) and an axonal marker (pan-neurofilament antibody, abbreviated as Pan-Neu, green) in d7 and d35 iNs. Scale bars are 20 µm. We will refer to ape iNs for results where we combined chimpanzee and bonobo iNs for analysis.
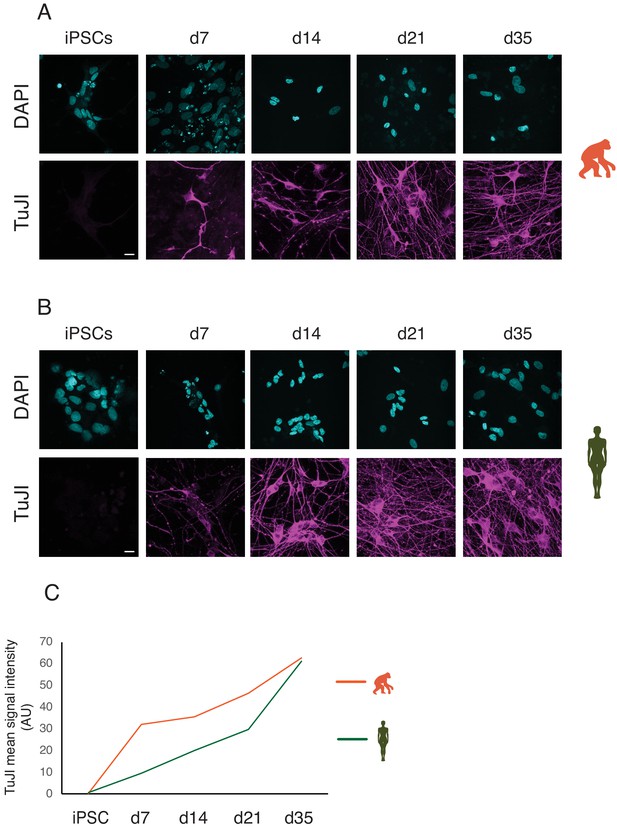
iNs express neuronal markers.
Time course of TUJI expression during differentiation of iNs from iPSCs in chimpanzee (A, SandraA) and human (B, 409B2). Magenta: TUJI; cyan: DAPI. Scale bars are 20 µm. (C) Quantification of TUJI signal intensity in chimpanzee (orange) and human (green) iNs. The intensity of the signal was quantified using FiJI and is expressed as AU (arbitrary unit; see Materials and methods).
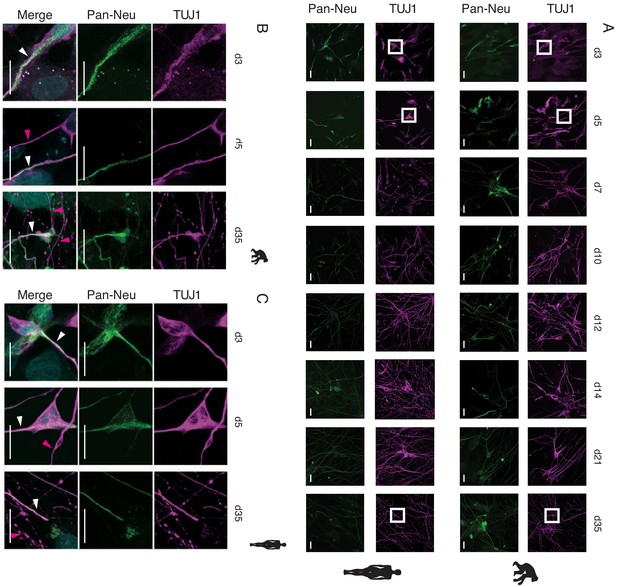
Polarity establishment in ape and human iNs. Axo-dendritic polarity is established around d7 in chimpanzee (SandraA) and human (409B2) iNs.
(A) Distribution of TUJI (TUJI, magenta) and an axonal marker (pan-neurofilament antibody, abbreviated as Pan-Neu, green) during iNs differentiation. Scale bars are 20 µm. (B) High magnification of the areas highlighted with a white box in (A).
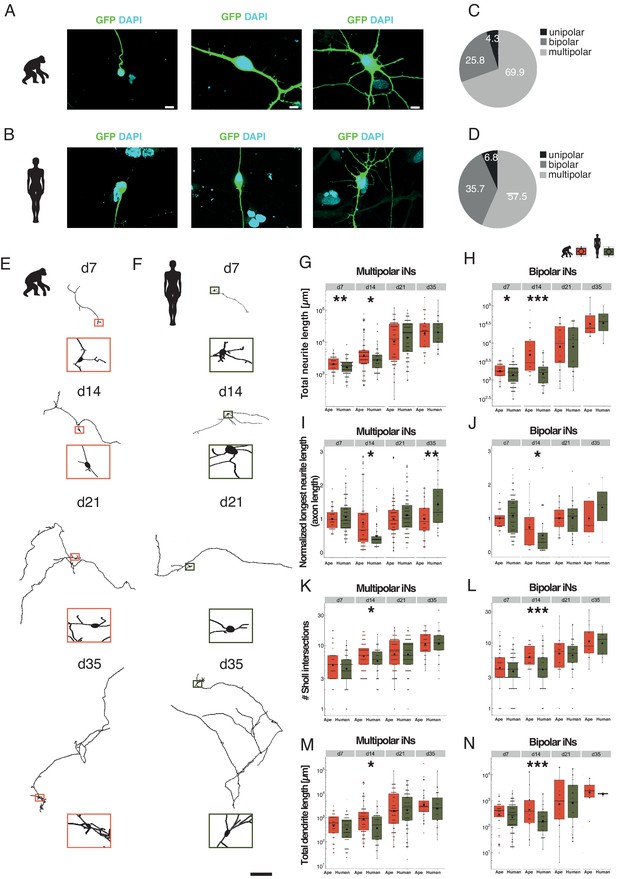
Slower morphological maturation of human iNs.
iNs were lipofected 4 days prior fixation with a plasmid-expressing cytosolic GFP and fixed at different time points. (A, B) Examples of monopolar (left), bipolar (middle) and multipolar (right) iNs from chimpanzee (SandraA, A) and human (409B2, B). Cell morphology is highlighted by GFP expression (green); the nuclei are stained with DAPI (cyan). Scale bars are 20 µm. (C, D) Monopolar (black), bipolar (dark gray), and multipolar (gray) iNs in ape (C) and human (D) expressed as % of total. (E, F) Chimpanzee (E, SandraA, orange) and human (F, 409B2, green) multipolar iNs development over time. Zoom-ins show the cell body. Scale bar is 1 mm. (G, H) Total neurite length, expressed in µm for multipolar and bipolar iNs. (G) Apes multipolar iNs show a higher total neurite length at d7 (p=0.0098) and d14 (p=0.0215) compared to human iNs. The scale is logarithmic. (H) Apes bipolar iNs show a higher total neurite length at d7 (p=0.0278) and d14 (p=0.0003). The scale is logarithmic. (I) Relative length of the longest neurite (axon) in apes and human multipolar iNs. Data representation as normalized data against the mean per batch of the ape data in logarithmic scale. Ape iNs show a higher axon length at d14 (p=0.0294) compared to human iNs, according to total neurite length. Human iNs show a higher axon length at d35 (p=0.0094) compared to ape iNs. (J) Relative longest neurite (axon) length in bipolar iNs. Ape iNs show a higher axon length at d14 (p=0.0426). Data representation as normalized data against the mean per batch of the ape data, the scale is logarithmic. (K, L) Total number of Sholl intersections for multipolar (E) and bipolar (F) iNs. Apes iNs show a higher number of Sholl intersections at d14 for both multipolar iNs (p=0.0469) and bipolar (p=0.0008) iNs. The scale is logarithmic. (M, N) Total dendrite length, expressed in microns for multipolar (M) and bipolar (N) iNs. Ape iNs show a higher total dendrite length at d14 for both multipolar (p=0.0009) and bipolar iNs (p=0.04471). The scale is logarithmic. For all graphs, black rhombs represent the mean and black lines the median. Significance score: p<0.05*, p<0.01**, p<0.001***, Mann–Whitney U test.
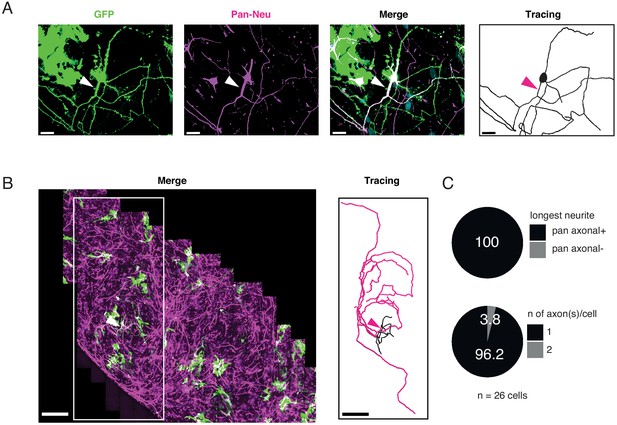
Characterization of axons in iNs.
iNs were lipofected with a plasmid-expressing cytosolic GFP (GFP, green), fixed at d35, and stained with an axonal marker (Pan-Neu, magenta). (A) Chimpanzee iN stained with a pan-neurofilament antibody. The corresponding Imaris tracing is on the right. The arrowheads indicate that the longest of the neurites -as judged by the complete tracing in Imaris- is also positive for axonal marker (white arrowhead). Scale bar is 30 µm. (B) Low-magnification maximum-intensity projection of GFP-labeled cells (green) stained with pan-neurofilament antibody (magenta). The Imaris tracing on the right corresponds to the white box. Magenta tracing corresponds to the longest neurite. Of note, ape and human iNs generate long axons, compared to dendrites. As the longest neurite (axon) encompasses a large part of the total neurite length. Scale bar is 300 µm. (C) Quantifications show that (1) for 100% of the stained cells, the longest neurite is positive for axonal marker (upper pie chart) and (2) the majority of the cell (96% of total) features one axon. 4% of the cells were found to have two axons (bottom pie chart). Of note, in our system the % of cells with two axons is lower than what reported for iNs autaptic culture (Rhee et al., 2019). This is possibly due to the NGN2-iNs system in conjunction with the culture conditions.
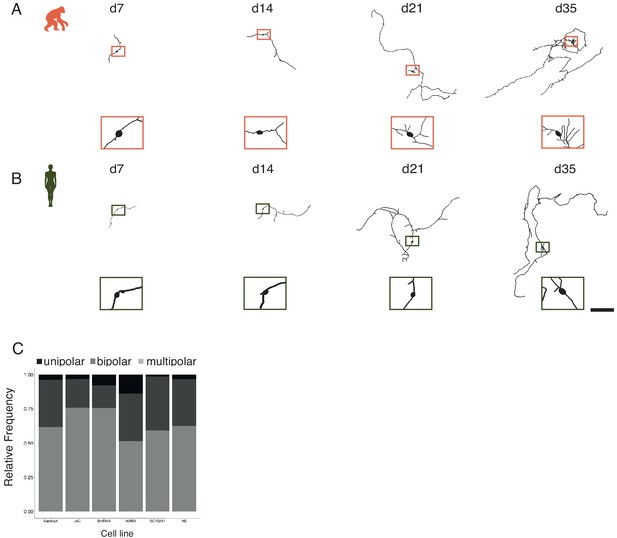
Dynamics of structural maturation in bipolar and multipolar iNs.
iNs were lipofected 4 days prior fixation with a plasmid-expressing cytosolic GFP and fixed at different time points to quantify their structural features. (A, B) Chimpanzee (A, SandraA, orange) and human (B, 409B2, green) bipolar iNs development over time. Zoom-ins show the cell body. Scale bar is 1 mm. (C) Relative frequency of monopolar (black), bipolar (dark gray), and multipolar (gray) iNs for all the cell lines used in this study: chimpanzee (SandraA and JoC) and bonobo (BmRNA) and human (409B2, SC102A, and H9).
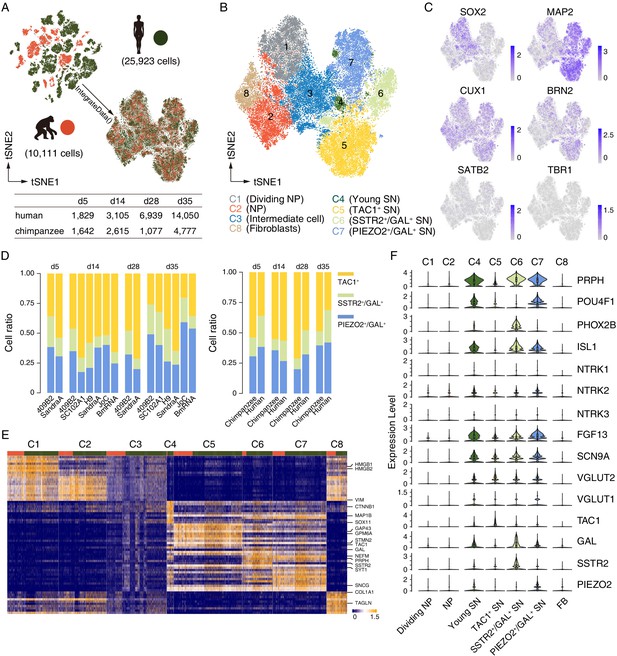
scRNAseq analysis reveals that Nng2-induced iNs are sensory neurons.
Cell identity and heterogeneity were assessed using tSNE combined for all time points for ape (orange) and human (green) cells. (A) Filtered cell number for ape and human cells at different time points. (B) Identification of 8 different cell clusters by marker gene expression. NP, neural progenitor; SN, sensory neuron. (C) Marker gene expression (blue) for progenitor cells (SOX2), mature neurons (MAP2), and cortical cells (CUX1, BRN2, SATB2, and TRB1). Scale bars: uncorrected normalized expression. (D) Cell ratios of the three neuronal sub-classes of TAC1 (yellow), SSTR2 and GAL (green), and PIEZO and GAL (blue) expressing neurons for all cell lines and species. TAC1+, SSTR2+/GAL+, and PIEZO2+/GAL+ neurons on d5: 54%/36% (chimpanzees/humans), 16%/26%, and 31%/38; TAC1+, SSTR2+/GAL+, and PIEZO2+/GAL+ neurons on d14: 55%/57%, 9%/17%, and 36%/27%; TAC1+, SSTR2+/GAL+, and PIEZO2+/GAL+ neurons on d28: 71%/48%, 9%/20%, and 20%/32%; TAC1+, SSTR2+/GAL+, and PIEZO2+/GAL+ neurons on d35: 49%/31%, 12%/27%, and 40%/42%. (E) Heatmap showing scaled expression levels of top 10 significantly higher expressed genes in seven cell populations in iNs culture. The main populations we identified are fibroblasts (FB), neural progenitors, and neurons. Top annotation bars: ape (orange) and human (green). (F) The expression levels of sensory neurons markers are shown as a violin plot for neural progenitor’s clusters C1 and C2 and for neuron’s clusters C4, C5, C6, and C7. Note that the POU homeodomain transcription factors BRN3A (POU4F1) and Islet1 (ISL1) are highly expressed in a subset of early iNs. All iN clusters show high expression of growth factor FGF13 and voltage-gated sodium channel Nav1.7 (SCN9A) that regulate mechano-heat sensation in vivo.
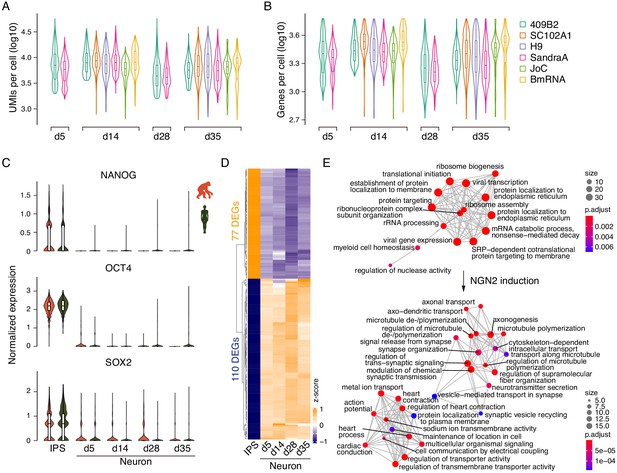
Downregulation of stem cell markers in iNs.
(A,B) The violin plots show number of UMIs (A) and genes (B) per cell per cell line and time point of iN differentiation. (C) The violin plots show the expression in iPSCs and iNs at d5, d14, d28, and d35 of three stem cell marker genes (NANOG, OCT4, and SOX2) in apes (orange) and humans (green). All stem cell genes show a reduction in their expression at the transition from iPSCs to neurons. (D) Differences in gene expression between the different time points of iN differentiation and the stem cells. (E) Networks of enriched GO biological processes in the stem cells and iNs are shown in the top and bottom panels, respectively.
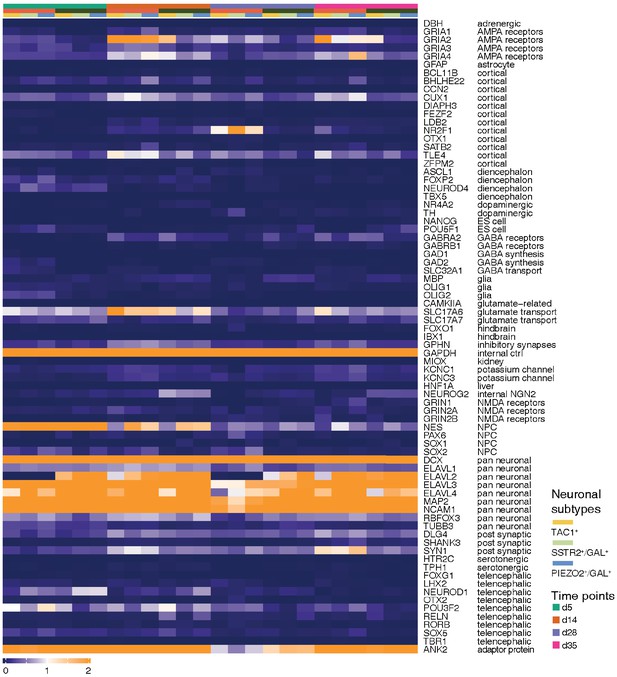
Expression of cortical excitatory neuron genes.
Heatmap of mRNA expression of all marker genes found in the single-cell quantitative RT-PCR analysis (Fluidigm) of Zhang et al. We could identify expression of almost all markers in all species and cell lines, with the absence of expression of few genes in our dataset including forebrain marker FOXG1 and GABA receptor GABRB2.
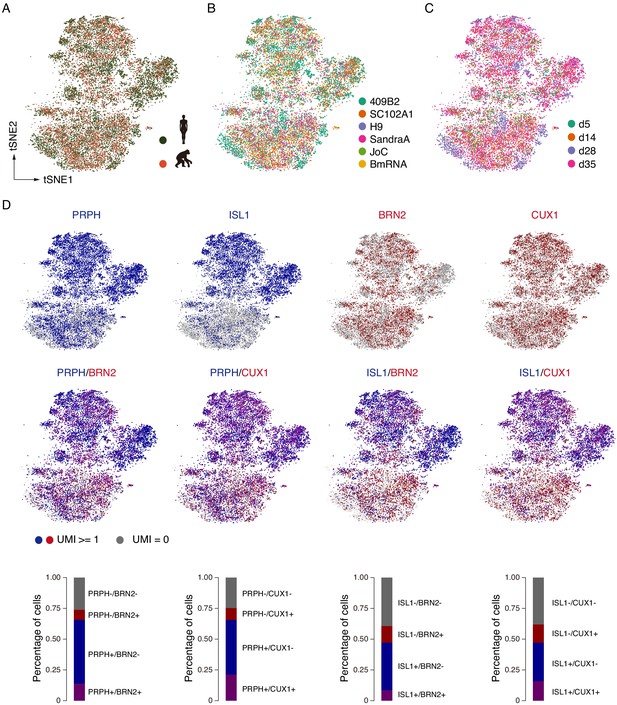
scRNAseq revealed that NGN2 induces cortical and sensory neuron fates.
(A–C) tSNE plots showing single iNs from each species (A) and cell line (B) at each time point (C). (D) scRNAseq analysis reveals that NGN2-induced iNs express both sensory neuron markers as well as cortical markers. Cell identity and heterogeneity was assessed using tSNE combined for all time points (d5, d14, d28 and d35) and ape and human cells together. Marker gene expression for single markers or a combination of sensory and cortical markers. Sensory neurons are shown in blue (PRPH and ISL1) and cortical markers are shown in red (BRN2 and CUX1). Quantifications show that around 66% of the neurons are PRPH positive: 22% of neurons are BRN2 positive and 13.7% of cells are PRPH and BRN2 positive; 31% of cells are CUX1 positive and 21.2% of cells are PRPH and CUX1 positive. Quantifications show that around 47% of neurons are ISL1 positive: 8.3% of cells are ISL1 and BRN2 positive; 15.9% of cells are ISL1 and CUX1 positive, suggesting that the system consists of sensory and cortical neurons and possibly of cortical sensory neurons.
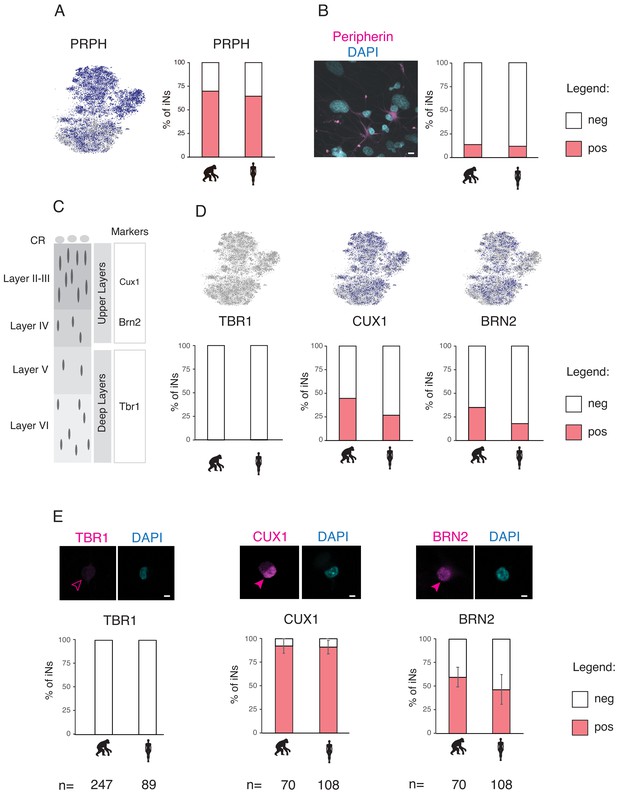
NGN2 induces cortical sensory neuron fates.
The cell identity and the culture heterogeneity of the iNs was assessed by immunofluoresce and mRNA expression analyses for peripheral, cortical, and sensory markers (A) mRNA expression of the peripheral marker Peripherin (PRPH) in iNs. 70% of the iNs were found the express PRPH mRNA. (B) 12% of iNs were found to be immuno-positive for PRPH protein (magenta). (C) Marker genes for cortical layers (CUX1 for layers II–IV, BRN2 for layers II–II, and TBR1 for layers V–VI). (D) mRNA expression of the cortical markers TBR1, CUX1, and BRN2 in iNs. iNs were negative for TBR1, 27–45% of the iNs expressed CUX1 mRNA, and 18–35% BRN2 mRNA. (E) Immunofluorescence of the cortical marker proteins TBR1, CUX1, and BRN2 (magenta) in iNs. iNs were negative for TBR1, 90% of the iNs were immune positive for CUX1, and 50–60% were immune positive for BRN2. Scale bars are 10 µm. A Tukey's post hoc test revealed no significant pairwise differences between the species (see Supplementary file 6).
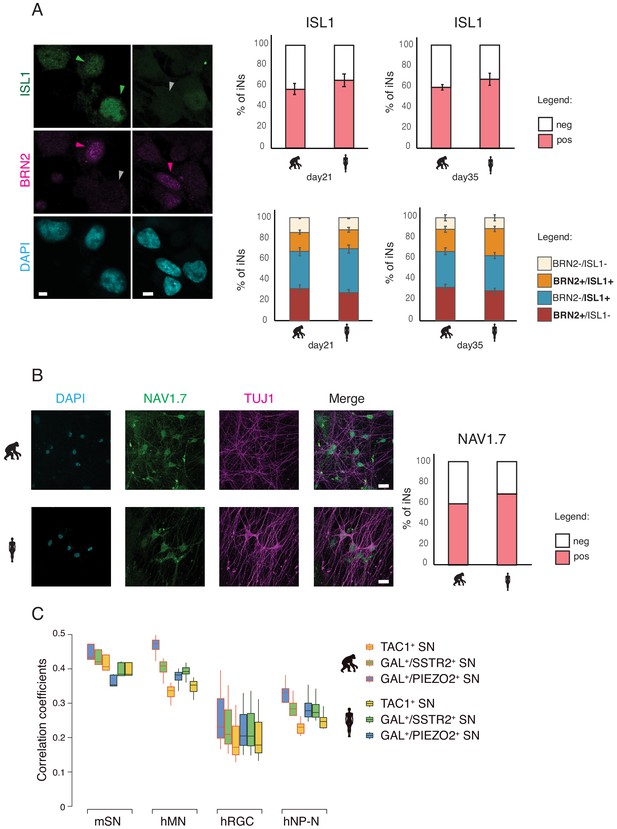
Sensory identity of iNs.
The sensory fate was assessed by immunostaining for the sensory neuron proteins ISLET1 and NAV1.7. (A) 60% of the iNs were positive for ISLET1 (green) at d21 and d35 of differentiation. A proportion of iNs co-expressed BRN2 (magenta) and ISLET1 (green) protein. (C) 60–65% of iNs were immune-positive for NAV1.7 protein at d21 of differentiation. A Tukey's post hoc test revealed no significant pairwise differences between the species (see Supplementary file 6).
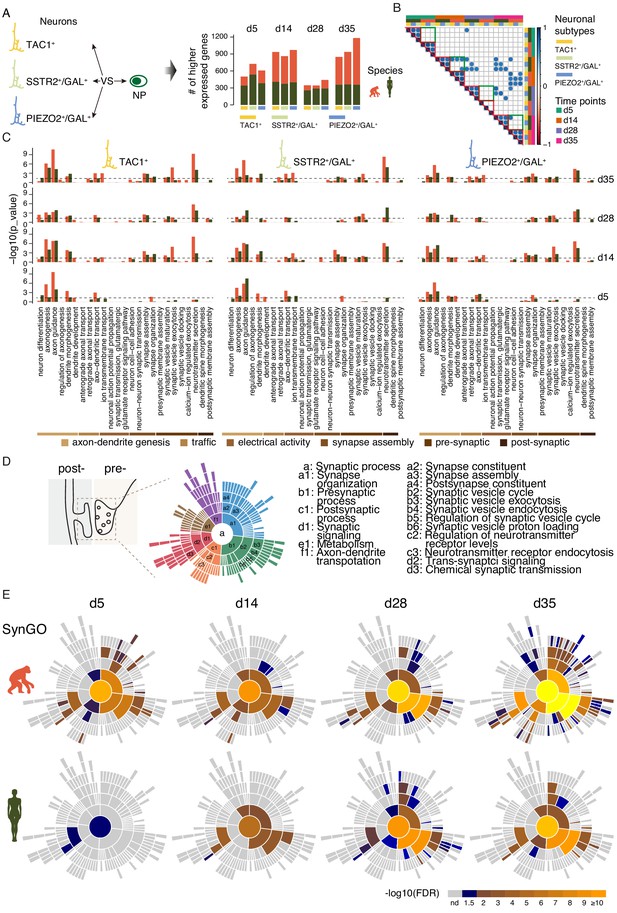
scRNAseq analysis of synaptic maturation in ape and human iNs.
(A) Higher expressed genes in ape and human iN sub-classes compared with corresponding NP of the same species at multiple time points. Left panel, schematics of cell populations for the identification of differentially expressed genes; right panel, bar plot of numbers of higher expressed genes in ape and human iNs. (B) Pearson correlation of enriched GO terms in iNs sub types. Circles represent correlation coefficient larger than 0.7. Red triangles indicate correlations between sub types of same species. Green boxes indicate correlations within the same sub types but between different species. (C) Categories of GO terms on biological processes (BP) enriched with higher expressed genes in ape (orange) and human (green) iN sub-populations at each time point. Reference dashed line (blue) represents p-value of 0.05 (binomial test). (D) Left, schematic representation of synaptic parts depicted by SynGO ontology analysis. Right, six top levels (a1–f1) and selected sub-classifiers of SynGO BP terms. (a, synaptic process). (E) Sunburst plots of enriched synaptic GO terms in developing ape (top) and human (bottom) iNs. Bottom, the log10-transformed FDR-corrected p value per ontology term is visualized for BP. Note that the bottom BP sunburst plots are aligned with the top-right one.
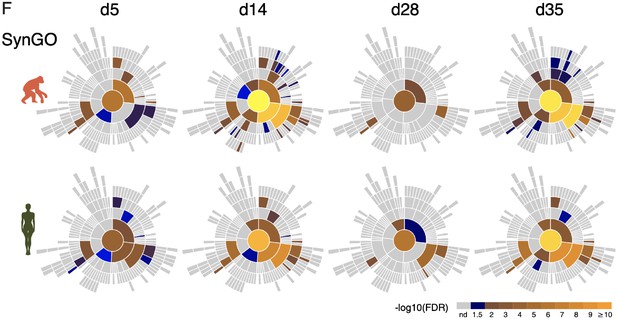
scRNAseq analysis of synaptic maturation in ape and human iNs.
F. Sunburst plots of enriched synaptic GO terms in developing ape (top) and human (bottom) iNs with higher expressed genes conserved in developing ape and human iNs sub-clusters at each time point. Bottom, the log10-transformed FDR-corrected p value per ontology term is visualized for BP. Note that the bottom BP sunburst plots are aligned with the top-right one.
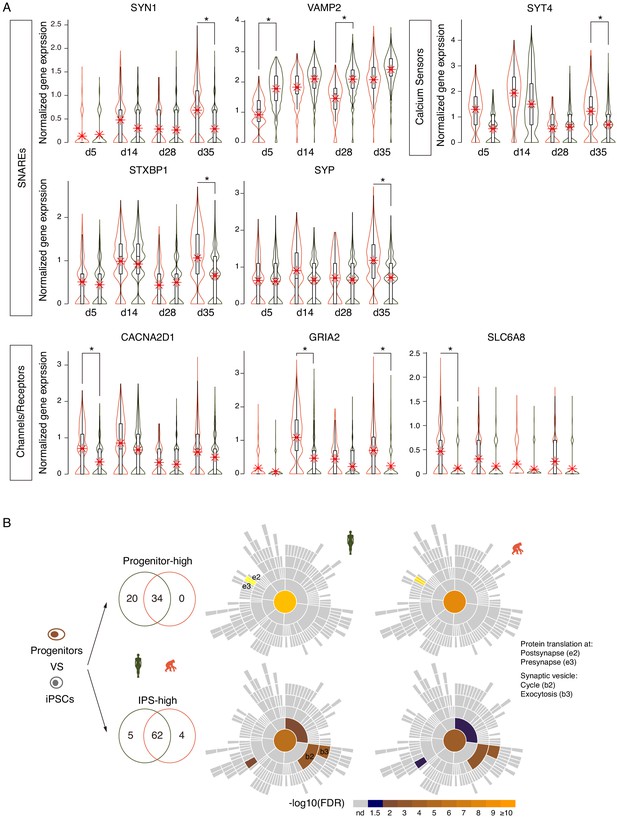
Transcriptional neoteny of synaptic genes in human iNs.
(A) Synaptic genes are differentially expressed in ape and human iNs. Expression over time of SNARE proteins (synapsin (SYN1), VAMP2, STXBP1, and the syntaxin-binding protein SYP), calcium sensors for regulated secretion (Synaptotagmin, SYT4), and channels/receptors (the voltage-operated calcium channel’s subunit CACNA2D1, the glutamate receptor’s subunits GRIA2, and the creatine transporter SLC6A8). (B) SynGO enrichment analysis with significantly differentially expressed genes between NP and iPS cells of the same species. Left panel, higher expressed genes in human and ape NP (top panel) compared with corresponding iPSCs (bottom panel). Right panel, sunburst plots of enriched synaptic GO terms in human and ape NP and iPS cells. The log10-transformed FDR-corrected p value per ontology term is visualized for BP. Note that the bottom BP sunburst plots are aligned with the top-right one.
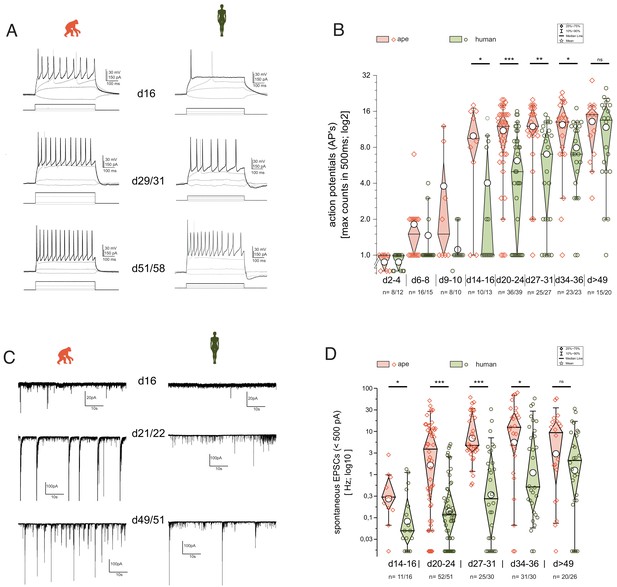
Functional development is delayed in human iNs.
The development of repetitive action potentials (APs) and spontaneous excitatory post-synaptic potentials (sEPSCs) during iNs differentiation was detected using patch-clamp recordings. (A) Voltage responses of individual cells of ape and human to depolarization by current injection at d16, d29–31, and d51–58. Given are the traces with the maximal inducible numbers of APs (black; in gray the responses to −10 to +10 pA of current injection and the trace with the first AP occurring are shown as comparisons). (B) Boxplots of the maximal AP counts (log2 scale). The maximal inducible number of APs in each cell was determined and grouped according to their developmental stage representing app. <1, >1, 2, 3, 4, 5, and more than 7 weeks after induction of NGN2. iNs at d2–4 hardly showed action potentials (APs) and if at all only single APs with reduced amplitude. At d6–d10, iNs showed single APs, with some ape iNs showing double or the very first repetitive APs. After 2 weeks of differentiation Ape cells can maintain repetitive APs with a further increase during the following weeks. Human cells after 2 weeks of differentiation fire mainly single APs. Their ability to maintain repetitive firing developed only in the following weeks. (C) Current responses at −80 mV over a time period of 60 s reveal spontaneous synaptic activity. Spontaneous synaptic activity is observed in ape cell lines regularly after 3 weeks of differentiation and cells without are rarely observed at later time points. In humans however, we observed at early differentiation only rarely spontaneous activity which came up mainly following 4 weeks of differentiation. (D) Boxplots of the observed frequencies of sEPSCs in ape and human. The number of spontaneous events were counted over 60 s and used for further analysis after log10 transformation to account for their distribution. Significance score: p<0.05*, p<0.01**, p<0.001***, unpaired T-test with Welch correction.
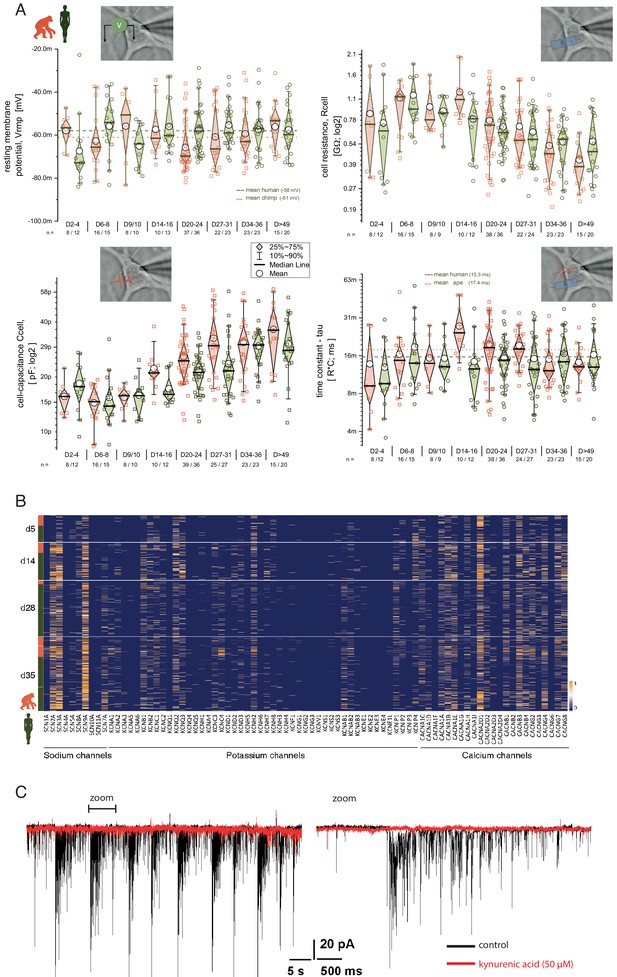
Basic electrical properties of apes and human iNs.
Comparison of basic electrical properties between ape and human iNs 2–8 weeks post DOX-induced expression of NGN2 to verify the maturation of iNs. (A) Basic electrical properties: resting-membrane potential (Vrmp), cell-input resistances (Rcell), cell capacitance (Ccell), and time constant (tau = R*C; C; a measure of the cell response to stimulus). (B) Heatmap showing the expression of 81/88 voltage-gated sodium, potassium, and calcium channels in ape and human iNs. Each row represents gene expression in one cell. (C) Application of kynurenic acid to verify the glutamatergic identity of iNs.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (Homo sapiens) | Human iPS cell line | Riken BRC Cellbank | hiPS-409-B2 | Female |
| Cell line (Homo sapiens) | Human iPS cell line stable transfected with Ngn2 and rttA | This paper | hiPS-409-B2_Ngn2 | Generated for this study. See Materials and methods. |
| Cell line (Homo sapiens) | Human iPS cell line | Systems Biosciences | SC102A-1 | Male |
| Cell line (Homo sapiens) | Human iPS cell line stable transfected with Ngn2 and rttA | This paper | SC102A-1_Ngn2 | Generated for this study. See Materials and methods. |
| Cell line (Homo sapiens) | Human iPS cell line | This paper | HmRNA | Generated for this study. See Materials and methods. Female Reprogramming by mRNA |
| Cell line (Homo sapiens) | Human iPS cell line stable transfected with Ngn2 and rttA | This paper | HmRNA_Ngn2 | Generated for this study. See Materials and methods. |
| Cell line (Pan troglodytes) | Chimpanzee iPS cell line | Generated in a previous study (Mora-Bermúdez et al., 2016) | Sandra_A | Female |
| Cell line (Pan troglodytes) | Chimpanzee iPS cell line stable transfected with Ngn2 and rttA | This paper | Sandra_A_Ngn2 | Generated for this study. See Materials and methods. |
| Cell line (Pan troglodytes) | Chimpanzee iPS cell line | Generated in a previous study (Mora-Bermúdez et al., 2016) | Jo_C | Male |
| Cell line (Pan troglodytes) | Chimpanzee iPS cell line stable transfected with Ngn2 and rttA | This paper | Jo_C_Ngn2 | Generated for this study. See Materials and methods. |
| Cell line (Pan troglodytes) | Chimpanzee iPS cell line | Provided by the Max-Delbrück-Centrum für Molekulare Medizin | ciPS01 Chimp male iPSC Sendai CL5 | Male |
| Cell line (Pan troglodytes) | Chimpanzee iPS cell line stable transfected with Ngn2 and rttA | This paper | ciPS01_Ngn2 | Generated for this study. See Materials and methods. |
| Cell line (Pan paniscus) | Bonobo iPS cell line | Generated for a previous study (Kanton et al., 2019) | BmRNA | Female Reprogramming by mRNA |
| Cell line (Pan paniscus) | Bonobo iPS cell line stable transfected with Ngn2 and rttA | This paper | BmRNA_Ngn2 | Generated for this study. See Materials and methods. |
| Antibody | MAP2 chicken polyclonal antibody | Invitrogen | PA1-16751 | Dilution (1:1000) |
| Antibody | SYN1/2 guinea pig polyclonal antibody | Synaptic Systems | 106004 | Dilution (1:1000) |
| Antibody | TUBB3 mouse monoclonal antibody | BioLegend | 801202 | Dilution (1:1000) Coupled to AlexaFlour 488 or Alexa Flour 555 |
| Antibody | Purified anit-Neurofilament Marker (pan-axonal, cocktail) mouse monoclonal antibody | BioLegend | 8379074 | Dilution (1:400) Clone: SMI312 |
| Antibody | ISL1 mouse monoclonal antibody | Invitrogen | MA5-15515 | Dilution (1:1000) |
| Antibody | PRPH chicken polyclonal antibody | Abcam | ab39374 | Dilution (1:1000) |
| Antibody | BRN2 goat polyclonal antibody | Santa Cruz | sc-6029 | Dilution (1:100) |
| Antibody | TBR1 rabbit polyclonal antibody | Abcam | Ab31940 | Dilution (1:200) |
| Antibody | CUX1 mouse monoclonal antibody | Santa Cruz | sc-514008 | Dilution (1:500) |
| Recombinant DNA reagent | pmax GFP (plasmid) | Lonza | D-00072 | |
| Recombinant DNA reagent | pLVX-EF1α-(Tet-On-Advanced)-IRES-G418(R) (plasmid) | Provided by Nael Nadif Kasri (Frega et al., 2017) | This vector encodes a Tet-On Advanced transactivator under control of a constitutive EF1α promoter and confers resistance to the antibiotic G418. | |
| Recombinant DNA reagent | pLVX-(TRE-thight)-(MOUSE)Ngn2-PGK-Puromycin(R) (plasmid) | Provided by Nael Nadif Kasri (Frega et al., 2017) | This vector encodes the gene for murine neurogenin-2 under control of a Tet-controlled promoter and the puromycin resistance gene under control of a constitutive PGK promoter. | |
| Commercial assay or kit | Stem MACS mRNA transfection Kit | Miltenyi Biotec | 130-104-463 | |
| Commercial assay or kit | Human Pluripotent Stem Cell 3 Colour Immunohistochemistry Kit | R and D Systems1 | SC021 | |
| Commercial assay or kit | Human Pluripotent Stem Cell Functional Identification Kit | R and D Systems | SC027B | |
| Commercial assay or kit | StemMACS Trilineage Differentiation Kit | Miltenyi Biotec | 130-115-660 | |
| Software, algorithm | Imaris 9.5 | Bitplane | ||
| Software, algorithm | Origin | OriginLab version 2018-2019b | ||
| Software, algorithm | Patch and Fitmaster software | HEKA version 2.9x | ||
| Software, algorithm | Custom script to analyse neuron morphology based on Imaris raw files (.hoc) | This paper (Peter and Schörnig, 2020) | For quantification, we exported Sholl intersections and .hoc files, as they contain information about both the neurite length and the branching pattern. HOC files were analyzed using a custom script. See Materials and methods. (Peter and Schörnig, 2020) | |
| Software, algorithm | R 3.5.1 | R |
Additional files
-
Supplementary file 1
N number of iNs used for morphological analysis.
- https://cdn.elifesciences.org/articles/59323/elife-59323-supp1-v2.docx
-
Supplementary file 2
Materials.
- https://cdn.elifesciences.org/articles/59323/elife-59323-supp2-v2.docx
-
Supplementary file 3
Statistics on the electrical properties.
- https://cdn.elifesciences.org/articles/59323/elife-59323-supp3-v2.docx
-
Supplementary file 4
Statistics on the morphological analysis.
- https://cdn.elifesciences.org/articles/59323/elife-59323-supp4-v2.docx
-
Supplementary file 5
Overview on iNs batches and number of cells analyzed for scRNAseq, electrophysiological and morphological experiments.
- https://cdn.elifesciences.org/articles/59323/elife-59323-supp5-v2.xlsx
-
Supplementary file 6
Statistics on the neuronal subtypes.
- https://cdn.elifesciences.org/articles/59323/elife-59323-supp6-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59323/elife-59323-transrepform-v2.docx





