Establishment and maintenance of motor neuron identity via temporal modularity in terminal selector function
Figures
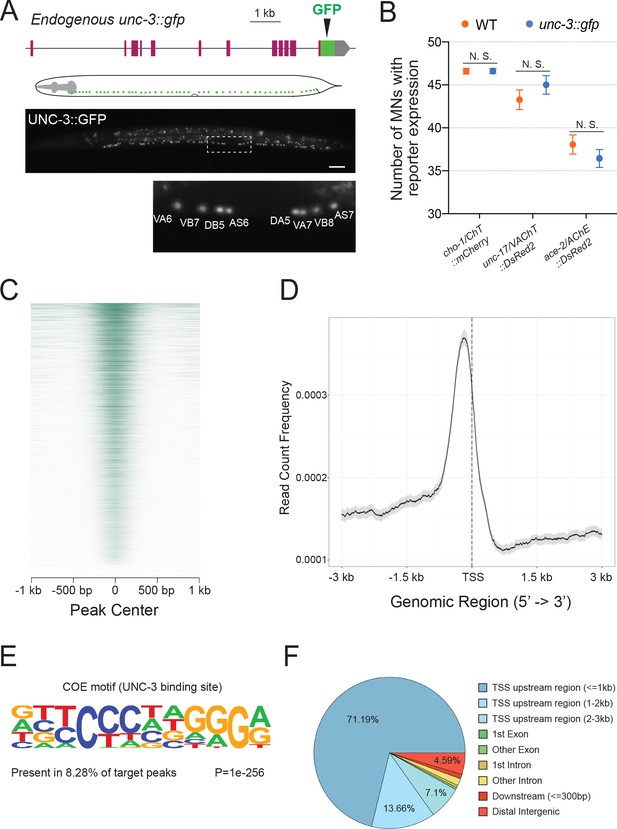
Mapping UNC-3 binding genome-wide with ChIP-Seq.
(A) Diagram illustrating the endogenous reporter allele of UNC-3. GFP is inserted immediately upstream of unc-3’s stop codon. Below, a representative image at L2 stage showing expression of UNC-3::GFP fusion protein in cholinergic MN nuclei. Region highlighted in dashed box is enlarged. Scale bar, 20 μm. (B) Quantification of terminal identity gene markers that report expression of known UNC-3 targets (cho-1/ChT, unc-17/VAChT, ace-2/AChE) in WT and ot839 [unc-3::gfp] animals at the L4 stage (N = 15). N. S.: not significant. (C) Heatmap of UNC-3 ChIP-Seq signal around 1.0 kb of the center of the binding peak. (D) Summary plot of UNC-3 ChIP-Seq signal with a 95% confidence interval (gray area) around 3.0 kb of the TSS. The average signal peak is detected at ~200 bp upstream of the TSS. (E) de novo motif discovery analysis of 6,892 UNC-3 binding peaks identifies a 12 bp-long UNC-3 binding motif. (F) Pie chart summarizes genomic distribution of UNC-3 ChIP-Seq signal.
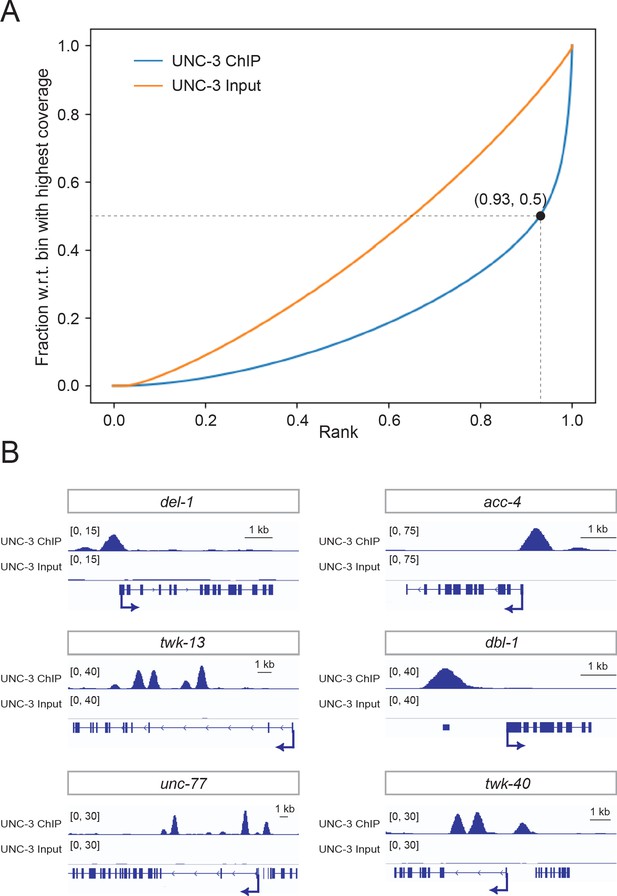
UNC-3 ChIP-Seq results yield genome-wide enrichment of UNC-3.
(A) Fingerprint plot indicating localized, strong enrichment of UNC-3 in the genome. Specifically, when counting the reads contained in 93% of all genomic bins (data point 0.93, 0.5 on UNC-3 ChIP curve), only 50% of the maximum number of reads are reached, which indicates 7% of the genome contains half of total sequencing reads from the ChIP sample. (B) Snapshots of UNC-3 ChIP-Seq and input (negative control) signals at the cis-regulatory regions of known UNC-3 targets (del-1, acc-4, twk-13, dbl-1, unc-77, twk-40).
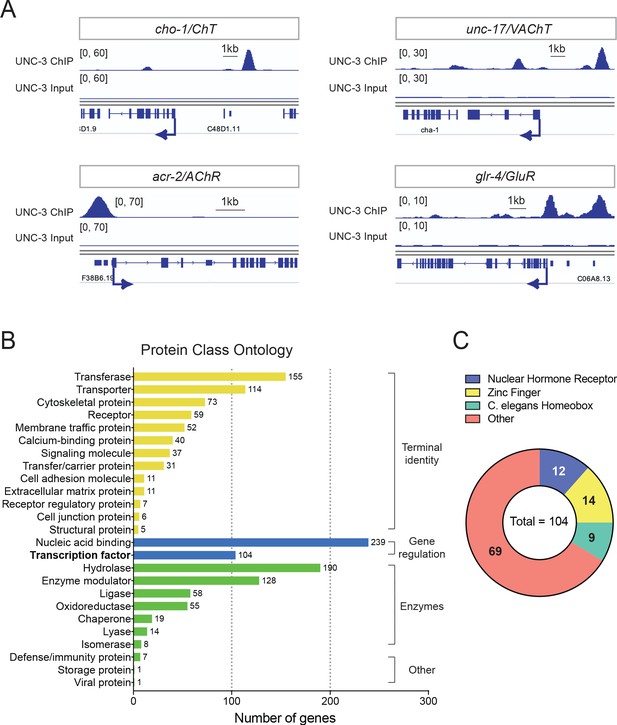
Global analyses of UNC-3 ChIP-Seq data.
(A) Snapshots of UNC-3 ChIP-Seq and input (negative control) signals at the cis-regulatory regions of known UNC-3 targets (cho-1/ChT, unc-17/VAChT, acr-2/AChR, glr-4/GluR). (B) Graph summarizing protein class ontology analysis of putative target genes of UNC-3 identified by ChIP-Seq. Out of the 3502 protein-coding UNC-3 targets, 1425 encode for proteins with known protein class terms and these were the ones considered by PANTHER. This analysis classifies UNC-3 targets into three broad categories: terminal identity genes, gene expression regulators, and enzymes. (C) Pie chart breaking down TF families that show UNC-3 binding.
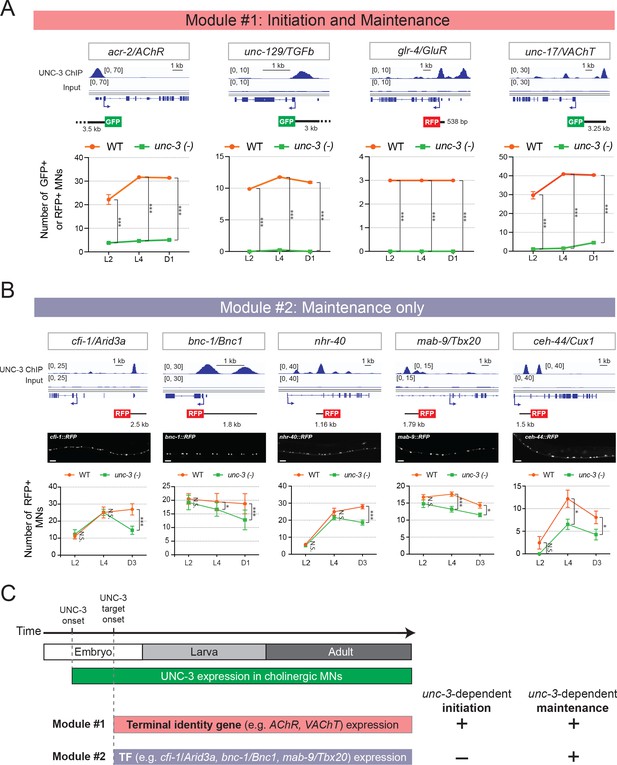
Terminal identity genes and transcription factors display distinct temporal requirements for UNC-3.
(A) Top: snapshots of UNC-3 ChIP-Seq and input (negative control) signals at the cis-regulatory regions of four cholinergic terminal identity genes (acr-2/AChR, unc-129/TFGb, glr-4/GluR, and unc-17/VAChT). The length of DNA elements included in each reporter is shown. Bottom: quantification of terminal identity gene reporters in WT and unc-3 (n3435) animals at three different developmental stages – L2, L4, and day 1 adults (N ⩾ 12). UNC-3 is required for both initiation and maintenance of all four terminal identity genes. ***p<0.001. (B) Top: snapshots of UNC-3 ChIP-Seq and input (negative control) signals at the cis-regulatory regions of 5 transcription factors (cfi-1/Arid3a, bnc-1/Bnc1, nhr-40, mab-9/Tbx20, and ceh-44/Cux1). The length of DNA elements included in each reporter is shown. Middle: representative images of WT L4 animals showing expression of the transgenic reporters in MNs. Scale bar, 20 μm. Bottom: quantification of transcription factor reporters in WT and unc-3 (n3435) animals at three different developmental stages – L2, L4, and young adults (day 1 or day 3) (N ⩾ 12). UNC-3 is required for maintenance, but not initiation of the 5 TFs. N.S.: not significant, *p<0.05, ***p<0.001. (C) Schematic summarizing the phenomenon of temporal modularity in UNC-3 function. The first module consists of terminal identity genes and TFs (Figure 3—figure supplement 2), which require UNC-3 for both initiation and maintenance of gene expression. The second module consists exclusively of TFs that require UNC-3 only for maintenance.
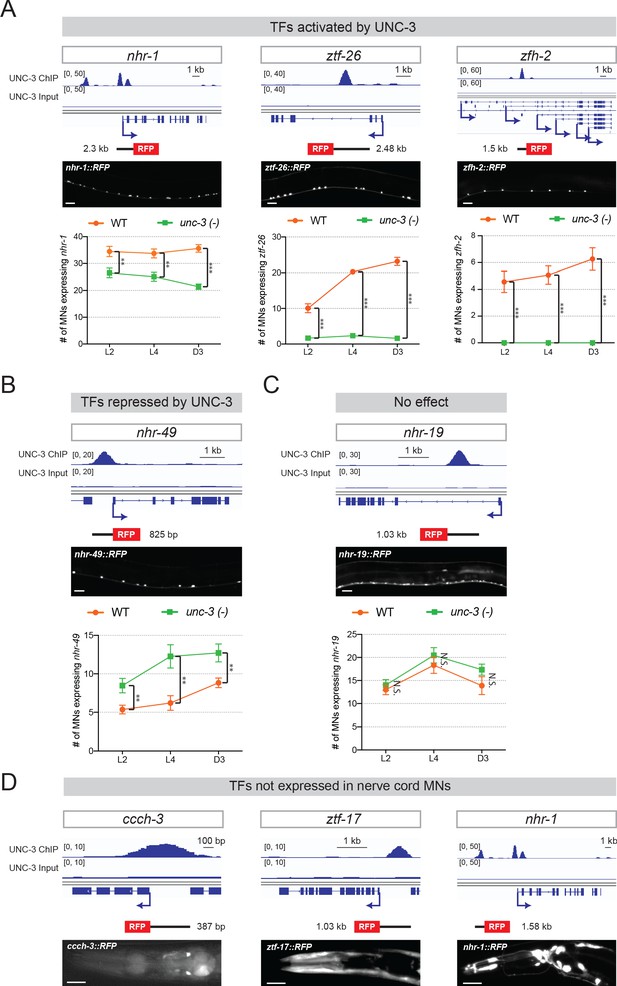
UNC-3 directly controls the expression of several TF reporters in MNs.
(A–C) Top: snapshots of UNC-3 ChIP-Seq and input (negative control) signal at the cis-regulatory regions of 5 TF-encoding genes (nhr-1, ztf-26, zfh-2, nhr-49, nhr-19). Transgenic RFP reporters contain the cis-regulatory regions bound by UNC-3, as well as flanking sequences. Middle: representative images of WT L4 animals showing expression of the transgenic reporters in nerve cord MNs. Scale bar, 20 μm. Bottom: quantification of TF reporters in WT and unc-3 (n3435) animals at three different developmental stages – L2, L4, and day 3 adult. nhr-1, ztf-26, and zfh-2 are activated by UNC-3 (A), while nhr-49 is repressed by UNC-3 (B) and nhr-19 does not appear to be controlled by UNC-3 (C). N ⩾ 15. N.S.: not significant, *p<0.05, **p<0.01, ***p<0.001. (D) Three TF reporters (ccch-3, ztf-17, nhr-1) are not expressed in nerve cord MNs, but show expression in head neurons. Top: snapshots of UNC-3 ChIP-Seq and input (negative control) signals at the cis-regulatory regions of ccch-3, ztf-17, and nhr-1. Transgenic RFP reporters contain the cis-regulatory regions bound by UNC-3. Bottom: representative images of WT L4 animals showing expression of the transgenic reporters in some unidentified neurons of the head. It is known that UNC-3 is expressed in some head neurons.
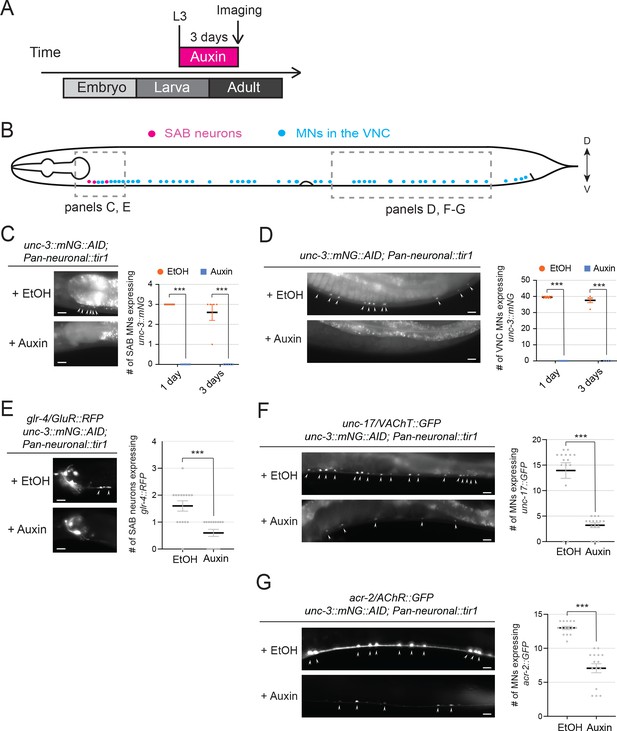
UNC-3 is required to maintain the expression of glr-4/GluR, unc-17/VAChT, and acr-2/AChR in cholinergic motor neurons.
(A) Diagram illustrating the design of UNC-3 knock-down experiments. Auxin (or EtOH, as control) was applied to L3 worms carrying unc-3::mNG::AID and a pan-neuronal tir-1 allele (with reporters of target genes to be tested). Examination of reporter expression was performed following 3 days of continuous auxin/EtOH administration. (B) Diagram illustrating the worm body with highlights of the locations of the SAB neurons (in the anterior) and the MNs along the VNC. Two boxed regions focus on the SAB neurons and the MNs in the posterior half of the worm body, which are shown in the representative images in the next panels. C-D: Demonstration of UNC-3 knock-down efficiency. Left: representative images showing expression of unc-3::mNG::AID after 3 days of auxin or EtOH administration in the SAB neurons (C) or MNs in the posterior VNC (D); Right: Quantification of the number of SAB neurons (C) or MNs in the posterior VNC (D) showing expression of unc-3::mNG::AID following auxin/EtOH treatment. To get a preliminary idea of the efficiency, nine worms from the auxin group and 10 worms from the EtOH group were checked one day after starting the treatment. To demonstrate the knock-down is persistent, five worms from each group were checked 3 days after starting the treatment. ***p<0.001. E-G: glr-4/GluR, unc-17/VAChT, and acr-2/AChR require UNC-3 for both initiation and maintenance of their expression. Left: representative images showing the expression of glr-4::RFP (E), unc-17::GFP (F), and acr-2::GFP (G) following 3 days or auxin/EtOH treatment. Downregulation of these reporters is apparent in terms of both the number of cells showing positive expression and the fluorescent intensity of the positive cells; Right: quantification of the number of SAB neurons expressing glr-4::RFP (E), and the number of MNs in the posterior VNC expressing unc-17::GFP (F) and acr-2::GFP from worms shown on the left (N = 15). ***p<0.001.
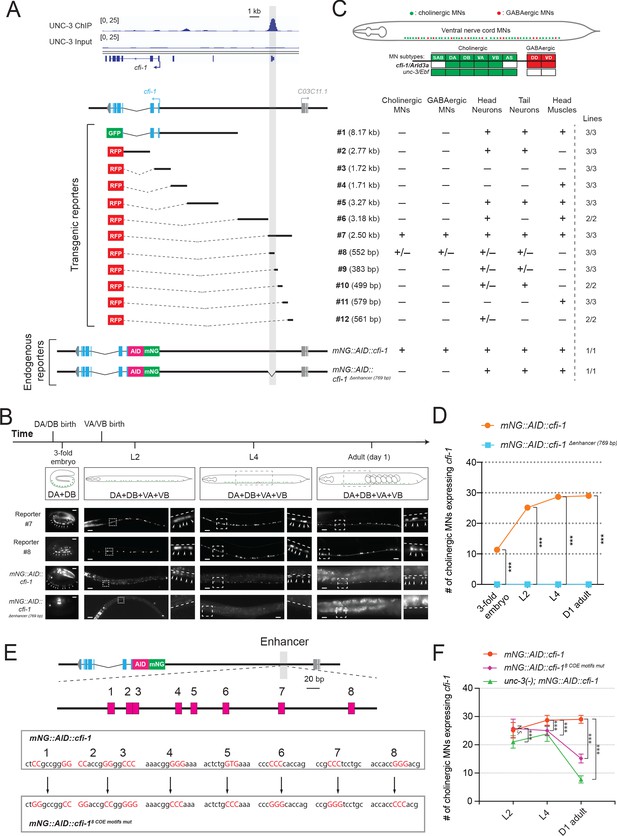
UNC-3 acts through a distal enhancer to maintain cfi-1 expression in cholinergic motor neurons.
(A) Top: Snapshots of UNC-3 ChIP-Seq and input (negative control) signals at the cis-regulatory region of cfi-1. The gray bar highlights an UNC-3 binding peak located ~12 kb upstream of the TSS of cfi-1 (−11,391 bp to −12,146 bp). Bottom: schematic showing the strategy of constructing cfi-1 reporters. Twelve transcriptional fusion reporters ([−1 bp to −8,170 bp], #2 [993 bp to 3,764 bp], #3 [547 bp to −1,173 bp], #4 [-1,164 bp to −2,875 bp], #5 [−2,865 bp to −6,141 bp], #6 [-8,162 bp to −11,346 bp], #7 [−11,329 bp to −13,824 bp], #8 [−11,329 bp to −11,881 bp], #9 [−11,851 bp to −12,234 bp], #10 [−12,223 bp to −12,722 bp], #11 [−12,705 bp to −13,284 bp], and #12 [-13,263 bp to −13,824 bp]) carry cis-regulatory regions fused to fluorescent reporters (GFP or RFP). The endogenous reporter alleles (mNG::AID::cfi-1 and mNG::AID::cfi-1 Δenhancer (769 bp)) have an in-frame fluorescent protein mNeonGreen (mNG) insertion immediately after the ATG of cfi-1. The enhancer KO allele mNG::AID::cfi-1 Δenhancer (769 bp) carries a 769 bp deletion (−11,329 bp to −12,097 bp). Table on the right summarizes the expression pattern of each reporter allele at L4 stage. N ⩾ 12. +: reporter expressed, –: reporter not expressed, +/–: reporter partially expressed in the respective neurons. Number of independent transgenic lines tested for each reporter is shown on the right. (B) Representative images showing the expression of reporter #7, reporter #8, mNG::AID::cfi-1, and mNG::AID::cfi-1 Δenhancer (769 bp) at specific life stages. Areas highlighted in dashed boxes are enlarged and presented on the right side of each picture. The onset of cfi-1 expression occurs at the 3-fold embryonic stage. mNG+ MNs are annotated with arrowheads. Scale bars, 5 μm (3-fold embryos); 20 μm (larvae and adults). (C) Schematic summarizing the expression pattern of cfi-1 and unc-3 in nerve cord MNs. (D) Quantification of the number of cholinergic MNs expressing endogenous cfi-1 (mNG::AID::cfi-1) in WT and animals carrying the enhancer deletion (mNG::AID::cfi-1 Δenhancer (769 bp)). Deletion of the enhancer element located ~12 kb upstream of the TSS of cfi-1 completely abolishes cfi-1 expression in MNs at all tested stages. A red fluorescent marker (ttr-39::mCherry) for GABAergic MNs was used to exclude these neurons from the quantification. Cholinergic MNs expressing cfi-1 were positive for mNG and negative for mCherry. (E) Bioinformatic analysis predicted 8 UNC-3 binding sites (COE motifs, shown as pink boxes) in the cfi-1 enhancer region, which displays UNC-3 binding (−11,391 bp to −12,146 bp). Using CRISPR/Cas9, these eight motifs were mutated by substituting duplets or triplets of nucleotides as shown below, thereby generating the strain cfi-1 (syb1856 [mNG::AID::cfi-18 COE motifs mut]). (F) Quantification of the number of cholinergic MNs expressing the endogenous cfi-1 reporter (mNG::AID::cfi-1) in WT and unc-3 (n3435) animals, as well as in animals with mutated COE motifs (mNG::AID::cfi-18 COE motifs mut) at L2, L4, and day 1 adult stages (N ⩾ 12). N.S.: not significant, ***p<0.001.
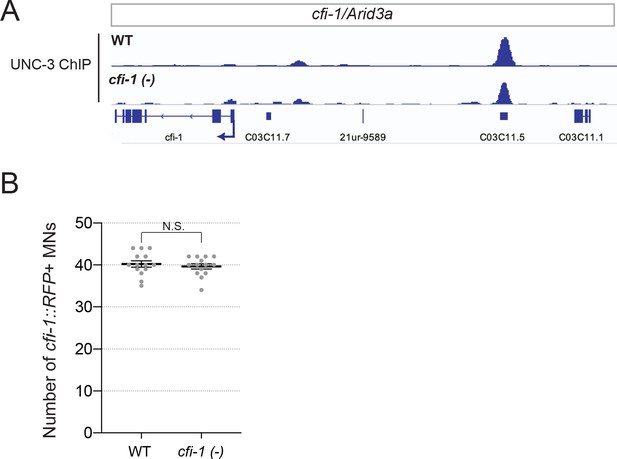
CFI-1 does not auto-regulate its expression in motor neurons.
(A) Snapshots of UNC-3 ChIP-Seq signal at the cfi-1 locus in WT and cfi-1 (ot786) animals. UNC-3 binds to the cfi-1 enhancer normally in cfi-1 (ot786) mutants. (B) Quantification of the number of MNs expressing the transcriptional fusion reporter #7 (cfi-12.5kb::RFP) in WT worms and cfi-1 (ot786) mutants (N = 14). No significant difference was detected between WT and cfi-1 (ot786), suggesting that the expression of reporter #7 (cfi-12.5kb::RFP) is not altered upon genetic removal of cfi-1. N.S.: not significant.
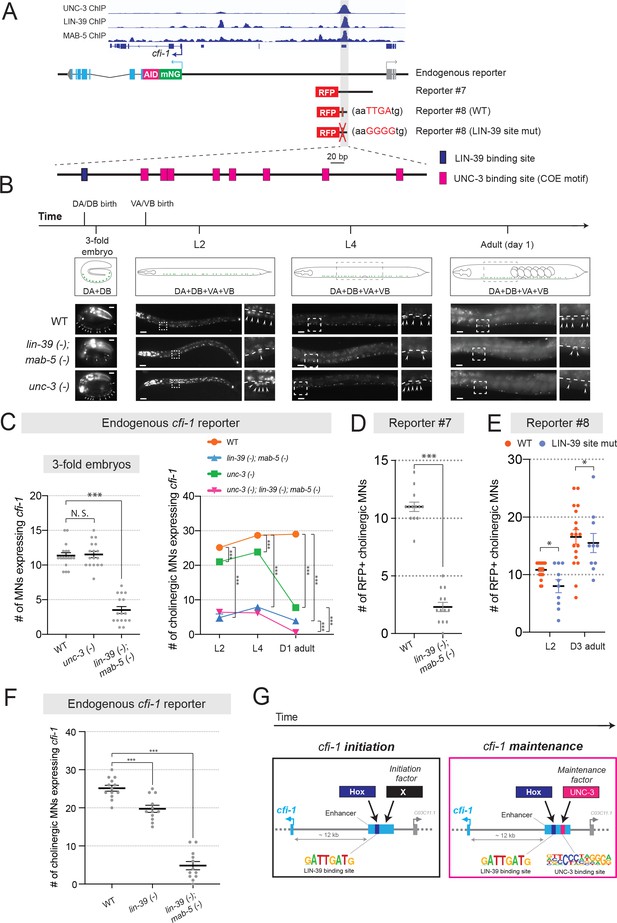
UNC-3 and Hox control cfi-1 expression in cholinergic MNs.
(A) A snapshot of UNC-3 (L2 stage), LIN-39 (L3 stage), and MAB-5 (L2 stage) ChIP-Seq signals at the cfi-1 locus. UNC-3, LIN-39, and MAB-5 bind to the same cfi-1 enhancer (highlighted in gray). Below: Schematics illustrating the reporters used in the rest of the figure. (B) Representative images showing the expression of the mNG::AID::cfi-1 in WT (same images shown in Figure 4B), unc-3 (n3435), and lin-39 (n1760); mab-5 (e1239) animals during 3-fold embryonic, L2, L4, and day 1 adult stages. cfi-1 is expressed in four cholinergic MN subtypes (DA, DB, VA, and VB). DA and DB are born embryonically, while VA and VB are born post-embryonically. Areas highlighted in dashed boxes are enlarged and presented on the right side of each picture. mNG+ MNs are annotated with arrowheads. Scale bars, 5 μm (3-fold embryos); 20 μm (larvae and adults). (C) Quantification of the number of cholinergic MNs expressing the endogenous cfi-1 reporter (mNG::AID::cfi-1) in WT animals, unc-3 (n3435) mutants, lin-39 (n1760); mab-5 (e1239) double mutants, and unc-3 (n3435); lin-39 (n1760); mab-5 (e1239) triple mutants during 3-fold embryonic, L2, L4, and day 1 adult stages (N ⩾ 12). N.S.: not significant, ***p<0.001. A red fluorescent marker (ttr-39::mCherry) for GABAergic MNs was used to exclude these neurons from the quantification. Cholinergic MNs expressing cfi-1 were positive for mNG and negative for mCherry. (D) Quantification of the expression of transgenic cfi-1 reporter #7 in WT and lin-39 (n1760); mab-5 (e1239) animals at L2 stage (N = 13). Reporter expression is strongly affected in lin-39 (-); mab-5 (-) double mutants. ***p<0.001. (E) Quantification of the WT transgenic reporter #8 and the same reporter with the LIN-39 binding site mutated (point mutations) at larval (L2) and adult (D3) stages (N ⩾ 13). *p<0.05. (F) Quantification of the expression of the mNG::AID::cfi-1 allele in WT animals, lin-39 (n1760) single mutants, and lin-39 (n1760); mab-5 (e1239) double mutants at the L2 stage (N ⩾ 12). While the number of cholinergic MNs with cfi-1 expression is mildly decreased in lin-39 single mutants, more severe effects are observed in double mutants. ***p<0.001. (G) Schematic summarizing the mechanisms underlying initiation and maintenance of cfi-1 expression in cholinergic MNs.
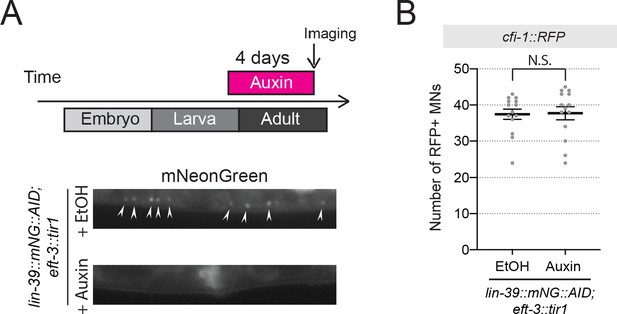
Auxin-inducible depletion of LIN-39 at larval stage 4 (L4) does not affect cfi-1 expression in nerve cord MNs.
(A) Representative images showing expression of LIN-39::mNG::AID after treatment with ethanol and auxin (negative control). LIN-39::mNG::AID is degraded and mNG fluorescent signal becomes undetectable in MNs, when worms are imaged after 4 days of auxin treatment. Arrowheads indicate MN nuclei in the nerve cord. (B) Knock-down of lin-39::mNG::AID starting at the L4 stage (onset of auxin treatment at L4) did not affect cfi-12.5kb::RFP expression in adult animals 4 days later (N ⩾ 13). N.S.: not significant. The number of MNs expressing cfi-12.5kb::RFP was quantified.
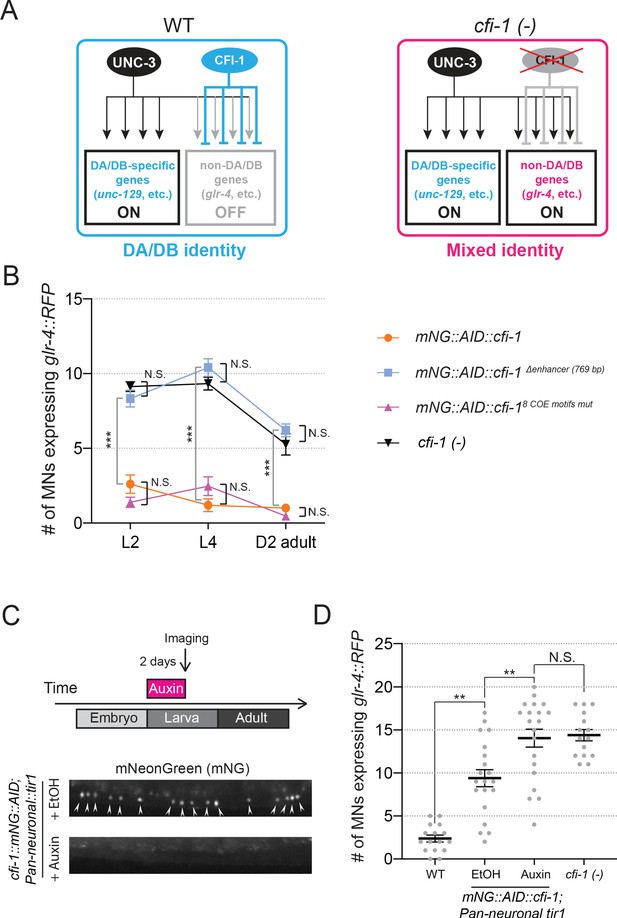
CFI-1 is required post-embryonically to maintain DA and DB neuronal identities.
(A) CFI-1 regulates DA and DB MN identity by repressing the glutamate receptor subunit glr-4/GluR and possibly other genes. In cfi-1 (-) mutants, glr-4/GluR becomes ectopically activated by UNC-3 in DA and DB MNs, and these neurons adopt a mixed molecular identity. (B) Quantification of the number of MNs expressing glr-4::RFP in mNG::AID::cfi-1 worms, worms carrying the cfi-1 enhancer deletion (mNG::AID::cfi-1 Δ enhancer (769 bp)), worms with mutated COE motifs (mNG::AID::cfi-18 COE motifs mut), as well as in cfi-1 null mutants at L2, L4, and day 2 adult stages (N = 15). N.S.: not significant, *** p < 0.001. The remaining expression of cfi-1 in ~ 15 MNs of mNG::AID::cfi-18 COE motifs mut animals (Figure 4F) is likely the reason for not observing glr-4/GluR ectopic expression in MNs of these animals. (C) Representative images showing expression of mNG::AID::CFI-1 after treatment with ethanol and auxin. Upon two days of continuous auxin treatment (onset of treatment at L1), mNG::AID::CFI-1 is degraded and mNG expression in MNs (arrowheads) becomes undetectable. (D) Quantification of MNs expressing glr-4::RFP was performed on L4 worms (2 days after the onset of auxin treatment). Some mild hypomorphic effects in the expression of glr-4::RFP was observed in the EtOH group (negative control), potentially due to mild reduction in CFI-1 levels triggered by TIR1 even in the absence of auxin. A significant increase in the number of MNs expressing glr-4::RFP was evident in auxin-treated worms in comparison to control animals. WT and cfi-1 (-) data are also provided for comparison. N ⩾ 20. N.S.: not significant, **p<0.01.
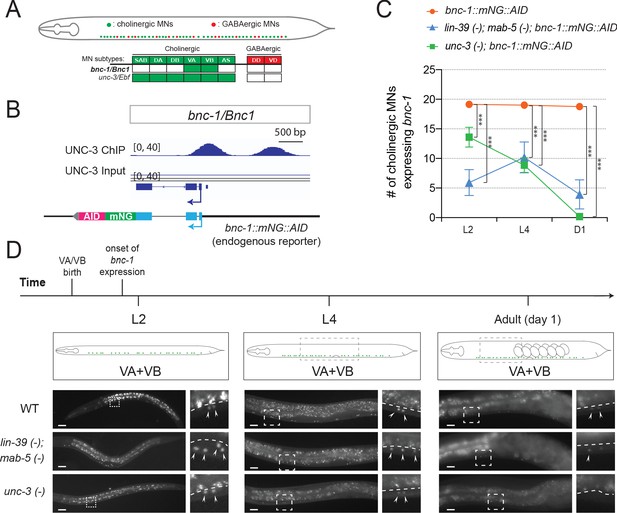
Hox proteins and UNC-3 control bnc-1 expression in VA and VB neurons.
(A) Schematic summarizing the expression of bnc-1/Bnc1 and unc-3/Ebf in MN subtypes of the C. elegans ventral nerve cord. (B) A snapshot of UNC-3 ChIP-Seq and input (negative control) signals at the cis-regulatory region of bnc-1. (C) Quantification of expression of the endogenous reporter bnc-1::mNG::AID in WT, unc-3 (n3435), and lin-39 (n1760); mab-5 (e1239) animals during L2, L4, and day 1 adult stages (N ⩾ 12). ***p<0.001. (D) Representative images of the endogenous reporter bnc-1::mNG::AID in WT, unc-3 (e151), and lin-39 (n1760); mab-5 (e1239) animals during L2, L4, and day 1 adult stages. bnc-1 is expressed in two cholinergic MN subtypes (VA and VB). Areas highlighted in dashed boxes are enlarged and presented on the right side of each picture. Arrowheads point to nuclei of VA and VB neurons that express the reporter. Above the white dashed line lies the intestine, which is autofluorescent in the green channel. Scale bar, 20 μm.
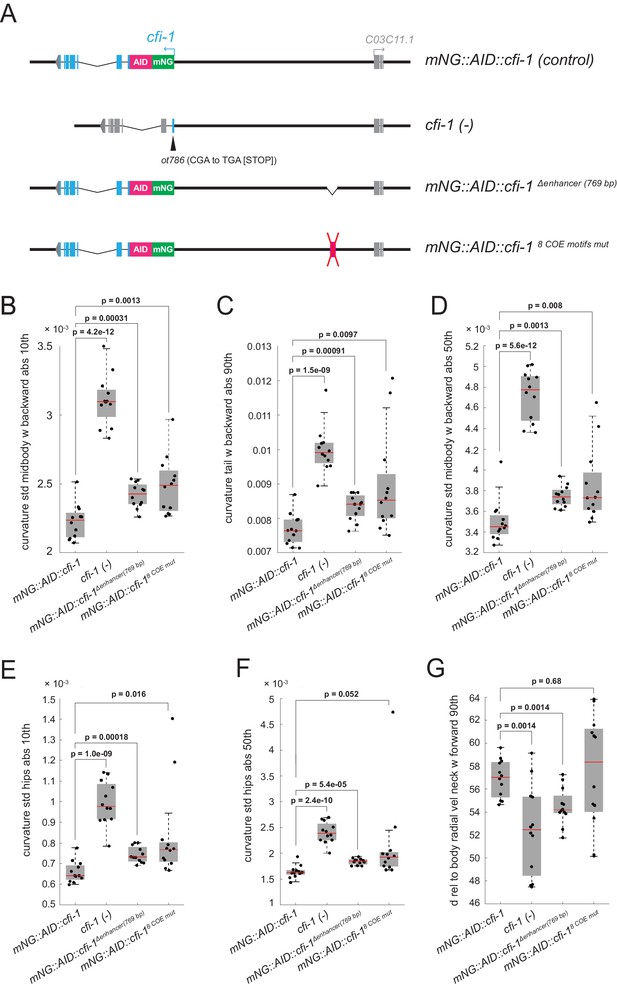
Minimal disruption of temporal modularity in UNC-3 function leads to locomotion defects.
(A) Schematics illustrating four cfi-1 alleles tested for behavioral analysis. (B–G) Examples of six locomotion features significantly disrupted in animals carrying a putative null (ot786) allele for cfi-1. Locomotion analysis was performed on day 2 adult worms. Animals lacking cfi-1 expression (initiation and maintenance) specifically in MNs (mNG::AID::cfi-1 Δenhancer (769 bp)) and animals unable to maintain cfi-1 expression in cholinergic MNs (mNG::AID::cfi-18 COE motifs mut) display locomotion defects when compared to control mNG::AID::cfi-1 animals. As expected, these defects are milder when compared to animals carrying the cfi-1 (ot786) allele. Panels B-F show locomotion features related to body curvature, whereas panel G shows radial velocity of the neck. A detailed description of each locomotion feature is provided below. (B) curvature_std_mid-body_w_backward_abs_10th: 10th percentile of the absolute value of the standard deviation of the curvature of the mid-body, while worm is moving backwards. (C) curvature_tail_w_backward_abs_90th: 90th percentile of the absolute value of the curvature of the tail, while worm is moving backwards. (D) curvature_std_mid-body_w_backward_50th: 50th percentile of the standard deviation of the curvature of the mid-body, while worm is moving backwards. (E) curvature_std_hips_abs_10th: 10th percentile of the absolute value of the standard deviation of the curvature of the hips. (F) curvature_std_hips_abs_50th: 50th percentile of the absolute value of the standard deviation of the curvature of the hips. (G) d_rel_to_body_radial_vel_neck_w_forward_90th: 90th percentile of the derivative of radial velocity of the neck relative to the centroid of the mid-body points, while worm is moving forwards.
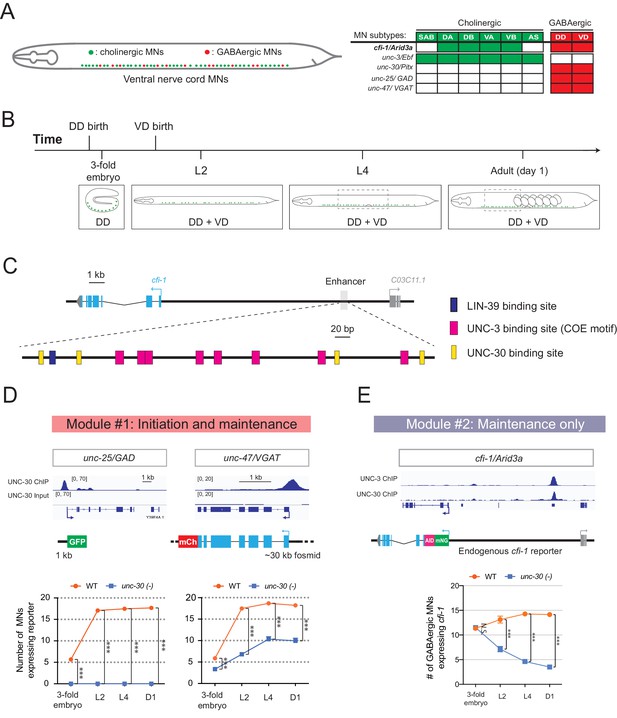
Temporal modularity in UNC-30/Pitx function in GABAergic MNs.
(A) Schematic summarizing the expression of cfi-1/Arid3a, unc-3/Ebf, unc-30/Pitx, unc-25/GAD, and unc-47/VGAT in MN subtypes of the C. elegans ventral nerve cord. (B) Schematic showing time of birth and cell body position of GABAergic nerve cord MNs. DD neurons are born embryonically. VD neurons are born post-embryonically. (C) Bioinformatic analyses predict 4 UNC-30 binding sites (yellow boxes) in the cfi-1 enhancer. The location of UNC-3 and LIN-39 binding sites are also shown. (D) Top: snapshots of UNC-30 ChIP-Seq and input (negative control) signals at the cis-regulatory regions of 2 GABAergic terminal identity genes (unc-25/GAD, unc-47/VGAT). Bottom: quantification of the expression of transgenic reporters in WT and unc-30 (e191) animals at four different developmental stages – 3-fold embryo, L2, L4, and day 1 adults (N = 15). UNC-30 is required for both initiation and maintenance of unc-25/GAD and unc-47/VGAT. ***p<0.001. (E) Top: a snapshot of UNC-3 ChIP-Seq and UNC-30 ChIP-Seq signals at the cfi-1 locus. Bottom: quantification of the number of MNs expressing the endogenous reporter mNG::AID::cfi-1 in WT and unc-30 (e191) animals. All cfi-1-expressing MNs in the ventral cord (cholinergic and GABAergic MNs) were counted in 3-fold embryos due to a lack of a specific marker that labels GABAergic MNs in embryos. Expression of cfi-1 specifically in GABA neurons was quantified at L2, L4, and day 1 adult stages (N ⩾ 12). At those stages, cholinergic MNs were identified based on a fluorescent marker (cho-1::mChOpti), which are ruled out during scoring. GABAergic MNs were scored positive for cfi-1 expression when the mNG::AID::cfi-1 (green) expression co-localized with ttr-39::mCherry (red). N.S.: not significant, ***p<0.001.
Tables
Summary of cis-regulatory analysis to identify novel transcription factors controlled by UNC-3.
Transgenic reporter (RFP) animals for each TF were generated and examined for neuronal expression. GFP markers for cholinergic and GABAergic MNs were used to define TF reporter expression. TF reporters expressed in MNs were further tested for UNC-3 dependency. Eighteen reporters were generated that correspond to 16 TFs (Two reporters were generated for nhr-1 and nhr-40 because two distinct UNC-3 ChIP-Seq peaks were found in the cis-regulatory region of these genes). Not applicable (N/A).
| Number of reporters | Gene | TF family | DNA region included relative to ATG | unc-3 dependence | RFP expression | Expression in cholinergic MNs | Expression in GABAergic MNs | # of lines |
|---|---|---|---|---|---|---|---|---|
| 1 | nhr-1 (second peak) | Nuclear hormone receptor | −838 bp to +1,535 bp | Positively regulated | VNC, Head and Tail Neurons | YES | YES | 2 |
| 2 | nhr-40 (second peak) | Nuclear hormone receptor | +4,356 bp to +5,525 bp | Positively regulated | VNC, Head and Tail Neurons | YES | YES | 2 |
| 3 | mab-9 | T-box | −5,561 bp to −3,773 bp | Positively regulated | VNC, Head and Tail Neurons | YES | YES | 2 |
| 4 | ztf-26 | Zinc finger | +910 bp to 3,394 bp | Positively regulated | VNC and Head Neurons | YES | NO | 2 |
| 5 | ceh-44 | C. elegans homeobox | +1,605 bp to 3,111 bp | Positively regulated | VNC, Head and Tail Neurons | YES | Not determined | 2 |
| 6 | zfh-2 | Zinc finger | +12,048 bp to +13,549 bp | Positively regulated | VNC, Head and Tail Neurons | YES | NO | 2 |
| 7 | cfi-1 | AT-rich interaction domain | −13,824 bp to −11,329 bp | Positively regulated | VNC, Head and Tail Neurons | YES | YES | 2 |
| 8 | bnc-1 | Zinc finger | −1,800 bp to −1 bp | Positively regulated | VNC and Tail Neurons | YES | NO | 2 |
| 9 | nhr-19 | Nuclear hormone receptor | +1,261 bp to +2,294 bp | NO | VNC, Head and Tail Neurons | YES | YES | 2 |
| 10 | nhr-40 (first peak) | Nuclear hormone receptor | +919 bp to +1,839 bp | NO | VNC, Head and Tail Neurons | YES | YES | 2 |
| 11 | nhr-49 | Nuclear hormone receptor | −750 bp to 75 bp | Negatively regulated | VNC, Head and Tail Neurons | NO | YES | 2 |
| 12 | nhr-1 (first peak) | Nuclear hormone receptor | −7,452 bp to −5,871 bp | Not determined | Head and Tail Neurons | N/A | N/A | 2 |
| 13 | ccch-3 | Zinc finger | −385 bp to −1 bp | Not determined | Head Neurons | N/A | N/A | 2 |
| 14 | ztf-17 | Zinc finger | −1,031 bp to +3 bp | Not determined | Head Neurons, Muscles, and Intestine | N/A | N/A | 2 |
| 15 | nhr-47 | Nuclear hormone receptor | −675 bp to +171 bp | N/A | No RFP expression | N/A | N/A | 2 |
| 16 | unc-55 | Nuclear receptor | −2,105 bp to −1,327 bp | N/A | No RFP expression | N/A | N/A | 2 |
| 17 | ztf-13 | Zinc finger | −1,392 bp to −1 bp | N/A | No RFP expression | N/A | N/A | 2 |
| 18 | ztf-14 | Zinc finger | −2,087 bp to −531 bp | N/A | No RFP expression | N/A | N/A | 2 |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Caenorhabditis elegans) | unc-3 | Wormbase | WBGene00006743 | |
| Gene (C. elegans) | cfi-1 | Wormbase | WBGene00000476 | |
| Gene (C. elegans) | unc-30 | Wormbase | WBGene00006766 | |
| Gene (C. elegans) | lin-39 | Wormbase | WBGene00003024 | |
| Gene (C. elegans) | mab-5 | Wormbase | WBGene00003102 | |
| Strain, strain background (C. elegans) | unc-3 (n3435) | Bob Horvitz (MIT, Cambridge MA) | MT10785 | Null Allele: deletion |
| Strain, strain background (C. elegans) | unc-3 (e151) | Caenorhabditis Genetics Center | CB151 | Allele: substitution |
| Strain, strain background (C. elegans) | lin-39 (n1760) mab-5 (e1239)/ht2 III; lgIs58 [[gcy-32::gfp]] V | Caenorhabditis Genetics Center | LE4023 | |
| Strain, strain background (C. elegans) | lin-39 (n1760)/dpy-17 (e164) unc-32 (e189) III | Caenorhabditis Genetics Center | MT4009 | Null Allele: substitution |
| Strain, strain background (C. elegans) | unc-30 (e191) | Caenorhabditis Genetics Center | CB845 | Allele: substitution |
| Strain, strain background (C. elegans) | cfi-1 (ot786) | This paper. Kratsios lab (University of Chicago, IL, USA). | KRA464 | Null Allele: substitution - R to STOP (39); Strain was 3x backcrossed |
| Strain, strain background (C. elegans) | unc-3 (ot839 [unc-3::gfp]) X | Oliver Hobert (Columbia University, New York NY) | OH13990 | CRISPR-generated allele |
| Strain, strain background (C. elegans) | cfi-1 (kas16 [mNG::AID::cfi-1]) I | This paper. Kratsios lab (University of Chicago, IL, USA). | KRA345 | CRISPR-generated allele; See Materials and methods - Targeted genome editing |
| Strain, strain background (C. elegans) | bnc-1 (ot845 [bnc-1::mNGAID]) V | Kerk et al., 2017 | OH14070 | CRISPR-generated allele |
| Strain, strain background (C. elegans) | lin-39 (kas9 [lin-39::mNG::AID]) III | Feng et al., 2020 Jan 3. doi:10.7554/eLife.50065 | KRA467 | CRISPR-generated allele |
| Strain, strain background (C. elegans) | unc-3 (ot837 [unc-3::mNG::AID]) X; ieSi57 [Peft-3::TIR1:: mRuby::unc-54 3’ UTR, cb-unc-119(+)] II | This paper. Kratsios lab (University of Chicago, IL, USA). | KRA376 | CRISPR-generated allele |
| Strain, strain background (C. elegans) | cfi-1 (syb1812 [A4e_enhancer deletion −13,264–12,495 from cfi-1 ATG in kas16] kas16[cfi-1::mNG::AID]) I | This paper. Kratsios lab (University of Chicago, IL, USA). | PHX1812 | See Materials and methods - Targeted genome editing, and Figure 4A legend |
| Strain, strain background (C. elegans) | cfi-1 (syb1856 [8 COE motifs mut in A4e] kas16 [cfi-1::mNG::AID]) I | This paper. Kratsios lab (University of Chicago, IL, USA). | PHX1856 | See Materials and methods - Targeted genome editing, and Figure 4E legend |
| Strain, strain background (C. elegans) | otIs544 [cho-1(fosmid):: SL2::mCherry::H2B + pha-1(+)] | Oliver Hobert (Columbia University, New York NY) | OH13646 | |
| Strain, strain background (C. elegans) | juIs14 [acr-2p::GFP + lin-15(+)] IV. | Caenorhabditis Genetics Center | CZ631 | |
| Strain, strain background (C. elegans) | otIs426 [Punc-17_1 kb:: YFP, Pmyo-2::GFP] | Oliver Hobert (Columbia University, New York NY) | OH11454 | |
| Strain, strain background (C. elegans) | evIs82b [unc-129:: GFP + dpy-20(+)] IV. | Oliver Hobert (Columbia University, New York NY) | OH1894 | |
| Strain, strain background (C. elegans) | otIs476 [glr-4 prom:: TagRFP + pha-1(+)] | Kerk et al., 2017 | OH12052 | |
| Strain, strain background (C. elegans) | otIs477 [glr-4 prom:: TagRFP + pha-1(+)] | Oliver Hobert (Columbia University, New York NY) | OH12053 | |
| Strain, strain background (C. elegans) | vsIs48 [unc-17::GFP] | Caenorhabditis Genetics Center | LX929 | |
| Genetic reagent (C. elegans) | Pcfi-1_8.17 kb::GFP | Shaham and Bargmann, 2002 | nsEx37 | See Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_−11,329 bp −13,824 bp::RFP | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx12 kasEx182 kasEx183 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_2.77 kb(993 bp to 3,764 bp)::rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx184 kasEx185 kasEx186 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_1.72 kb (547 bp to −1,173 bp)::rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx187 kasEx188 kasEx189 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_1.71 kb (−1,164 bp to −2,875 bp): :rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx190 kasEx191 kasEx192 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_3.27 kb(−2,865 bp to −6,141 bp)::rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx193 kasEx194 kasEx195 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_3.18 kb(−8,162 bp to −11,346 bp): :rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx196 kasEx197 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_552 bp(−11,329 bp to −11,881 bp): :rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx198 kasEx199 kasEx200 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_383 bp(−11,851 bp to −12,234 bp): :rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx201 kasEx202 kasEx203 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_499 bp(−12,223 bp to −12,722 bp)::rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx204 kasEx205 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_579 bp(−12,705 bp to −13,284 bp)::rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx206 kasEx207 kasEx208 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | cfi-1_561 bp(−13,263 bp to −13,824 bp)::rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx209 kasEx210 | See Materials and methods, and Figure 4A legend |
| Genetic reagent (C. elegans) | nhr-1 (peak 1,–7455 to −5853)::RFP:: unc-54 3'UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx211 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | nhr-1 (peak 2,–893 to +1535)::RFP:: unc-54 3'UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx212 kasEx213 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | nhr-40 (peak 1, +938 to +1846)::RFP:: unc-54 3'UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx214 kasEx215 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | nhr-40 (peak 2, +4360 to +5522)::RFP:: unc-54 3'UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx216 kasEx217 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | zfh-2::rfp (12,048 bp to 13,549 bp) | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx218 kasEx219 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | nhr-49 (−803 to +58):: RFP::unc-54 3'UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx220 kasEx221 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | nhr-19 (+1250 to +2302)::RFP::unc-54 3'UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx222 kasEx223 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | ccch-3 (Peak 1, 0 to −387)::RFP:: unc-54 3’UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx224 kasEx225 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | ztf-17 (Peak 1, 0 to −1034)::RFP:: unc-54 3’UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx226 kasEx227 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | ztf-26 (Peak 1,–1341 to +753)::RFP:: unc-54 3’UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx228 kasEx229 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | ztf-26 (Peak 2, +909 to +3394)::RFP:: unc-54 3’UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx230 kasEx231 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | mab-9 (−5569 to −3768)::RFP::unc-54 3'UTR | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx232 kasEx233 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Genetic reagent (C. elegans) | ceh-44 (1,605 bp to 3,111 bp): :rfp | This paper. Kratsios lab (University of Chicago, IL, USA). | kasEx234 kasEx235 | See Materials and methods - Generation of transgenic animals carrying transcriptional fusion reporters |
| Antibody | anti-GFP (Rabbit polyclonal) | Abcam | Ab290 | Dilution: 1:1000; RRID:AB_303395 |
| Commercial assay or kit | Dynabeads Protein G | Invitrogen | 1004D | |
| Commercial assay or kit | Gibson Assembly Cloning Kit | NEB | #5510S | |
| Commercial assay or kit | QIAquick PCR Purification Kit | QIAGEN | #28104 | |
| Commercial assay or kit | Ampure XP beads | Beckman Coulter Life Sciences | A63881 | |
| Chemical compound, drug | cOmplete ULTRA Protease Inhibitor Cocktail | Roche | #05892970001 | |
| Chemical compound, drug | Auxin (indole-3-acetic acid) | Alfa Aesar | #10196875 | |
| Software, algorithm | ZEN | ZEISS | Version 2.3.69.1000, Blue edition | RRID:SCR_013672 |
| Software, algorithm | Image J | Image J | Version 1.52i | RRID:SCR_003070 |
| Software, algorithm | Adobe Photoshop CS6 | Adobe | Version 13.0 × 64 | |
| Software, algorithm | Adobe Illustrator CS6 | Adobe | Version 16.0.0 × 64 |
Additional files
-
Supplementary file 1
Table summarizing UNC-3 ChIP-Seq signal distribution at the cis-regulatory regions of previously identified UNC-3 targets.
- https://cdn.elifesciences.org/articles/59464/elife-59464-supp1-v1.docx
-
Supplementary file 2
Table summarizing the results of protein class ontology analysis on novel target genes of UNC-3.
In total, 1425 genes are classified into 25 protein classes.
- https://cdn.elifesciences.org/articles/59464/elife-59464-supp2-v1.xlsx
-
Supplementary file 3
UNC-3 binds to the cis-regulatory region of numerous genes expressed in cholinergic MNs.
- https://cdn.elifesciences.org/articles/59464/elife-59464-supp3-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59464/elife-59464-transrepform-v1.docx





