Sorting nexin-27 regulates AMPA receptor trafficking through the synaptic adhesion protein LRFN2
Figures
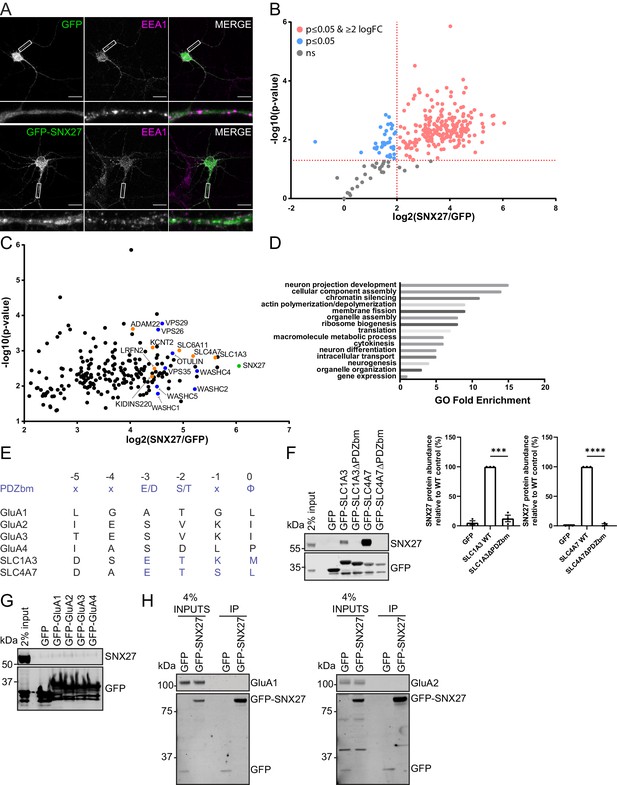
SNX27 interactome reveals new neuronal cargoes.
(A) Immunofluorescence staining of endogenous early endosomal marker EEA1 (pseudocoloured in magenta) in DIV21 rat cortical neurons transduced with GFP or GFP-SNX27 expressing sindbis virus. Scale bars, 20 µm. White boxes indicate the 20 µm section zoomed in. (B) TMT Interactome of SNX27 compared to the GFP control quantified across three independent experiments (n = 3) in DIV21 rat cortical neurons. Plotted proteins were present in all three data sets and analysed using a one-sample t-test and Benjamini–Hochberg false-discovery rate (276 proteins). Vertical red line represents the threshold for enriched proteins in the SNX27 interactome (≥2) compared to the GFP control. Horizontal red line represents the threshold for statistical analysis (p≤0.05). Pink circles are the protein interactors that are enriched and meet statistical significance. Blue circles are the proteins that meet statistical significance but are not over twofold enriched. Grey circles are interactions that are not statistically significant. (C) Filtered TMT interactome (212 proteins) showing only those proteins that were statistically significant and over two log fold change. SNX27 is shown in green with retromer, and the WASH complex shown in blue. Transmembrane proteins that contain a Type I PDZbm with the acidic residue at the −3 position are shown in orange. (D) Gene ontology analysis using the PANTHER classification system of the filtered SNX27 interactome. (E) Schematic highlighting the last six amino acids of the C-terminal tails of isoform 1 of human GluA1, GluA2, GluA3, GluA4, SLC1A3, and SLC4A7 (sequences are conserved in rat). Only SLC1A3 and SLC4A7 possess the optimal PDZ binding motif (PDZbm) consensus sequence for high-affinity binding to SNX27 (highlighted in blue, φ; hydrophobic amino acid). (F–H) Fluorescence-based western analysis after GFP-Trap immunoprecipitation of the: (F) C-terminal tails of GFP-SLC1A3 and GFP-SLC4A7 (+/- PDZbm) with endogenous SNX27 in HEK293T cells. Quantification from three independent experiments (n = 3). Data expressed as a percentage of the WT condition and analysed by an unpaired t-test. Error bars represent mean ± SEM. ****, p≤0.0001; ***, p≤0.001. (G) C-terminal tails of AMPA receptors (GFP-GluA1-4) with endogenous SNX27 in HEK293T cells. (H) Full-length GFP-SNX27 expressing sindbis virus with endogenous GluA1 or GluA2 in DIV20 rat cortical neurons.
-
Figure 1—source data 1
Original immunoblots.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig1-data1-v2.ai
-
Figure 1—source data 2
Data for Figure 1F.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig1-data2-v2.pzfx
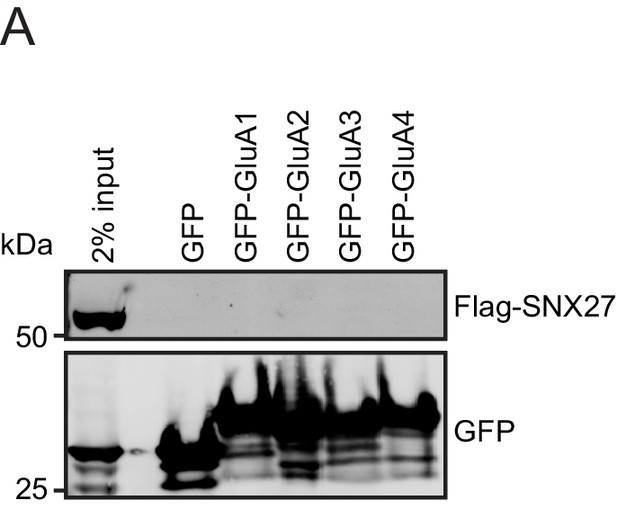
SNX27 does not directly interact with AMPA receptors.
(A) Fluorescence-based western analysis after GFP-Trap immunoprecipitation of the C-terminal tails of AMPA receptors (GFP-GluA1-4) with rat Flag-SNX27 in HEK293T cells.
-
Figure 1—figure supplement 1—source data 1
Original immunoblots.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig1-figsupp1-data1-v2.ai
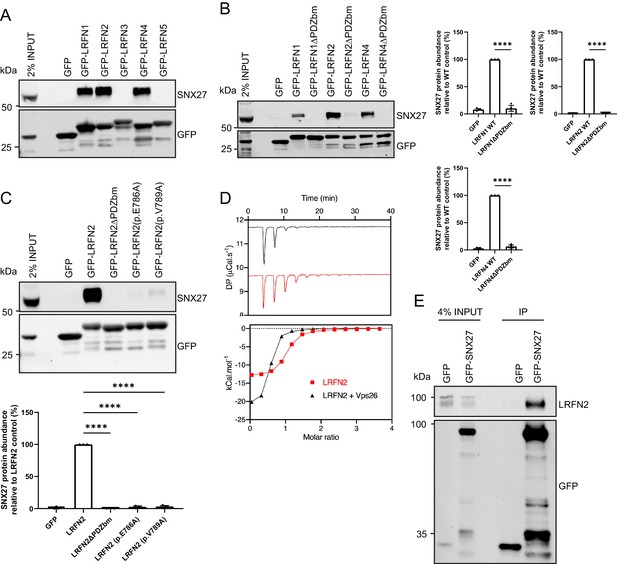
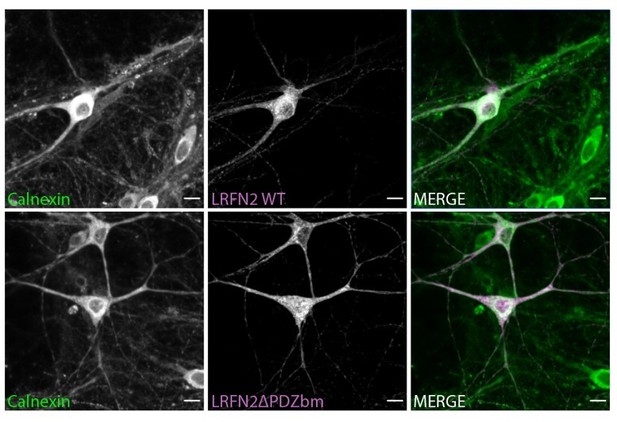
LRFNs interact with SNX27 through their PDZ binding motifs.
(A–C) Fluorescence-based western analysis after GFP-Trap immunoprecipitation of: (A) the C-terminal tails of GFP-LRFN1-5 with endogenous SNX27 in HEK293T cells; (B) the C-terminal domains of LRFN1, LRFN2, and LRFN4 (+/- PDZ binding motif (PDZbm)) with endogenous SNX27 in HEK293T cells. Quantification from three independent experiments (n = 3). Data expressed as a percentage of the wild-type condition and analysed by an unpaired t-test; (C) the C-terminal domain of LRFN2 (+/- PDZbm and mutants pE786A, pV789A) with endogenous SNX27 in HEK293T cells. Quantification from three independent experiments (n = 3). Data expressed as a percentage of the wild-type condition and analysed by an unpaired t-test. (D) Binding of the LRFN2 peptide to the SNX27 PDZ domain, measured by ITC either to SNX27 alone or in the presence of the retromer component VPS26. Top panel shows raw data and bottom panel shows integrated and normalised data. (E) Fluorescence-based western analysis after GFP-Trap immunoprecipitation of full-length GFP-SNX27 with endogenous LRFN2 in DIV20 rat cortical neurons. In all figures error bars represent mean ± SEM. ****, p≤0.0001.
-
Figure 2—source data 1
Original immunoblots.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig2-data1-v2.ai
-
Figure 2—source data 2
Data for Figure 2B and C.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig2-data2-v2.pzfx
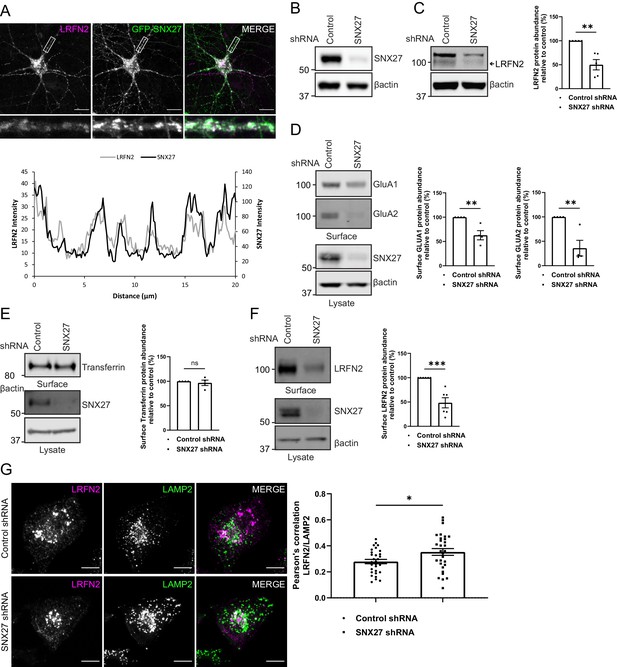
The membrane trafficking of LRFN2 is dependent on SNX27.
(A) Immunofluorescence staining of DIV20 rat hippocampal neurons co-transduced with mCherry-LRFN2 (pseudocoloured in magenta) and GFP-SNX27 expressing sindbis viruses. Scale bars, 20 µm. White boxes indicate the 20 µm section zoomed in. Representative fluorescence intensity plot shown of the 20 µm zoomed in section from (i). (B–C) Fluorescence-based western analysis of DIV19 rat cortical neurons transduced with either a control or SNX27 shRNA for: (B) endogenous SNX27 (C) endogenous LRFN2. Actin was used as a protein load control. Quantification from five independent experiments (n = 5). Data expressed as a percentage of the control shRNA and analysed by an unpaired t-test. (D–E) Fluorescence-based western analysis after surface biotinylation and streptavidin agarose capture of membrane proteins of DIV19 rat cortical neurons transduced with either a control or SNX27 shRNA for: (D) Endogenous surface GluA1 and GluA2. Total levels of endogenous SNX27 are also shown. Quantification from four independent experiments (n = 4). Data expressed as a percentage of the control shRNA and analysed by an unpaired t-test. (E) Endogenous surface Transferrin receptor. Total levels of endogenous SNX27 are also shown. Quantification from four independent experiments (n = 4). Data expressed as a percentage of the control shRNA and analysed by an unpaired t-test. (F) Endogenous surface LRFN2. Total levels of endogenous SNX27 are also shown. Quantification from six independent experiments (n = 6). Data expressed as a percentage of the control shRNA and analysed by an unpaired t-test. (G) Immunofluorescence staining of internalised LRFN2 in H4 cells transduced with either control or SNX27 shRNA. Cells were transfected with mCherry-LRFN2 and after 24 hr the surface LRFN2 labelled using an mCherry antibody. The labelled mCherry-LRFN2 was allowed to internalise for 2 hr before fixation and permeabilisation. Cells were co-stained with LAMP2 as a lysosome marker. Scale bars, 10 µm. Quantification of colocalisation of LRFN2 (pseudocoloured in magenta) and LAMP2 (pseudocoloured in green) from three independent experiments (n = 30 cells analysed). Data analysed by an unpaired t-test. In all figures error bars represent mean ± SEM. ****, P≤0.001; **, P≤0.01; *, P≤0.05.
-
Figure 3—source data 1
Original immunoblots.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig3-data1-v2.ai
-
Figure 3—source data 2
Data for Figure 3C – G.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig3-data2-v2.pzfx
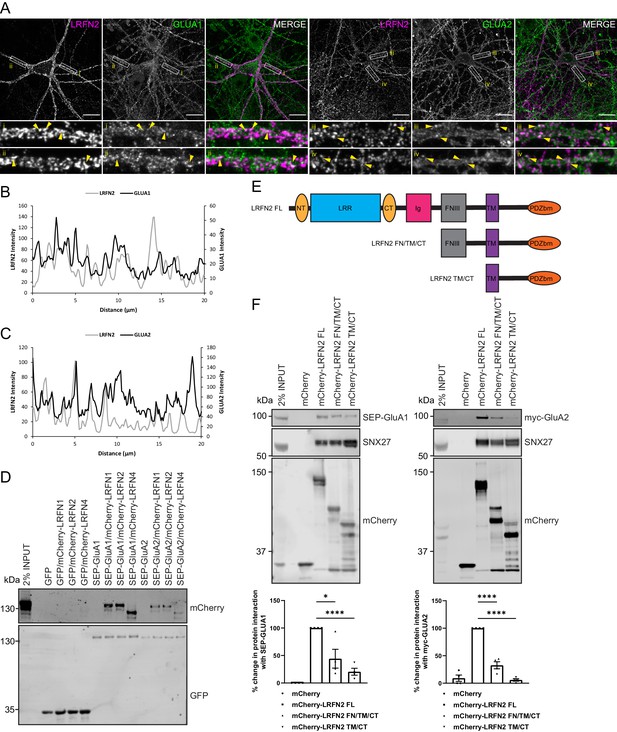
LRFNs interact with AMPA receptors.
(A) Immunofluorescence staining of endogenous surface GluA1 and GluA2 in DIV20 rat hippocampal neurons transduced with mCherry-LRFN2 expressing sindbis virus. An mCherry antibody was used to stain for surface LRFN2 expression followed by a far-red labelled secondary antibody (pseudocoloured in magenta). Cells were co-stained with antibodies against extracellular epitopes of the endogenous AMPA receptor subunits GluA1 and GluA2 (shown in green) Scale bars, 20 µm. White boxes indicate the 20 µm section zoomed in. Yellow arrows show points of overlap. (B) Representative fluorescence intensity plots of the 20 µm zoomed in sections for surface LRFN2 and GluA1 in A (region i). (C) Representative fluorescence intensity plots shown of the 20 µm zoomed in sections for surface LRFN2 and GluA2 in (A; region iii). (D) Fluorescence-based western analysis after GFP-Trap immunoprecipitation of full-length SEP-GluA1 or SEP-GluA2 co-expressed with full-length mCherry-LRFN1, mCherry-LRFN2 or mCherry-LRFN4 in HEK293T cells. (E) Schematic of LRFN2 constructs mCherry-LRFN2 FL (full-length) and N-terminal mutants. LRR, leucine-rich repeat; NT/CT, N/C-terminal domains of LRR; Ig, immunoglobulin domain; FNIII, fibronectin type-III; TM, transmembrane; PDZbm, PDZ binding motif. (F) Fluorescence-based western analysis after RFP-Trap immunoprecipitation of mCherry-LRFN2 wild-type (LRFN2 FL) or N-terminal mutants co-expressed with full-length SEP-GluA1 or myc-GluA2 in HEK293T cells. Quantification from four independent experiments (n = 4). Data expressed as a percentage of the full-length mCherry-LRFN2 and analysed by an unpaired t-test. In all figures error bars represent mean ± SEM. ****, P≤0.0001; *, P≤0.05.
-
Figure 4—source data 1
Original immunoblots.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig4-data1-v2.ai
-
Figure 4—source data 2
Data for Figure 4F.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig4-data2-v2.pzfx
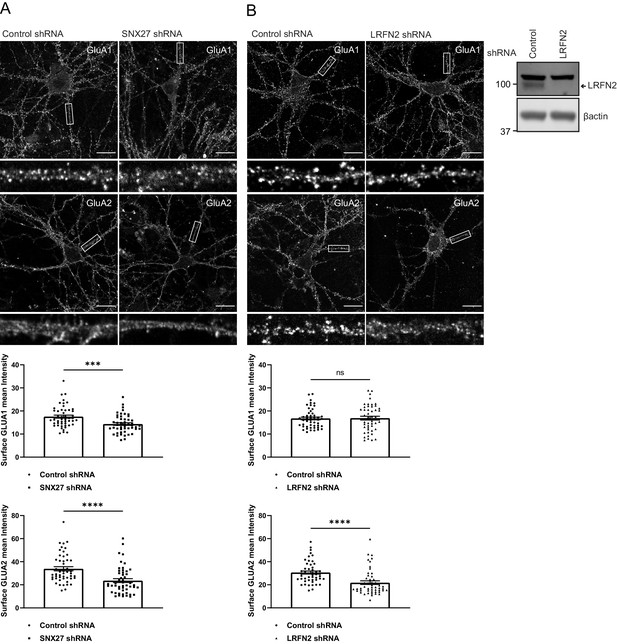
SNX27 and LRFN2 suppression affects surface AMPA receptor expression.
(A) Immunofluorescence staining of endogenous surface GluA1 and GluA2 in DIV19 rat hippocampal neurons transduced with either control or SNX27 shRNA. Scale bars, 20 µm. White boxes indicate the 20 µm section zoomed in. Quantification of surface GluA1 and GluA2 from five independent experiments (n = 50 neurons analysed). Data analysed by a Mann-Whitney U test. (B) Immunofluorescence staining of endogenous surface GluA1 and GluA2 in DIV19 rat hippocampal neurons transduced with either control or LRFN2 shRNA. Scale bars, 20 µm. White boxes indicate the 20 µm section zoomed in. Quantification of surface GluA1 and GluA2 from five independent experiments (n = 50 neurons analysed). Data analysed by a Mann-Whitney U test. In all figures error bars represent mean ± SEM. ****, p≤0.0001; ***, p≤0.001; ns, not significant. Fluorescence-based western analysis shown of DIV19 rat cortical neurons transduced with either control or LRFN2 shRNA for endogenous LRFN2. Actin was used as a protein load control.
-
Figure 5—source data 1
Original immunoblots.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig5-data1-v2.ai
-
Figure 5—source data 2
Data for Figure 5A and B.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig5-data2-v2.pzfx
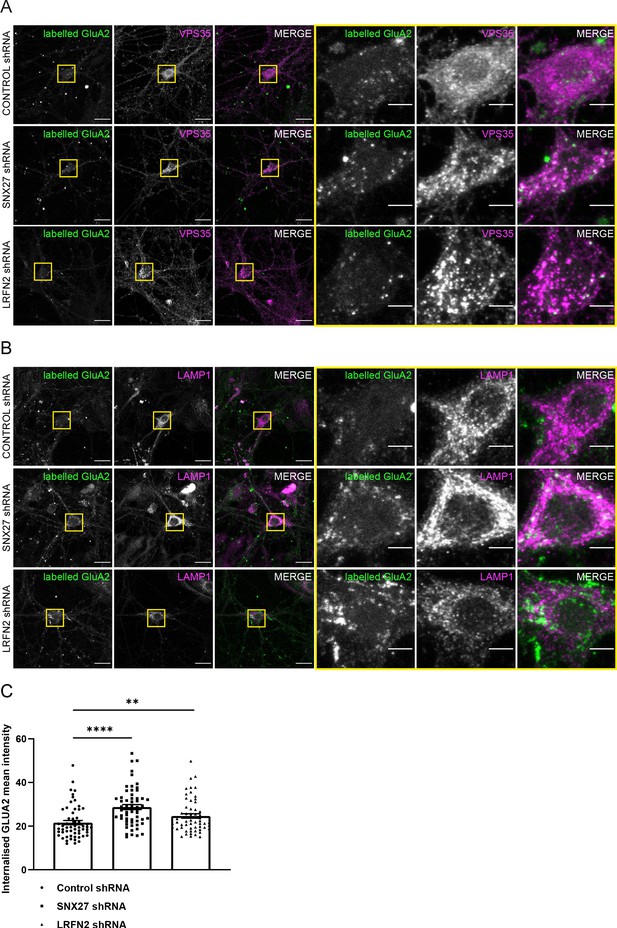
Suppression of SNX27 or LRFN2 increases AMPA receptors in the endo-lysosomal system.
(A–B) Immunofluorescence staining of endogenous internalised GluA2 (pseudocoloured in green) in DIV19 rat hippocampal neurons transduced with either control, SNX27 or LRFN2 shRNA. Surface labelled GluA2 was allowed to internalise for 1 hr before fixation and permeabilisation. Scale bars, 20 µm. Zoomed in images of the cell body (Yellow boxes). Scale bars, 5 µm. For clarity only the internalised GluA2 images are shown. The surface GluA2 expression can be found in Figure 6—figure supplement 1. (A) Cells were co-stained with VPS35 (pseudocoloured in magenta) as an endosomal marker. (B) Cells were co-stained with LAMP1 (pseudocoloured in magenta) as a lysosome marker. (C) Quantification of internalised GluA2 intensity from five independent experiments (n = 58 neurons analysed). The mean intensity of GluA2 was measured from the cell body and proximal dendrites. Data analysed by a Mann-Whitney U test. In all figures error bars represent mean ± SEM. ****, P≤0.0001; **, P≤0.01.
-
Figure 6—source data 1
Data for Figure 6C.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig6-data1-v2.pzfx
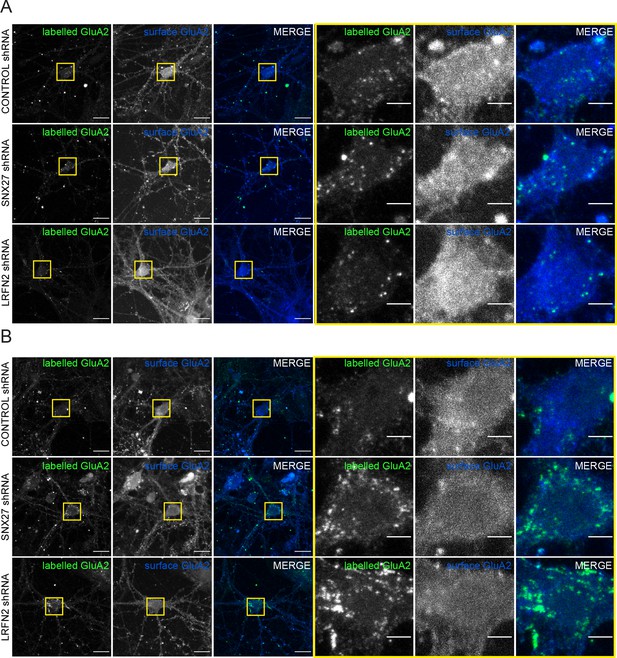
Suppression of SNX27 or LRFN2 increases AMPA receptors in the endo-lysosomal system.
(A) Immunofluorescence staining showing the surface expression of GluA2 (blue) with the labelled internalised GluA2 (pseudocoloured in green) in DIV19 rat hippocampal neurons transduced with either control, SNX27 or LRFN2 shRNA. The GluA2 was allowed to internalise for 1 hr before fixation. The antibody remaining on the surface was labelled with a secondary Alexa Fluor 405 antibody. Internalised receptors were then detected by permbealisation followed by staining with a secondary Alexa Fluor 647 antibody to distinguish from surface expression. Scale bars, 20 µm. Zoomed in images of the cell body (Yellow boxes). Scale bars, 5 µm. (A) Images for Figure 6A. (B) Images for Figure 6B.
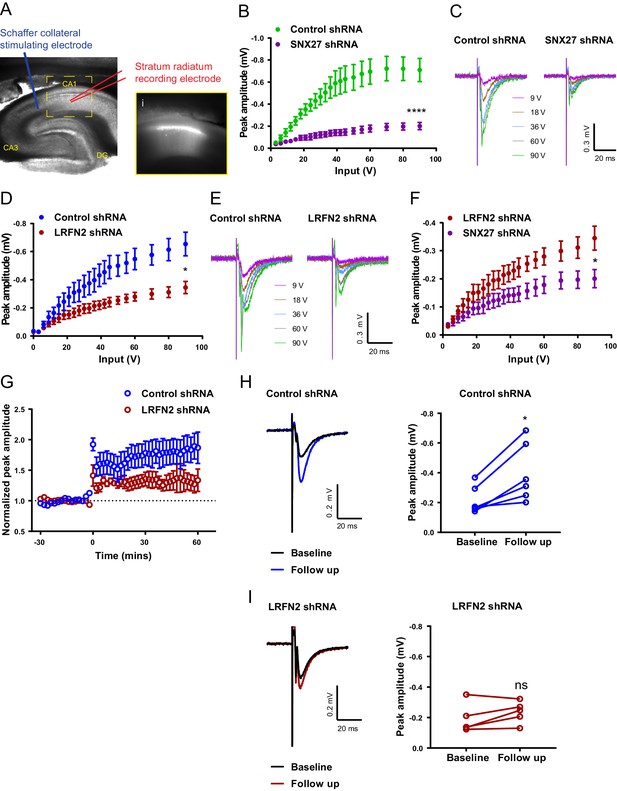
In vivo suppression of SNX27 and LRFN2 affects ex vivo AMPA receptor activity in the hippocampus.
(A) Representative infrared image of a transduced rat dorsal hippocampal slice expressing the control shRNA lentivirus showing placement of electrodes to record Schaffer collateral responses. Inset (i) shows GFP fluorescence of CA1 region. Animals were left for 6–8 weeks with the control shRNA injected into one hemisphere and either SNX27 or LRFN2 shRNA into the other hemisphere. (B) Input-output curves from SNX27 and control shRNA treated slices. Quantification from six animals (n = 10 recordings) for the control and five animals (n = 11 recordings) for the SNX27 shRNA. Data analysed by a two-way repeated-measures ANOVA. (C) Representative traces of the recordings from (B). (D) Input-output curves from LRFN2 and control shRNA-treated slices. Quantification from five animals (n = seven recordings) for the control and five animals (n = seven recordings) for the LRFN2 shRNA. Data analysed by a two-way repeated-measures ANOVA. (E) Representative traces of the recordings from (D). (F) Input-output curves comparing SNX27 and LRFN2 shRNA treatment. (G) LTP response following high frequency stimulation (HFS) at t = 0. Quantification from five animals (n = six recordings) for the control and four animals (n = five recordings) for the LRFN2 shRNA. (H) Representative traces and quantification of the LTP response after treatment with the control shRNA (follow-up is the final 10 min of the recording). Control slices underwent LTP with data analysed by a paired t-test. (I) Representative traces and quantification of the LTP response after treatment with the LRFN2 shRNA. LRFN2 shRNA treated slices were not significantly potentiated. In all figures error bars represent mean ± SEM. ****, p≤0.0001; *, p≤0.05; ns, not significant.
-
Figure 7—source data 1
Data for Figure 7B - I.
- https://cdn.elifesciences.org/articles/59432/elife-59432-fig7-data1-v2.pzfx
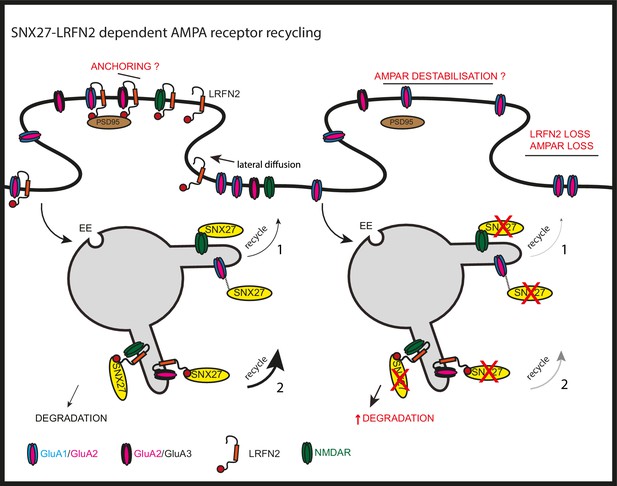
SNX27 regulation of AMPA receptor trafficking through LRFN2 It is well established that SNX27 suppression results in a loss of AMPA receptor expression and activity through a mechanism that evokes the direct sequence-dependent recognition of internalised AMPA receptors by endosome associated SNX27 (1).
In the absence of SNX27, internalised AMPA receptors fail to be recycled and enter the lysosomal degradative compartment. This missorting leads to an overall reduction in AMPA receptor expression at the cell surface. Our data proposes that LRFN2 acts as a ‘bridging’ factor in SNX27-mediated AMPA receptor trafficking (2). Here, internalised LRFN2 directly associates with SNX27 through its PDZ binding motif (PDZbm) and is a classical cargo for SNX27-dependent retrieval and recycling to the synaptic cell surface. Upon SNX27 suppression, internalised LRFN2 is missorted to lysosomes where it undergoes degradation, leading to a reduction in LRFN2 levels at the synaptic surface. LRFN2 associates with AMPA receptors and LRFN2 suppression leads to AMPA receptor missorting into the degradative pathway. Taken together, we propose that the association of LRFN2 with AMPA receptors is required for the SNX27-dependent endosomal recycling of GluA1-lacking AMPA receptors. LRFN2 may also display properties of an ‘auxiliary’ protein in aiding the retrieval and recycling of internalised AMPA receptors through the SNX27-positive endosomal compartment and, in addition, may function as an adhesion molecule to anchor and stabilise AMPA receptor expression at the cell surface. EE, early endosome; LE, late endosome.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H. sapiens) | Human kidney cells (Hek293T) | AATC | CRL-3216 RRID:CVCL_0063 | Authentication was from AATC. We did not independently authenticate the cell line. |
| Cell line (M. auratus) | Baby Hamster Kidney cells (BHK-21) | AATC | [C-13] CCL-10 RRID:CVCL_1915 | Authentication was from AATC. We did not independently authenticate the cell line. |
| Cell line (H. sapiens) | Neuroglioma (H4) | A gift from Dr Helen Scott and Professor James Uney | Authenticated (STR profiling) and mycoplasma tested (absent) (Eurofins) | |
| Biological sample (R. norvegicus) | Primary hippocampal and cortical neurons | University of Bristol Animal Services Unit | ||
| Antibody | ‘(mouse monoclonal)’ β–actin | Sigma-Aldrich | A1978 RRID:AB_476692 | WB ‘(1:1000)’ |
| Antibody | ‘(mouse monoclonal)’ EEA1 | BD Biosciences | 610457 RRID:AB_397830 | IF ‘(1:250)’ |
| Antibody | ‘(mouse monoclonal)’ FLAG | Sigma-Aldrich | F1804 RRID:AB_262044 | WB ‘(1:1000)’ |
| Antibody | ‘(mouse monoclonal)’ GFP | Roche | 11814460001 RRID:AB_390913 | WB ‘(1:1000)’ |
| Antibody | ‘(rabbit polyclonal)’ GluA1 | Merck Millipore | ab1504 RRID:AB_2113602 | WB ‘(1:1000)’ |
| Antibody | ‘(mouse monoclonal)’ GluA1 | Merck Millipore | MAB2263 RRID:AB_11212678 | IF ‘(1:100)’ |
| Antibody | ‘(mouse monoclonal)’ GluA2 | Merck Millipore | MAB397 RRID:AB_2113875 | WB ‘(1:1000)’ IF ‘(1:70)’ |
| Antibody | ‘(rabbit polyclonal)’ mCherry | Abcam | ab167453 RRID:AB_2571870 | WB ‘(1:1000)’ IF ‘(1:100)’ |
| Antibody | ‘(rabbit polyclonal)’ LRFN2 | Atlas | HPA07660 | WB ‘(1:500)’ |
| Antibody | ‘(mouse monoclonal)’ SNX27 | Abcam | ab77799 RRID:AB_10673818 | WB for human SNX27 ‘(1:500)’ |
| Antibody | ‘(rabbit polyclonal)’ SNX27 | A kind gift from Dr Martin Playford, NIH, U.S.A | WB for rat SNX27 ‘(1:500)’ | |
| Antibody | ‘(rabbit polyclonal)’ LAMP1 | Abcam | ab24170 RRID:AB_775978 | IF ‘(1:200)’ |
| Antibody | ‘(mouse monoclonal)’ LAMP2 | Hybridoma bank | H4B4 RRID:AB_2134755 | IF ‘(1:500)’ |
| Antibody | ‘(mouse monoclonal)’ Transferrin | Santa Cruz | Sc-65882 RRID:AB_1120670 | WB ‘(1:1000)’ |
| Antibody | ‘(rabbit polyclonal)’ VPS35 | Abcam | ab97545 RRID:AB_10696107 | IF ‘(1:200)’ |
| Antibody | ‘(rabbit)’ Alexa Fluor 405 | Invitrogen | A31556 RRID:AB_221605 | IF ‘(1:200–400)’ |
| Antibody | ‘(mouse)’ Alexa Fluor 488 | Invitrogen | A21202 RRID:AB_141607 | IF ‘(1:400)’ |
| Antibody | ‘(rabbit)’ Alexa Fluor 568 | Invitrogen | A10042 RRID:AB_2534017 | IF ‘(1:400)’ |
| Antibody | ‘(mouse)’ Alexa Fluor 568 | Invitrogen | A10037 RRID:AB_2534013 | IF ‘(1:400)’ |
| Antibody | ‘(rabbit)’ Alexa Fluor 647 | Invitrogen | A31573 RRID:AB_2536183 | IF ‘(1:400)’ |
| Antibody | ‘(mouse)’ Alexa Fluor 647 | Invitrogen | A31571 RRID:AB_162542 | IF ‘(1:400)’ |
| Antibody | ‘(mouse)’ Alexa Fluor 680 | Invitrogen | A21057 RRID:AB_141436 | WB ‘(1:10,000)’ |
| Antibody | ‘(rabbit)’ Alexa Fluor 800 | Invitrogen | SA535571 RRID:AB_2556775 | WB ‘(1:10,000)’ |
| Peptide, recombinant protein | LRFN2 peptides | Genscript | ||
| Chemical compound, drug | Sulpho NHS-SS-Biotin | Thermo Fisher | 21331 | |
| Chemical compound, drug | Streptavidin beads | GE Healthcare | 17-5113-01 | |
| Chemical compound, drug | GFP/RFP-Trap beads | ChromoTek | gta-20, rta-20 | |
| Recombinant DNA reagent | pEGFP-C3-GLUA1(CT) | This paper | available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pEGFP-C3-GLUA2(CT) | This paper | available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pEGFP-C3-GLUA3(CT) | This paper | available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pEGFP-C3-GLUA4(CT) | This paper | available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pEGFP-C1-LRFN1(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN2(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN3(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN4(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN5(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-SLC1A3(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-SLC4A7(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN1ΔPDZbm(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN2ΔPDZbm(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN4ΔPDZbm(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1- SLC1A3ΔPDZbm(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1- SLC4A7ΔPDZbm(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN2(pE786A)(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pEGFP-C1-LRFN2(pV789A)(CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pcDNA3.1-FLAG-SNX27 | This paper | available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pCMV2-SEP-GluA1 | Addgene | 64942 RRID:Addgene_64942 | Blanco-Suarez and Hanley, 2014 |
| Recombinant DNA reagent | pcDNA3-SEP-GluA2 | Addgene | 64941 RRID:Addgene_64941 | Ashby et al., 2004 |
| Recombinant DNA reagent | pCMV2-myc-GluA2 | Leuschner and Hoch, 1999 available on request from Kevin Wilkinson | ||
| Recombinant DNA reagent | pmCherryC1-LRFN2(FL) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pmCherryC1-LRFN2(FN/TM/CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pmCherryC1-LRFN2(TM/CT) | This paper | available on request from Peter Cullen | |
| Recombinant DNA reagent | pXLG3-GFP-H1-SNX27 shRNA (Rat) (+/- WPRE) | Binda et al., 2019 | Rat target sequence 5’-aagaacagcaccacagaccaa-3’ available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pXLG3-GFP-H1-SNX27 shRNA (Human) (+/- WPRE) | This paper | Human target sequence 5’- aagaacagtactacagaccaa-3’ available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pXLG3-GFP-H1-LRFN2 shRNA (+/- WPRE) | This paper | target sequence 5’-acgacgaggtactgattta-3’ available on request from Peter Cullen | |
| Recombinant DNA reagent | pXLG3-GFP-H1-control shRNA (+/-WPRE) | Binda et al., 2019 | non-targeting sequence 5’-aattctccgaacgtgtcac-3’ available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pSinRep5-GFP/ pSinRep5-GFP-SNX27 | This paper | available on request from Kevin Wilkinson | |
| Recombinant DNA reagent | pSinrep-mcherry/ pSinRep5-mCherry-LRFN2 | This paper | available on request from Peter Cullen |
Additional files
-
Supplementary file 1
A neuronal SNX27 interactome reveals new cargoes for SNX27-mediated trafficking.
(A) Raw data from TMT Interactome of SNX27 compared to the GFP control quantified across three independent experiments (N = 3) in DIV21 rat cortical neurons. (B) Filtered TMT interactome (212 proteins) showing only those proteins that were statistically significant (one-sample t-test and Benjamini–Hochberg false-discovery rate) and over two log fold change compared to the GFP control.
- https://cdn.elifesciences.org/articles/59432/elife-59432-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59432/elife-59432-transrepform-v2.docx