Laminar-specific cortico-cortical loops in mouse visual cortex
Figures
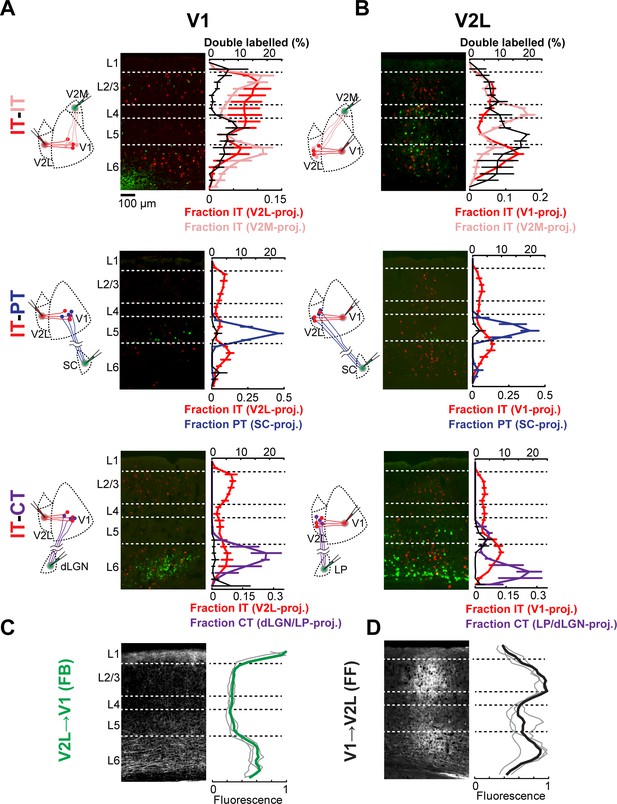
Cortical neurons projecting to different areas are intermingled and accessible to feedforward (FF) and feedback (FB) axons.
(A) Distribution of retrogradely labeled projection neurons in primary visual cortex (V1) after injection of a red-fluorescent tracer in lateral visual areas (V2L) and an infrared-fluorescent tracer in either medial visual areas (V2M), superior colliculus (SC), or visual thalamus. Left, experimental configuration; center, representative fluorescent histological section, with infrared fluorescence shown in green; right, colored traces show the mean laminar distribution of the different projection neurons binned in 50 μm increments, while the black trace shows the percentage of retrogradely labeled neurons that are double-labeled at each depth (n = 3 animals per group). Error bars, standard error; dashed lines, approximate layer boundaries. (B) Distribution of retrogradely labeled projection neurons in V2L after injection of a red-fluorescent tracer in V1 and an infrared-fluorescent tracer in either V2M, SC, or visual thalamus (n = 3 animals per group). (C) Distribution of anterogradely labeled V2L FB axons in V1. Left, representative fluorescent histological section; right, axonal fluorescence across cortical depth binned in 50 μm increments. Individual mice, thin gray traces; average, thick green trace (n = 3 animals). (D) Distribution of anterogradely labeled V1 FF axons in V2L (n = 3 animals).
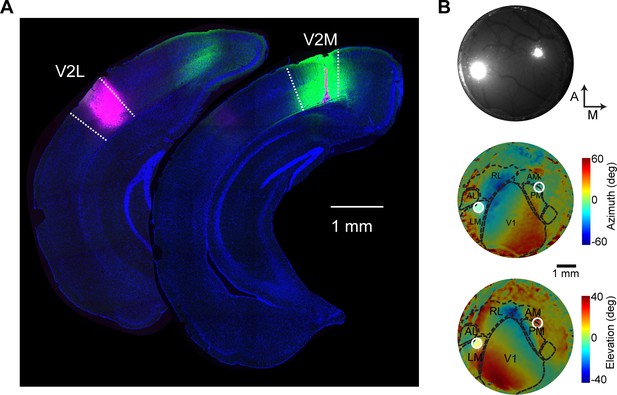
Histological and in vivo verification of lateral visual (V2L) and medial visual (V2M) area injection sites.
(A) Example coronal sections of a brain injected in V2L and V2M. (B) Example injections in V2L and V2M visualized in vivo. Top, image of the brain vasculature and injection sites. Middle, injection sites and area borders overlaid on the azimuth map determined by intrinsic signal imaging. Bottom, elevation map. White circles, injection sites.
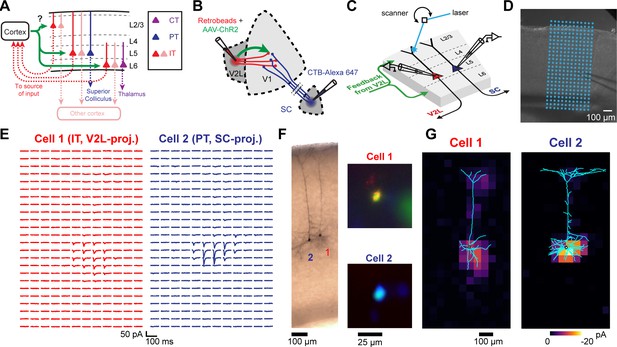
Measuring the strength and dendritic distribution of cortico-cortical (CC) inputs to different projection neurons.
(A) We probed the strength of CC inputs to looped and non-looped neurons in different cortical layers. (B) Example experiment configuration. Retrograde tracers are injected in two areas to label different projection neurons. One cortical area is also co-injected with adeno-associated virus (AAV)-channelrhodopsin-2 (ChR2) to express ChR2 in a specific CC projection. (C) Example of subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) experiment. Pairs of neighboring retrogradely labeled neurons in the same cortical layer were sequentially recorded. During each recording, a laser beam was scanned over the dendrites of the cell at different locations in a grid pattern. (D) Brightfield image of an acute coronal cortical slice showing the recording pipette and photostimulation grid. (E) Excitatory postsynaptic currents (EPSCs) recorded from a pair of neighboring L5 neurons, evoked by photostimulating ChR2+ V2L→V1 FB terminals on a grid. (F) Left, dendritic morphology staining of the recorded pair. Right, identity of the recorded projection neuron was confirmed by fluorescence in the soma of both a retrograde tracer and a different-colored dye introduced from the internal patch pipette solution. (G) sCRACM maps of the recorded pair overlaid on their reconstructed dendrites. Responsive locations are color-coded to represent mean amplitude.
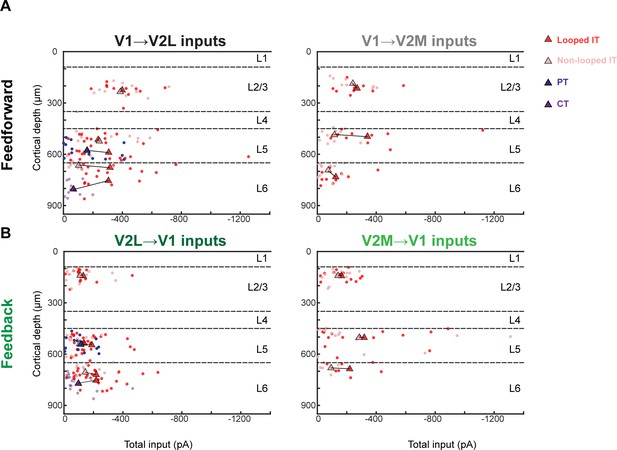
Total input vs. laminar depth across different projections and projection neuron classes.
(A) Total subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) input per neuron as a function of cortical depth for both feedforward (FF) projections. Circles, individual cells. Triangles, mean values per projection class for each experiment. Averages from paired data are joined by a line. Color indicates projection class. (B) Total sCRACM input per neuron as a function of cortical depth for both feedback (FB) projections.

Analysis of the incidence of retrograde infection of projection neurons by adeno-associated viruses (AAVs).
(A) Example of subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) traces from individual neurons (data from L2/3 looped intratelencephalic [IT] neurons). Each trace corresponds to the average excitatory postsynaptic current (EPSC) in the location eliciting the largest amplitude. Blue tick, laser pulse. The arrowhead indicates a single neuron (trace in blue) in which the laser pulse evoked an early-onset EPSC, suggestive of a non-synaptic response. Ten neurons with early-onset EPSCs were detected in the entire dataset and removed from further analysis. (B) Anti-green fluorescent protein (GFP) immunostained section of primary visual cortex (V1) showing fluorescent medial visual area (V2M) axons in an animal injected with AAV2/1-CAG-ChR2-Venus. (C) Higher magnification image of a region in (B). The arrow indicates an example of a retrogradely infected neuron in V1. (D) Configuration of experiment comparing strength of V2M feedback (FB) input to pairs of L6 looped and non-looped IT neurons in V1 using AAV5-CaMKIIa-hChR2(H134R)-EYFP. (E) sCRACM traces from 11 looped IT neurons recorded in L6 from the experiment in (D). (F) Left, paired comparisons of total FB input to looped vs. non-looped IT neurons from the experiment in (D). Inset traces represent group averages for each projection class. Blue tick, light pulse. Right, sCRACM Response Index (SRI) of the same data. *, p=0.0116.
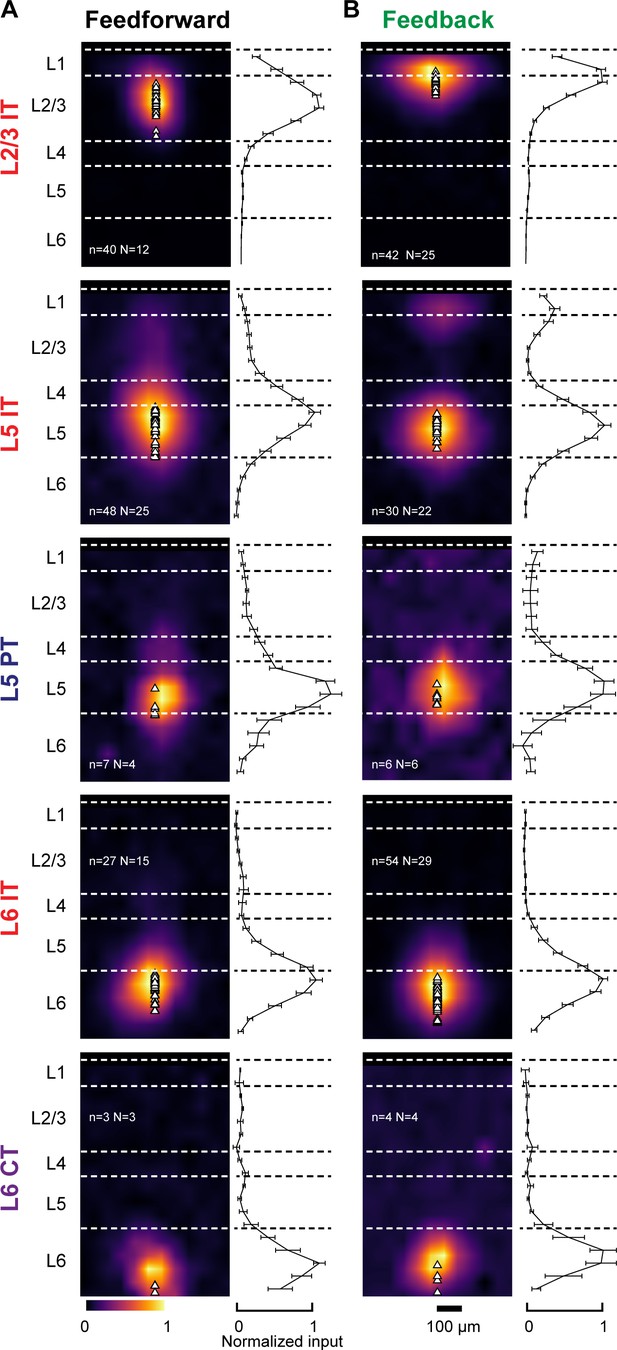
Dendritic distribution of feedforward (FF) and feedback (FB) inputs to different projection neuron classes.
(A) Left, group averages of subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) maps aligned by pia position showing primary visual cortex (V1) FF input to the different cell types (combining V1→V2L and V1→V2M inputs in the case of intratelencephalic [IT] neurons). Triangles, soma position. Right, vertical profiles of input strength. Error bars, s.e.m.; n, number of neurons; N, number of mice. (B) Group averages and vertical profiles of sCRACM maps showing FB input to the different cell types in V1 (combining V2L→V1 and V2M→V1 inputs in the case of IT neurons).

Soma-aligned dendritic distribution of feedforward (FF) and feedback (FB) inputs to different projection neuron classes.
(A) Left, group averages of subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) maps aligned by soma position showing primary visual cortex (V1) FF input to the different cell types (combining V1→V2L and V1→V2M inputs in the case of intratelencephalic [IT] neurons). Triangles, soma position. Right, vertical profiles of the mean distribution of inputs as a function of distance to soma. Error bars, s.e.m.; n, number of neurons; N, number of mice. (B) Group averages and vertical profiles of soma-aligned sCRACM maps showing FB input to the different cell types in V1 (combining V2L→V1 and V2M→V1 inputs in the case of IT neurons).
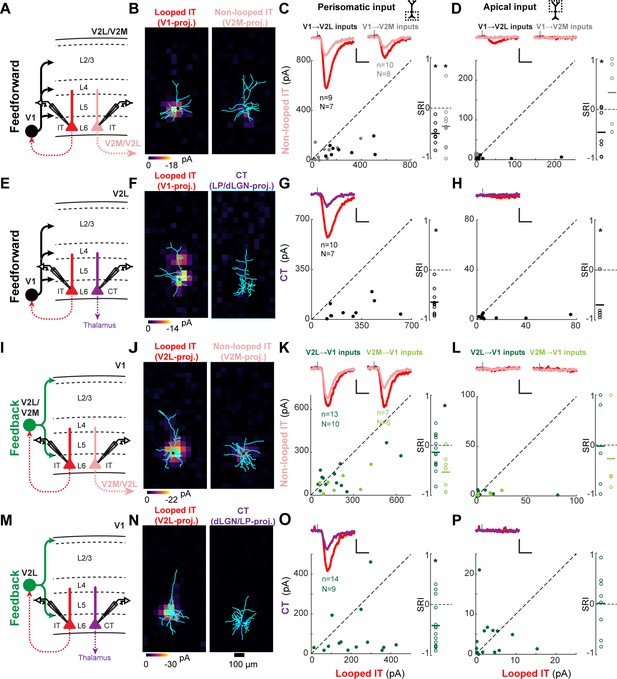
Most feedforward (FF) and feedback (FB) inputs are stronger in looped intratelencephalic (IT) neurons than in neighboring non-looped IT or corticothalamic (CT) neurons in L6.
(A) Configuration of experiments comparing strength of primary visual cortex (V1) FF input to pairs of L6 looped and non-looped IT neurons in lateral visual area (V2L) or medial visual area (V2M). (B) Example pair of subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) maps overlaid on reconstructed dendrites showing monosynaptic V1 FF inputs to a looped IT neuron (left) and an adjacent non-looped IT neuron (right) recorded in V2L. (C) Left, paired comparisons of perisomatic FF input to looped vs. non-looped IT neurons (n, number of cell pairs; N, number of mice); black dots, V1→V2L inputs; gray dots, V1→V2M inputs. Traces were generated by averaging the mean perisomatic excitatory postsynaptic current (EPSC) of each neuron across all neurons in the same projection class. Colors correspond to (A). Blue tick, laser pulse. Scale bars in all panels, 2 pA and 20 ms. Right, sCRACM Response Index (SRI) of the same data. Number of cell pairs and animals are the same as in the left plot unless otherwise specified. Horizontal line, mean. *, p<0.05, see text for exact value. (D) Same as C for apical inputs (SRI: V1→V2L, n = 7, N = 6; V1→V2M, n = 7, N = 6). (E) Configuration of experiment comparing strength of V1 FF input to pairs of L6 looped IT and CT neurons in V2L. (F) Example pair of sCRACM maps overlaid on reconstructed dendrites showing monosynaptic V1 FF inputs to a looped IT neuron (left) and an adjacent CT neuron (right) recorded in V2L. (G) Paired comparisons and SRI of perisomatic FF input to looped IT vs. CT neurons. (H) Paired comparisons and SRI (n = 5, N = 5) of apical FF input to looped IT vs. CT neurons. (I) Configuration of experiments comparing strength of V2L or V2M FB input to pairs of L6 looped and non-looped IT neurons in V1. (J) Example pair of sCRACM maps overlaid on reconstructed dendrites showing monosynaptic V2L FB inputs to a looped IT neuron (left) and an adjacent non-looped IT neuron (right) recorded in V1. (K) Paired comparisons and SRI of perisomatic FB input to looped vs. non-looped IT neurons. Dark green dots, V2L→V1 inputs; light green dots, V2M→V1 inputs. (L) Paired comparisons and SRI (V2L→V1, n = 5, N = 5; V2M→V1, n = 4, N = 4) of apical FB input to looped vs. non-looped IT neurons. (M) Configuration of experiment comparing strength of V2L FB input to pairs of L6 looped IT and CT neurons in V1. (N) Example pair of sCRACM maps overlaid on reconstructed dendrites showing monosynaptic V2L FB inputs to a looped IT neuron (left) and an adjacent CT neuron (right) recorded in V1. (O) Paired comparisons and SRI of perisomatic FB input to looped IT vs. CT neurons. (P) Paired comparisons and SRI (n = 8, N = 7) of apical FB input to looped IT vs. CT neurons.

Total subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) input to L6 neurons.
(A) Configuration of experiments comparing strength of primary visual cortex (V1) feedforward (FF) input to pairs of L6 looped and non-looped intratelencephalic (IT) neurons in lateral visual areas (V2L) or medial visual areas (V2M). (B) Left, paired comparisons of total FF input to looped vs. non-looped IT neurons. Black dots, V1→V2L inputs; gray dots, V1→V2M inputs. Traces were generated by averaging the mean excitatory postsynaptic currents (EPSC) of each neuron across all neurons in the same projection class. Blue tick, laser pulse. Scale bars in all panels, 2 pA and 20 ms. Right, sCRACM Response Index (SRI) of the same data. Number of cell pairs and animals are the same as in the left plot. Horizontal line, mean. *, p=0.0002. (C) Configuration of experiment comparing strength of V1 FF input to pairs of L6 looped IT and corticothalamic (CT) neurons in V2L. (D) Paired comparisons and SRI of total FF input to looped IT vs. CT neurons. *, p=1.2×10−5. (E) Configuration of experiments comparing strength of V2L or V2M FB input to pairs of L6 looped and non-looped IT neurons in V1. (F) Paired comparisons and SRI of total FB input to looped vs. non-looped IT neurons. Dark green, V2L→V1 inputs; light green, V2M→V1 inputs. *, p=0.0311. (G) Configuration of experiment comparing strength of V2L FB input to pairs of L6 looped IT and CT neurons in V1. (H) Paired comparisons and SRI of total FB input to looped IT vs. CT neurons. * p=0.0032.
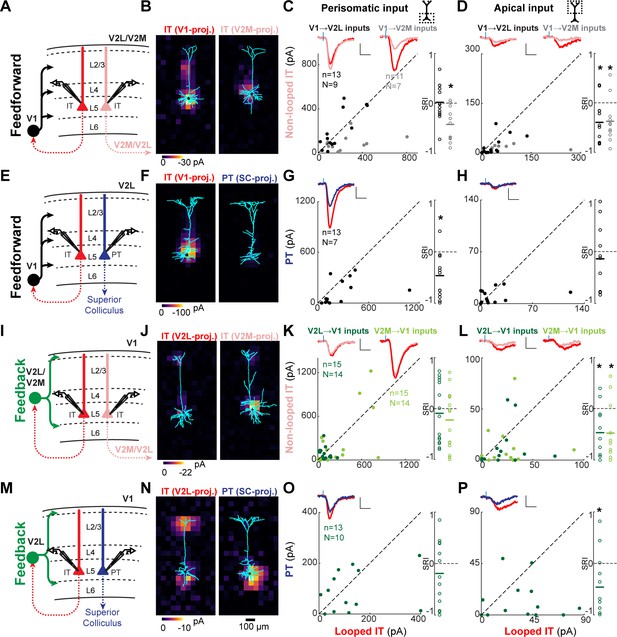
Feedforward (FF) and feedback (FB) inputs are stronger in looped intratelencephalic (IT) neurons than in neighboring non-looped IT or pyramidal tract (PT) neurons in L5.
(A) Configuration of experiments comparing strength of primary visual cortex (V1) FF input to pairs of L5 looped and non-looped IT neurons in lateral visual (V2L) or medial visual (V2M) areas. (B) Example pair of subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) maps overlaid on reconstructed dendrites showing monosynaptic V1 FF inputs to a looped IT neuron (left) and an adjacent non-looped IT neuron (right) recorded in V2L. (C) Left, paired comparisons of perisomatic FF input to looped vs. non-looped IT neurons; black dots, V1→V2L inputs; gray dots, V1→V2M inputs. Traces were generated by averaging the mean perisomatic excitatory postsynaptic current (EPSC) of each neuron across all neurons in the same projection class. Blue tick, laser pulse. Scale bars in all panels, 2 pA and 20 ms. Right, sCRACM Response Index (SRI) of the same data. Number of cell pairs and animals are the same as in the left plot unless otherwise specified. Horizontal line, mean. *, p<0.05, see text for exact value. (D) Same as C for apical inputs (SRI: V1→V2L, n = 12, N = 8; V1→V2M, n = 11, N = 7). (E) Configuration of experiment comparing strength of V1 FF input to pairs of L5 looped IT and PT neurons in V2L. (F) Example pair of sCRACM maps overlaid on reconstructed dendrites showing monosynaptic V1 FF inputs to a looped IT neuron (left) and an adjacent PT neuron (right) recorded in V2L. (G) Paired comparisons and SRI of perisomatic FF input to looped IT vs. PT neurons. (H) Paired comparisons and SRI (n = 11, N = 7) of apical FF input to looped IT vs. PT neurons. (I) Configuration of experiments comparing strength of V2L or V2M FB input to pairs of L5 looped and non-looped IT neurons in V1. (J) Example pair of sCRACM maps overlaid on reconstructed dendrites showing monosynaptic V2L FB inputs to a looped IT neuron (left) and an adjacent non-looped IT neuron (right) recorded in V1. (K) Paired comparisons and SRI of perisomatic FB input to looped vs. non-looped IT neurons. Dark green dots, V2L→V1 inputs; light green dots, V2M→V1 inputs. (L) Paired comparisons and SRI (V2L→V1, n = 11, N = 10; V2M→V1, n = 11, N = 10) of FB input in L1 to looped vs. non-looped IT neurons. (M) Configuration of experiment comparing strength of V2L FB input to pairs of L5 looped IT and PT neurons in V1. (N) Example pair of sCRACM maps overlaid on reconstructed dendrites showing monosynaptic V2L FB inputs to a looped IT neuron (left) and an adjacent PT neuron (right) recorded in V1. (O) Paired comparisons and SRI of perisomatic FB input to looped IT vs. PT neurons. (P) Paired comparisons and SRI (n = 12, N = 9) of FB input in L1 to looped IT vs. PT neurons.
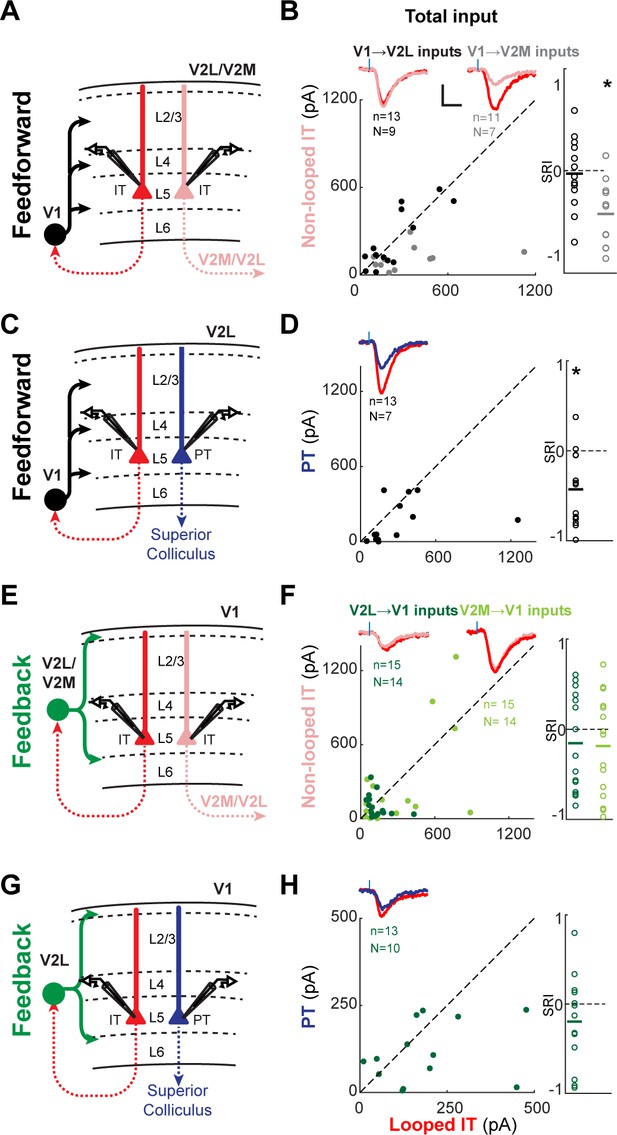
Total subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) input to L5 neurons.
(A) Configuration of experiments comparing strength of primary visual cortex (V1) feedforward (FF) input to pairs of L5 looped and non-looped intratelencephalic (IT) neurons in lateral visual areas (V2L) or medial visual areas (V2M). (B) Left, paired comparisons of total FF input to looped vs. non-looped IT neurons. Black dots, V1→V2L inputs; gray dots, V1→V2M inputs. Traces were generated by averaging the mean excitatory postsynaptic current (EPSC) of each neuron across all neurons in the same projection class. Blue tick, laser pulse. Scale bars in all panels, 2 pA and 20 ms. Right, sCRACM Response Index (SRI) of the same data. Number of cell pairs and animals are the same as in the left plot. Horizontal line, mean. *, p=0.0015. (C) Configuration of experiment comparing strength of V1 FF input to pairs of L5 looped IT and PT neurons in V2L. (D) Paired comparisons and SRI of total FF input to looped IT vs. PT neurons. *, p=0.0028. (E) Configuration of experiments comparing strength of V2L or V2M FB input to pairs of L5 looped and non-looped IT neurons in V1. (F) Paired comparisons and SRI of total FB input to looped vs. non-looped IT neurons. (G) Configuration of experiment comparing strength of V2L FB input to pairs of L5 looped IT and PT neurons in V1. (H) Paired comparisons and SRI of total FB input to looped IT vs. PT neurons.
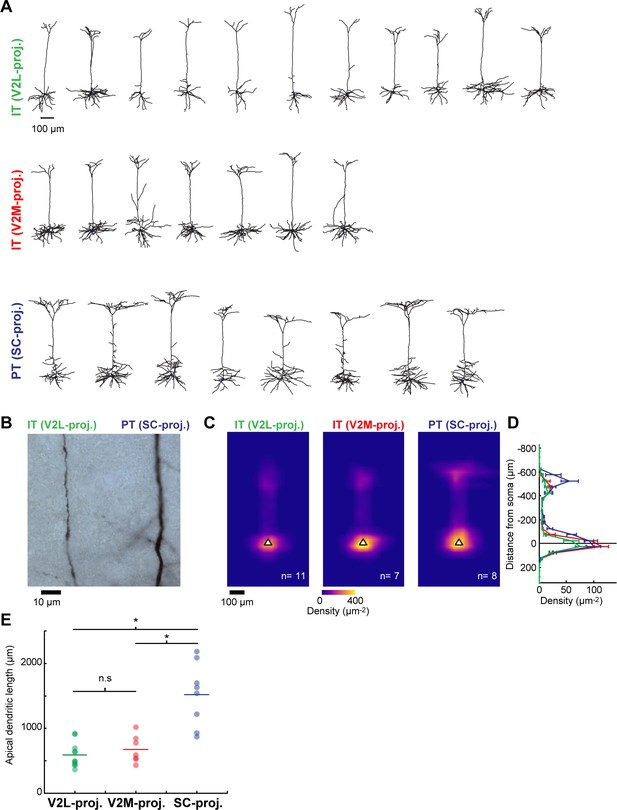
Dendritic morphology of the different L5 projection neuron types in primary visual cortex (V1).
(A) Reconstructed dendritic morphologies of the three different L5 projection neurons recorded in V1. Top and middle, intratelencephalic (IT) neurons projecting to lateral visual (V2L) or medial visual (V2M) areas; bottom, pyramidal tract (PT) neurons projecting to superior colliculus (SC). (B) Brightfield image showing representative example of apical shaft segments from a pair of biocytin-stained IT and PT L5 neurons. The apical dendrites of SC-projecting PT neurons were of larger diameter than those of same-layer V2L- and V2M-projecting IT neurons. (C) Average normalized dendritic length density of the three cell types, aligned by soma position (white triangle). (D) Mean vertical profiles of dendritic length density (error bars, s.e.m). (E) Total apical tuft dendritic length for the three cell types. Apical tuft branches of SC-projecting PT neurons are more extensive than those of V2L- and V2M-projecting IT neurons (Kruskal-Wallis test followed by Tukey-Kramer honestly significant difference (HSD) post-hoc test, SC-projecting vs. V2L-projecting, p=0.0006; SC-projecting vs. V2M-projecting, p=0.0168; V2L-projecting vs. V2M-projecting, p=0.8044).
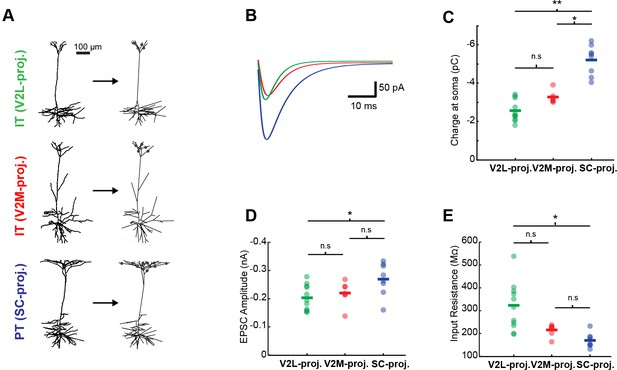
Simulations of the dendritic filtering of distal apical inputs.
(A) Example simulations of L5 neurons. Reconstructed dendritic arbors were imported into the NEURON environment. Synapses were randomly placed with constant density along apical tuft dendritic segments. (B) Simulated excitatory postsynaptic current (EPSC) at the soma evoked by L1 input under voltage-clamp conditions for the three example neurons shown in A. (C) Mean somatic charge per cell (based on 100 simulations) resulting from apical tuft input across the three projection neuron populations. Apical inputs lacking cell-type selectivity and exhibiting equal synaptic density across the different cell types generate larger somatic currents in superior colliculus (SC)-projecting neurons vs. medial visual area (V2M)- or lateral visual area (V2L)-projecting neurons (Kruskal-Wallis test followed by Tukey-Kramer HSD post-hoc test, SC-projecting vs. V2L-projecting, p=0.0001; SC-projecting vs. V2M-projecting, p=0.0325; V2L-projecting vs. V2M-projecting, p=0.3626). (D) ESPC amplitude resulting from apical tuft input across the three projection neuron populations. EPSC amplitudes are larger in pyramidal tract (PT) neurons than in V2L-projecting neurons (Kruskal-Wallis test followed by Tukey-Kramer HSD post-hoc test, SC-projecting vs. V2L-projecting, p=0.0262; SC-projecting vs. V2M-projecting, p=0.1495; V2L-projecting vs. V2M-projecting, p=0.8753). (E) Input resistance measured from a voltage step during simulated somatic voltage-clamp in the model L5 cells. PT neurons have lower input resistance than V2L-projecting neurons (Kruskal-Wallis test followed by Tukey-Kramer HSD post-hoc test, SC-projecting vs. V2L-projecting, p=0.0003; SC-projecting vs. V2M-projecting, p=0.2082; V2L-projecting vs. V2M-projecting, p=0.1356).

Feedforward (FB) input to looped L5 intratelencephalic (IT) neurons vs. pyramidal tract (PT) neurons in the presence of Ih blockers.
(A) Configuration of experiment comparing strength of lateral visual area (V2L) feedback (FB) input to pairs of looped IT and PT neurons in primary visual cortex (V1) L5. ZD7288 was added to the bath solution to block Ih currents. (B) Left, paired comparisons of FB inputs to L1 apical dendrites of looped IT neurons vs. PT neurons in L5. Traces represent group-averaged excitatory postsynaptic currents (EPSCs) in L1. Right, sCRACM Response Index (SRI) of the same data (n = 5, N = 5). *, p=0.0009.
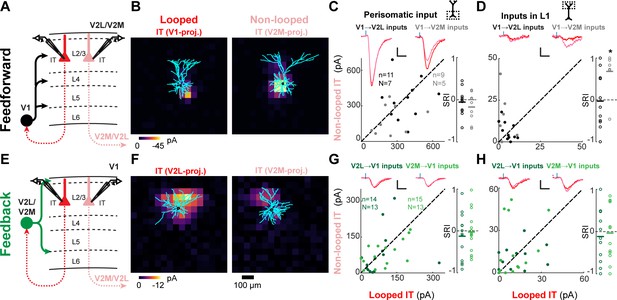
Feedforward (FF) and feedback (FB) connections are similar or weaker in looped L2/3 neurons.
(A) Configuration of experiments comparing strength of primary visual cortex (V1) FF input to pairs of L2/3 looped and non-looped intratelencephalic (IT) neurons in lateral visual area (V2L) or medial visual area (V2M). (B) Example pair of subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) maps overlaid on reconstructed dendrites showing monosynaptic V1 FF inputs to a looped IT neuron (left) and an adjacent non-looped IT neuron (right) recorded in V2L. (C) Left, paired comparisons of perisomatic FF input to looped vs. non-looped IT neurons; black dots, V1→V2L inputs; gray dots, V1→V2M inputs. Traces were generated by averaging the mean perisomatic excitatory postsynaptic current (EPSC) of each neuron across all neurons in the same projection class. Blue tick, laser pulse. Scale bars in all panels, 2 pA and 20 ms. Right, sCRACM Response Index (SRI) of the same data. Number of cell pairs and animals are the same as in the left plot unless otherwise specified. Horizontal line, mean. *, p<0.05, see text for exact value. (D) Same as C for inputs in L1 (SRI: V1→V2L, n = 11, N = 7; V1→V2M, n = 7, N = 5). (E) Configuration of experiments comparing strength of V2L or V2M FB input to pairs of L2/3 looped and non-looped IT neurons in V1. (F) Example pair of sCRACM maps overlaid on reconstructed dendrites showing monosynaptic V2L FB inputs to a looped IT neuron (left) and an adjacent non-looped IT neuron (right) recorded in V1. (G) Paired comparisons and SRI of perisomatic FB input to looped vs. non-looped IT neurons. Dark green dots, V2L→V1 inputs; light green dots, V2M→V1 inputs. (H) Same as G for inputs in L1 (SRI: V2L→V1, n = 11, N = 10; V2M→V1, n = 12, N = 11).
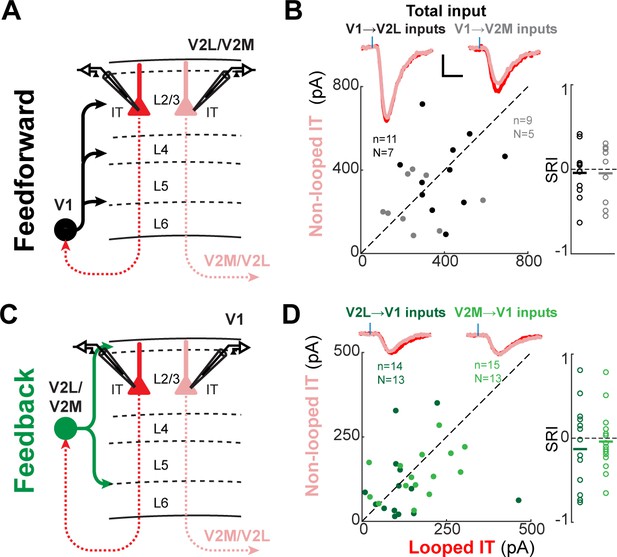
Total subcellular channelrhodopsin-2 (ChR2)-assisted circuit mapping (sCRACM) input to L2/3 neurons.
(A) Configuration of experiments comparing strength of primary visual cortex (V1) feedforward (FF) input to pairs of L2/3 looped and non-looped intratelencephalic (IT) neurons in lateral visual area (V2L) or medial visual area (V2M). (B) Left, paired comparisons of total FF input to looped vs. non-looped IT neurons. Black dots, V1→V2L inputs; gray dots, V1→V2M inputs. Traces were generated by averaging the mean excitatory postsynaptic current (EPSC) of each neuron across all neurons in the same projection class. Blue tick, laser pulse. Scale bars in all panels, 2 pA and 20 ms. Right, sCRACM Response Index (SRI) of the same data. (C) Configuration of experiments comparing strength of V2L or V2M FB input to pairs of L2/3 looped and non-looped IT neurons in V1. (D) Paired comparisons and SRI of total FB input to looped vs. non-looped IT neurons. Dark green dots, V2L→V1 inputs; light green dots, V2M→V1 inputs.
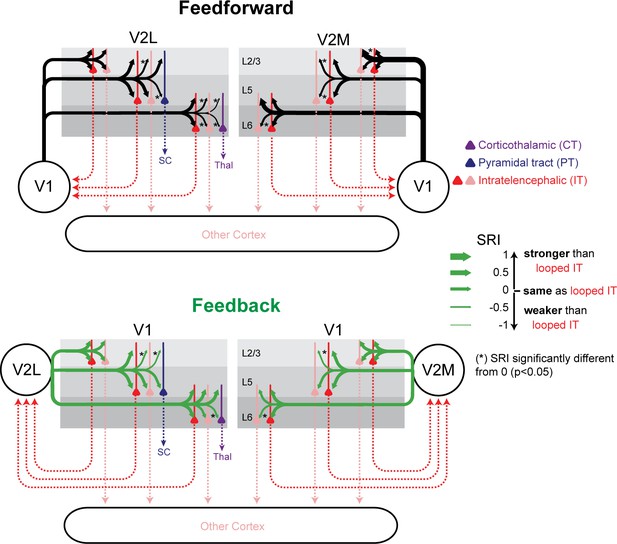
Summary of relative input strength across all experiments.
The sCRACM Response Index (SRI) of feedforward (FF) and feedback (FB) inputs to the different cell types is represented by arrow thickness. The top and bottom arrows to L5 neurons indicate inputs to apical and perisomatic domains, respectively. Inputs to looped intratelencephalic (IT) cells in each cortical layer are assigned an SRI of 0 (medium arrow thickness). * signifies significant difference (p<0.05) from the looped IT population.
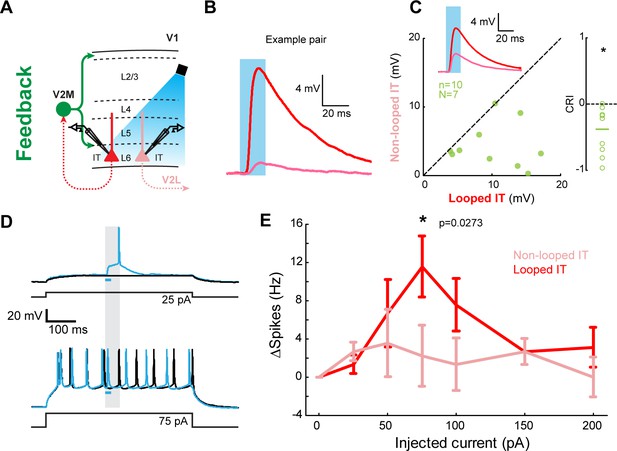
Feedback (FB) inputs in L6 can selectively modulate activity in looped intratelencephalic (IT) neurons.
(A) Experiment configuration. In the absence of channel blockers, V2M→V1 FB axons were photostimulated using an LED during current-clamp recordings of looped and non-looped IT neurons in L6. (B) Example of excitatory postsynaptic potentials (EPSPs) from an example pair. Blue shade, light pulse. (C) Left, paired comparisons of EPSP amplitudes evoked in looped vs. non-looped IT neurons. Inset traces represent group averages for each projection class. Blue shade, light pulse. Right, CRACM Response Index (CRI) of the same data (n = 10, N = 7). (D) Example traces of FB modulation in a looped IT neuron. Cells were driven by a sustained positive current injection. Black traces, LED OFF trials; blue traces, LED ON trials. Blue bar, duration of the LED light pulse. Gray shading, time period used to analyze spiking activity in (E). (E) Spike rate difference between LED-ON and LED-OFF trials in looped and non-looped IT neurons as a function of the amount of current injected during the depolarization step. *, p=0.0273, paired t-test with Bonferroni correction for seven comparisons.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus) | C57BL/6 | Jackson Laboratory | JAX:000664, RRID:IMSR_JAX:000664 | Bred in-house |
| Antibody | Anti-GFP (rabbit polyclonal) | Thermo Fisher | Catalog # A-6455, RRID:AB_221570 | (1:1000) |
| Antibody | Alexa Fluor 488-conjugated secondary antibody | Thermo Fisher | Catalog # A-11008, RRID:AB_143165 | (1:1000) |
| Recombinant DNA reagent | AAV-2/1-CAG-ChR2-Venus | Addgene | RRID:Addgene_20071 | |
| Recombinant DNA reagent | AAV5-CaMKIIa-hChR2(H134R)-EYFP | Addgene | RRID:Addgene_26969 | |
| Recombinant DNA reagent | AAV2/1-synapsin-EGFP | UPenn Vector Core | RRID:Addgene_105539 | |
| Peptide, recombinant protein | Cholera toxin B (Alexa Fluor 647) | Thermo Fisher | Catalog #: C34778 | 1 mg/ml |
| Chemical compound, drug | Red Retrobeads IX | Lumafluor | ||
| Software, algorithm | Ephus | Vidrio Technologies Suter et al., 2010 | PMID:21960959 |





