A low affinity cis-regulatory BMP response element restricts target gene activation to subsets of Drosophila neurons
Figures
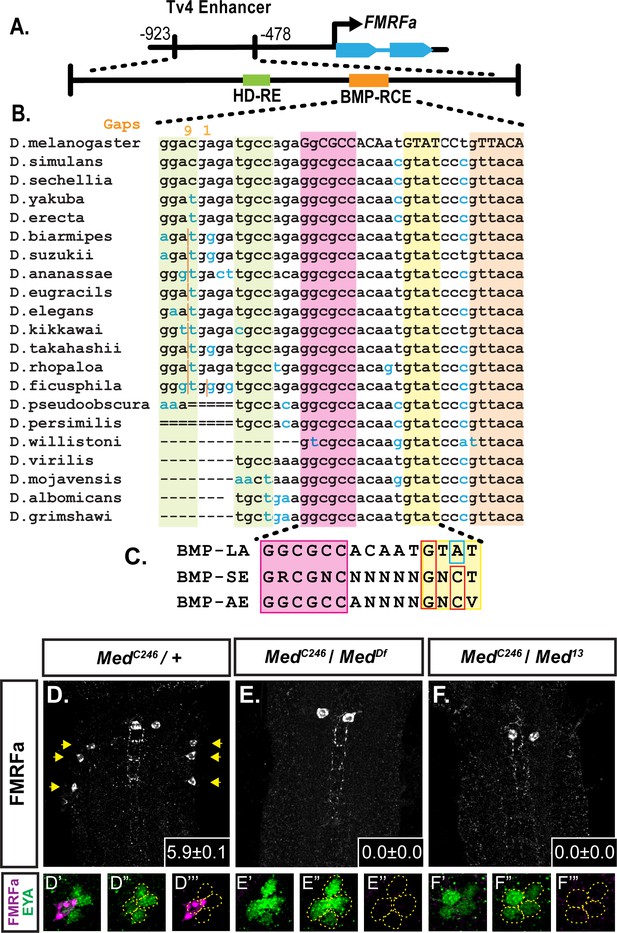
A novel BMP-Response element (BMP-RE) in the Tv4-neuron-specific enhancer of the FMRFa gene.
The 445 bp Tv4-enhancer depicted in (A) contains two cis-elements critical for FMRFa activation, the homeodomain response element (HD-RE that recruits Apterous), and the BMP response element (BMP-RCE) that binds pMad and mediates the BMP-dependence of the Tv4-enhancer. (B) Output from the UCSC Browser shows sequence conservation through the BMP-RE across 21 Drosophila species. Capitalized letters are conserved across all species. Highlighted sequences include the putative Mad/Brinker binding site (magenta), two 5’ optimal GNCV Medea binding sequences with non-canonical spacing (green), and three 3’ sequences that deviate from a non-stringent GNCN sequence in one critical nucleotide (yellow, orange). The yellow GTAT motif is ideally spaced but has a C > A switch. The orange GTTACA contains two motifs (GTTA with a C > T switch, and TACA with a G > T switch). (C) Comparison of a putative FMRFa BMP-RE with the well-defined BMP-SE and BMP-AE motifs. (D-F) FMRFa immunoreactivity in Tv4-neurons is lost in Medea nulls (E,F) compared to controls (D). Insets show mean number of FMRFa-positive Tv4-neurons per VNC ± SD. (D’-F’’) Fluorophore splits of single Tv -clusters in A-C, showing Tv-neurons (circled) labeled by anti-Eya (green) and anti-FMRFa (magenta) expression in each genotype.
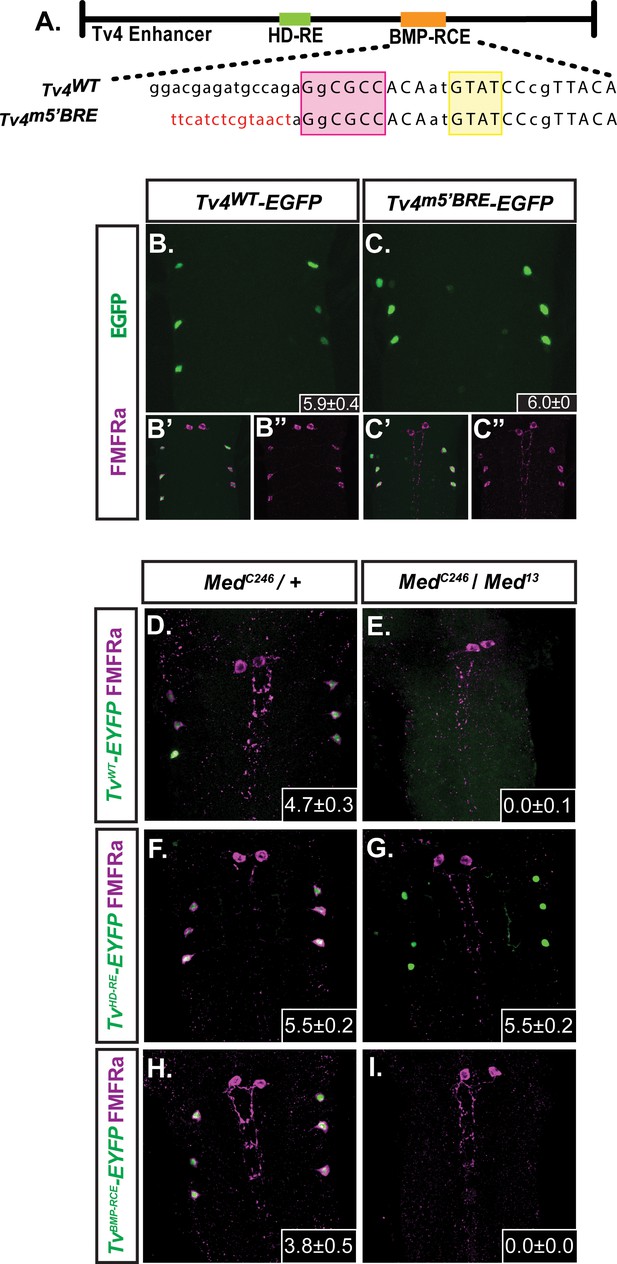
FMRFa expression in Tv4-neurons requires Medea.
(B,C) We compared the in vivo activity of the wildtype Tv4WT-enhancer (Tv4WT-EGFP) and a full-length Tv4-enhancer reporter that was substitution mutagenized to eliminate both GNCV sequences within the BMP-RE (Tv4m5’BMP-RE) (shown in A; sequences shown in Supplementary file 1a). Both the Tv4WT-EGFP (n = 14 animals) and Tv4m5’BMP-RE (n = 9 animals) reporters generated wildtype activity; therefore, neither of these canonical GNCV motifs are essential for BMP-dependent FMRFa expression. Insets show the mean ± SD of EGFP-positive Tv4-neurons. (D–I) Comparison of anti-FMRFa and FMRFa enhancer fragment reporter expression in controls (MedC246/+) and Medea nulls (MedC246/Med13 ). Panels show expression of the Tv4WT-EYFP reporter (D,E), the HD-RE-EYFP reporter (F,G) and the BMP-RCE-EYFP reporter (H,I). Medea is required for expression of the BMP-dependent Tv4WT-EYFP and BMP-RCE-EYFP reporters, but not for the expression of the BMP-independent HD-RE-EYFP reporter. Insets show the mean number of EYFP-positive Tv4-neurons per VNC ± SD.
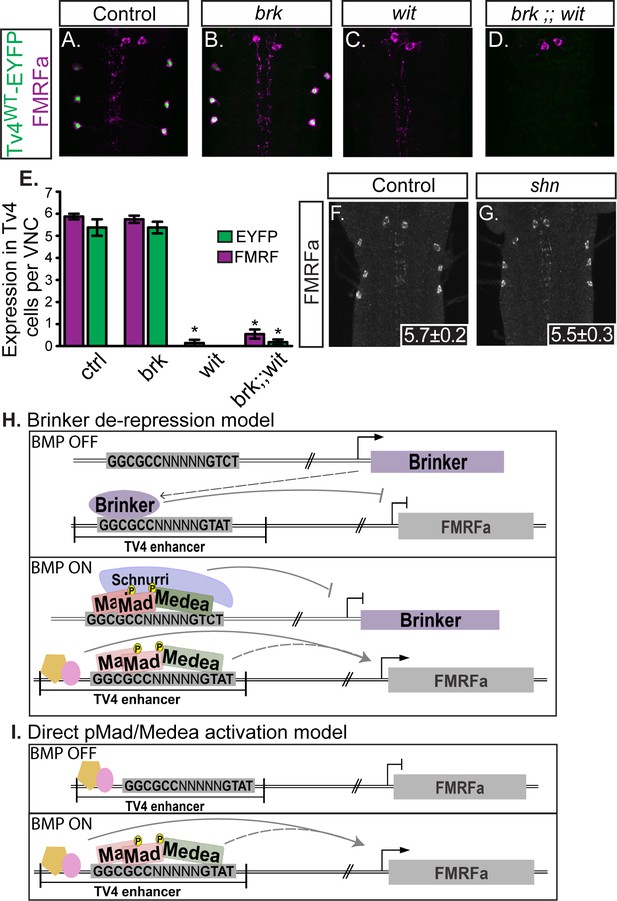
Neither brinker nor schnurri are required for BMP-dependent FMRFa.
(A,B) Expression of the BMP-dependent FMRFa prepropeptide and the Tv4WT-EYFP reporter were not affected in brk mutants. (C,D) Loss of FMRFa prepropeptide and Tv4WT-EYFP in wit nulls was not rescued by the loss of brk in the double mutant of brk and wit. Thus, FMRFa is not lost in wit mutants due to the de-repression of brk. (E) Quantification of data in A-D (n = 7–12 animals per group, *p<0.01 compared to controls using One-way ANOVA with Tukey HSD post-hoc). (F,G) No change in the number of FMRFa-positive Tv4-neurons was observed between control and shn nulls in late stage 17 embryos. Mean ± SD number of FMRFa-positive Tv4-neurons per VNC shown in inset, n = 5 per genotype. (H) In a Brinker de-repression model, brinker would act as an FMRFa repressor by binding to the BMP-LA element (at the GGCGCC motif). When the BMP signaling pathway is active, the activated pMad/Medea complex would translocate to the nucleus and bind to the brk BMP-SE element, recruiting Schnurri, and silencing brk expression. This would allows expression of FMRFa, activated by other transcription factors and/or direct binding of a pMad/Medea complex. (I) In a direct pMad/Medea complex activation model, FMRFa is only expressed when an activated pMad/Medea complex binds to the BMP-LA sequence. Our work indicates that this latter model is likely correct, as neither brk nor shn manipulation modulates BMP-dependent FMRFa expression. Genotypes: Control (TvWT-nEYFP). brk (brkXA/Y;;TvWT-nEYFP/TvWT-nEYFP). wit (TvWT-nEYFP,witA12/TvWT-nEYFP,witB11). brk;;wit (brkXA/Y;; TvWT-nEYFP,witA12/TvWT-nEYFP,witB11). shn control in F (w;shn1/+). shn null in G (shn1/shn1).
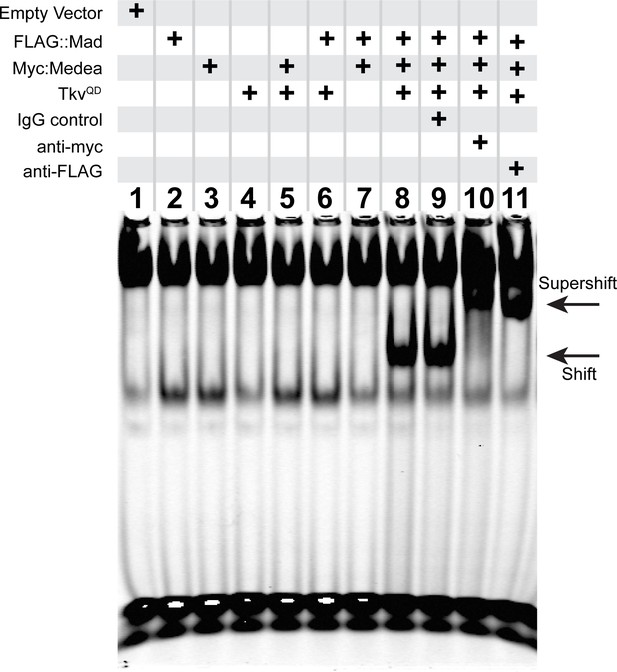
An activated pMad/Medea complex binds to the FMRFa BMP-RE.
EMSA using IRDye700-tagged DNA oligonucleotides of the FMRFa BMP-RE incubated with S2 cell extracts transfected with FLAG::Mad and/or Myc::Medea, and/or activated BMP-receptor TkvQD. Lanes 2–7 showed no specific band shift that differed from empty vector transfected cells (lane 1). Co-transfection of FLAG::Mad, Myc::Medea and TkvQD together generated a strong band shift (lane 8). Addition of antibodies to Myc (lane 10) or FLAG (lane 11) caused a super-shifted band that was not seen upon addition an IgG antibody control (lane 9). Thus, Mad, Medea, and activated Tkv receptor were capable of generating a band-shift of the FMRFa BMP-RE.
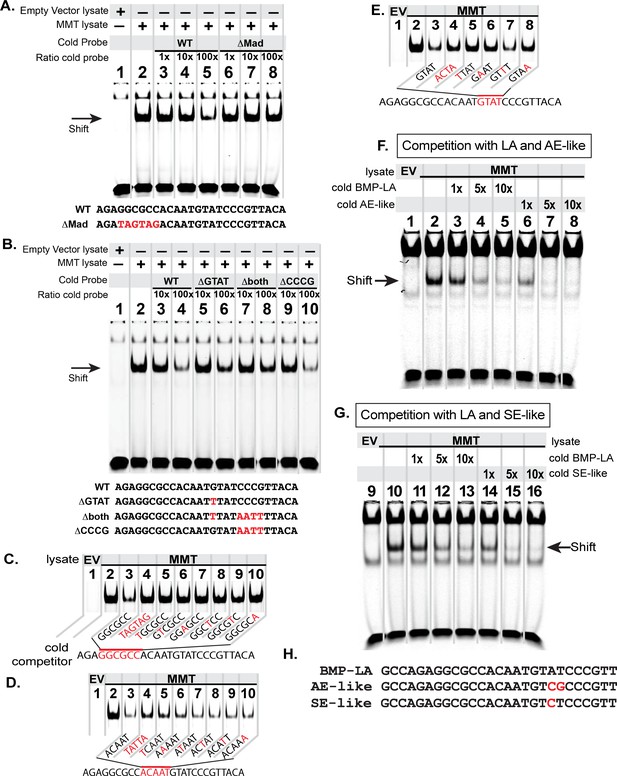
The FMRFa BMP-LA is required for pMad/Medea recruitment but at a lower affinity compared to BMP-AE and BMP-SE motifs.
We performed EMSA gels in which we ran IRDye700-tagged FMRFa BMP-RE DNA oligonucleotides (of sequence AGAGGCGCCACAATGTATCCCGTTACA) pre-incubated with lysates from S2 cells transfected with either empty vectors (EV; lane 1) or FLAG::Mad, Myc::Medea and activated BMP-receptor, TkvQD (MMT lysate; lanes 2–8 or 2–10) (A-B). The MMT lysate generated a band shift indicative of pMad/Medea binding to the tagged probe (lane 2 in A,B). In lanes 3–8 (A) or 3–10 (B), we ran MMT lysates pre-incubated with tagged probe and a stoichiometric excess of untagged (cold) DNA oligonucleotides with either wildtype or mutated sequence (shown below each gel). Loss of a band shift indicates that the untagged probe is capable of binding activated pMad/Medea. (A) We pre-incubated with untagged (cold) competitors of wildtype (WT) sequence or GGCGCC >TAGTAG mutated sequence (ΔMad) in 1×, 10×, 100× excess. At 100× excess, competition by the WT cold probe reduced the band shift (lane 5). In contrast, the mutated cold probe failed to reduce the band shift at 100× excess (lane 8). (B) We pre-incubated with 10x and 100x excess of cold probes of wildtype (WT) or mutated sequences, including of G and C nucleotides within candidate Medea-binding sequences GTAT (>TTAT, termed ΔGTAT), CCCG (>AATT, termed ΔCCCG), and a mutant to both of these sequences (Δboth). At 100× excess, the wildtype (lane 4) and ΔCCCG (lane 10) greatly reduced the band shift to the same extent; thus, the CCCG sequence does not contribute to pMad/Medea binding. By contrast, the ΔGTAT (lane 6) and Δboth (lane 8) cold probes only partially reduced the band shift, indicating that their ability to bind pMad/Medea was compromised. These data suggest that the minimal BMP-RE comprises a GGCGCC(N5)GTAT sequence. (C-D) The MMT lysate generated a band shift indicative of pMad/Medea binding to the tagged probe (lane 2). We added untagged (cold) BMP-RE DNA oligonucleotides at 100× stoichiometric excess with mutations in the sequences shown in red. (C) Nucleotide mutations are shown as red within the pMad-binding site. (D) Nucleotide mutations are shown as red within the linker region. (E) Nucleotide mutations are shown as red within the Medea-binding site. Examining the ability of each cold competitor to compete with tagged FMRFa BMP-RE, we reveal a necessary BMP-RE sequence of GGCGGGacaatGTaT, where capitalized nucleotides are found most necessary for pMad/Medea recruitment. (F,G) In these EMSA, we additionally transfected S2 cell extracts with a 1×, 5×, and 10× stoichiometric excess of untagged competitors (sequences shown in H). (F) Competition for the tagged BMP-LA by the untagged wildtype (BMP-LA) or the AE-like mutant. A 10× excess (lane 5) of untagged wildtype BMP-LA reduced but did not eliminate the band shift (lane 5). By contrast, the BMP-AE-like mutant totally out-competed the tagged BMP-LA at a 5× excess (lane 7 compared to lane 4) and significantly out-competed the tagged BMP-LA even at equimolar ratio (lane 6 compared to lane 3). (G) Competition for the tagged BMP-LA by the untagged wildtype or the SE-like mutant. The SE-like mutant totally out-competed the tagged BMP-LA at a 5× excess (lane 15 compared to lane 12) and significantly out-competed the tagged BMP-LA even at equimolar ratio (lane 14 compared to lane 11).
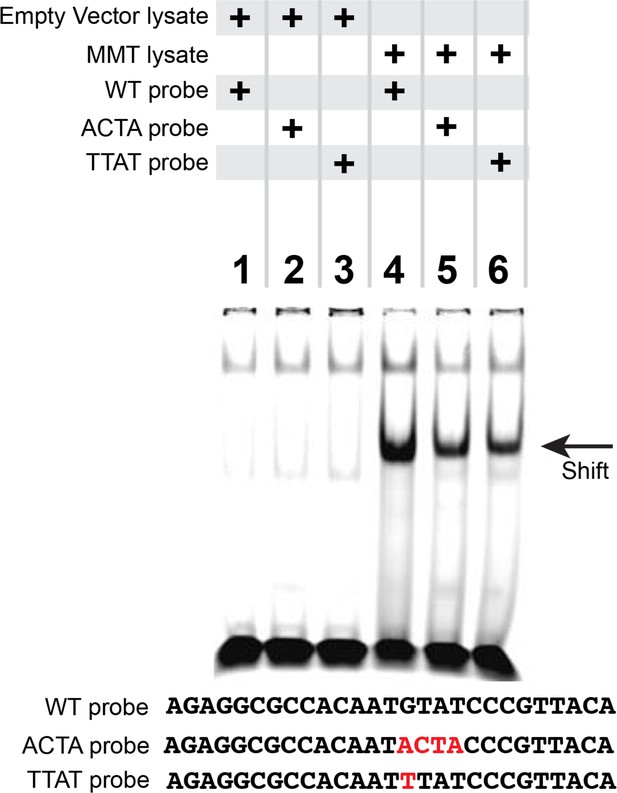
Reduced pMad/Medea recruitment upon GTAT sequence mutation in the FMRFa BMP-RE.
EMSA analysis in which we ran lysates from S2 cells transfected with either empty vectors (lanes 1–3) or with FLAG::Mad, Myc::Medea and activated BMP-receptor, TkvQD (MMT lysate) (lanes 4–6) that were pre-incubated with IRDye700-tagged FMRFa BMP-RE DNA oligonucleotides of wildtype sequence (lanes 1, 4), or GTAT mutated sequence (lanes 2, 3, 5, 6). Only S2 cell lysates with activated pMad/Medea band shifted any of the tagged DNA probes. Comparing lane 4 with lanes 5 and 6, we find that mutation of the GTAT sequence to either ACTA or TTAT reduced binding.
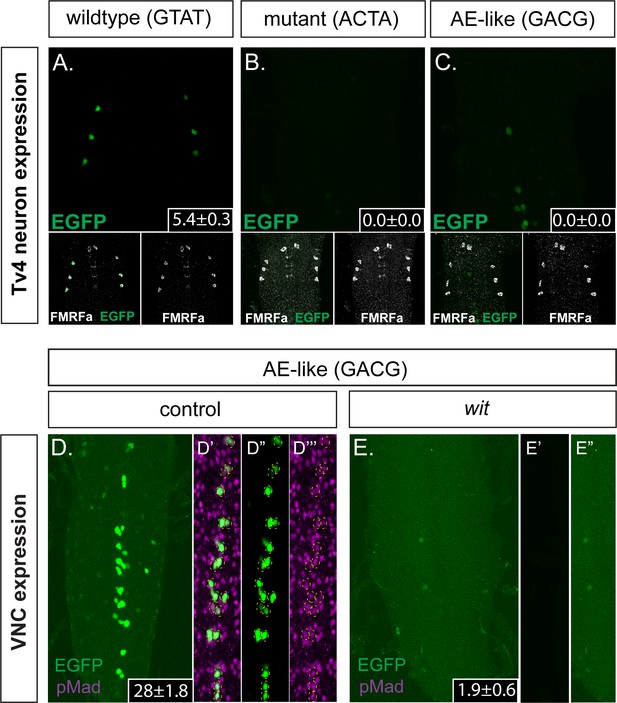
The FMRFa BMP-LA motif has a necessary but low affinity Medea binding site that specifies selective neuronal subtype activity.
(A–C) Conversion of the Medea-binding GTAT site in the wildtype 445 bp Tv4-neuron-specific FMRFa enhancer to a mutant version, ACTA (that reduces pMad/Medea recruitment; termed Tv4 mGTAT>ACTA–nEGFP), resulted in a complete loss of reporter gene expression in Tv4-neurons (B). Conversion of the Medea-binding GTAT site in the wildtype 445 bp Tv4-neuron-specific FMRFa enhancer to an optimal BMP-AE-like sequence (GACG; termed Tv4 mGTAT>GACG–nEGFP) also resulted in a total loss of reporter expression in Tv4-neurons (C). Numbers in insets indicate the mean ± SD number of EGFP-positive Tv4-neurons per VNC, out of the possible six Tv neurons. (D,E) The Tv4mGTAT>GACG–nEGFP reporter generated strong ectopic reporter activity in VNC midline cells (D) that is lost in the absence of neuronal BMP signaling (E; in wit mutants, witA12/witB11). Full z-projections though the whole VNC are shown. Numbers in insets indicate the mean ± SD number of EGFP-positive neurons per VNC. (D’–D’’’) Images of the midline ectopic EGFP expression generated from Tv4mGTAT>GACG–nEGFP. EGFP expression (green) was exclusively expressed in pMad-immunoreactive cells (magenta); all cells are yellow circled.
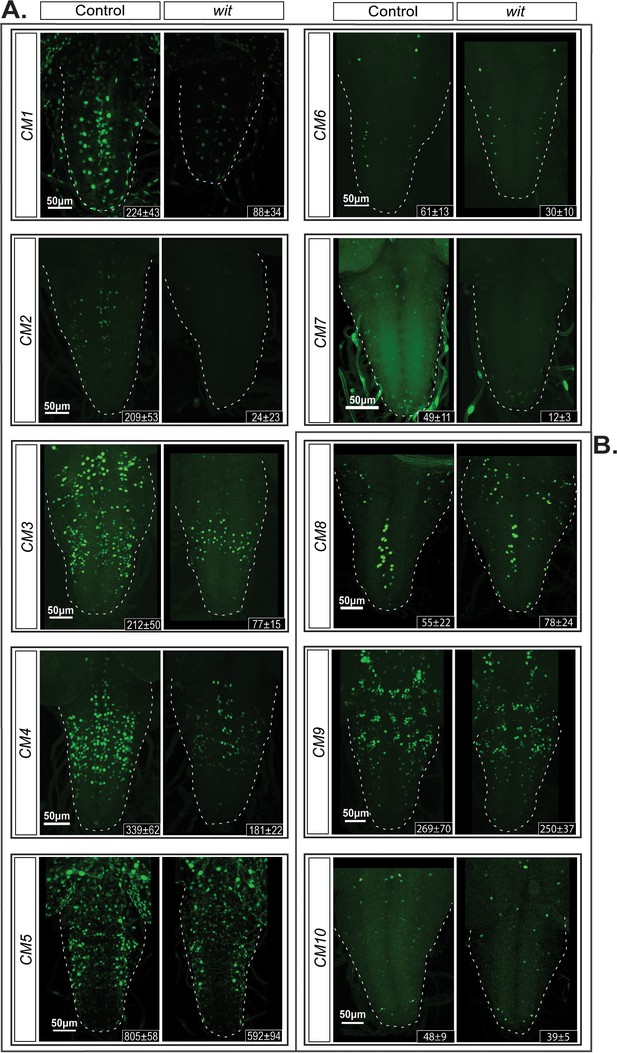
Identification of additional wit-responsive genomic fragments containing the BMP-LA motif.
We identified 10 genomic fragment reporters that exhibited expression in subsets of pMad-positive cells in the VNC and tested their wit-responsiveness. (A) EGFP reporter patterns of three genomic fragments that exhibit no wit-responsive loss of reporter expression in wit mutants (witA12/witB11) compared to controls (witA12/+) in late third Instar larval VNCs. (B) Nuclear EGFP expression patterns driven from seven genomic fragments containing conserved BMP-LAs that were down-regulated in wit mutants (witA12/witB11), compared to controls (witA12/+) in late third Instar larval VNCs. The observed down-regulation ranged from a near-total loss of all neuronal expression to loss of expression in a subset of neurons. Full z-projections though the whole VNC are shown. Genotypes: All control lines shown here were heterozygous (w;;CM#/+); wit mutants (w;;CM#,witA12/witB11).
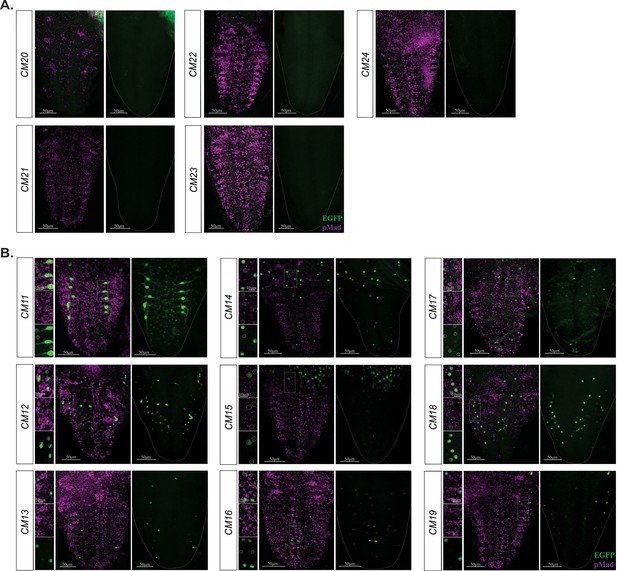
BMP-LA containing reporter fragments with no reporter and pMad co-expression.
(A) No reporter expression was observed in the VNC of these reporter lines. (B) These nine reporter lines exhibited a range of reporter patterns in the VNC but lacked EGFP reporter and pMad stain overlap. Areas of high reporter expression were magnified and showed in insets as separate channels and overlay; reporter expressing cells were circled. No overlap in expression of EGFP reporter and pMad stain was observed. Full z-projections though the whole VNC are shown. Side panels indicate nuclei with GFP reporter (green) and/or pMad stain (magenta). Genotypes: All control lines shown here were heterozygous (w;;CM#/+); wit mutants (w;;CM#,witA12/witB11).
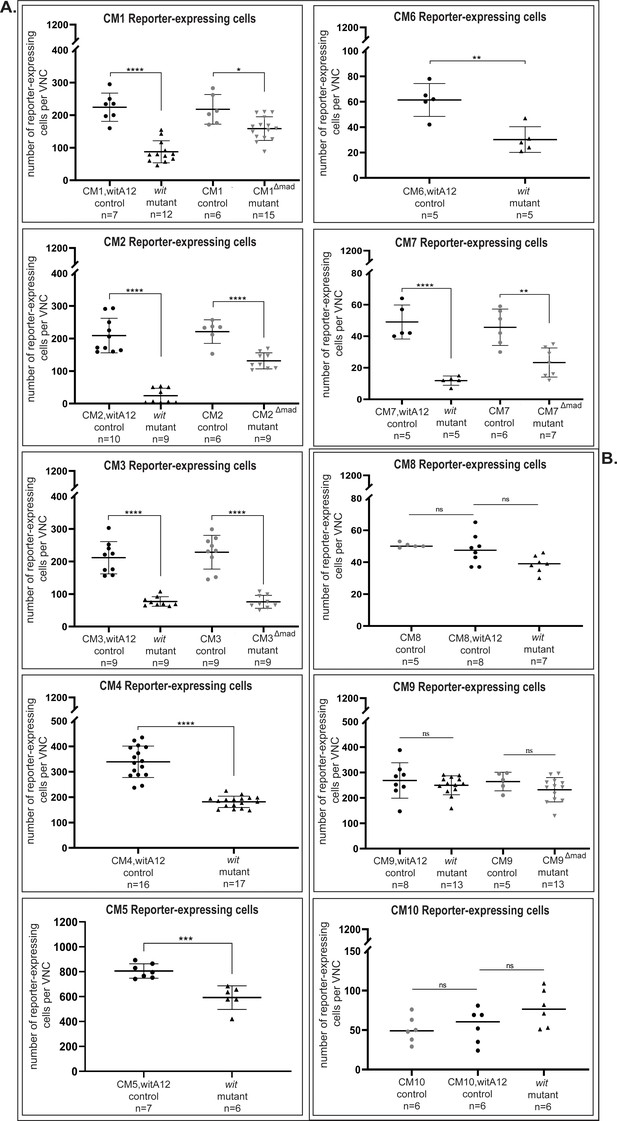
Quantification of reporter expressing cells reveals significant downregulation in wit mutants or pMad-binding site mutants compared to their respective controls.
(A) None of the three reporter fragments (CM8, CM9, CM10) showed a significant change in number of reporters expressing cells between controls and their respective mutants. (B) In the VNC, the CM1 (CM1,witA12/+ controls) reporter was expressed in 224 ± 43 nuclei, of which 56 ± 16 (25%) nuclei are pMad-positive (n = 7). In wit mutants (witA12/witB11), reporter activity was reduced by 61% to 88 ± 34 nuclei (n = 12). Once the pMad-binding site was mutated (CM1Δmad), reporter activity was reduced by 27% to 159 ± 36 nuclei (n = 15) compared to controls that had reporter expression in 218 ± 46 nuclei (n = 6). CM2 (CM2,witA12/+ controls) reporter was expressed in 209 ± 53 nuclei, of which 111 ± 33 (53%) nuclei are pMad-positive (n = 10). In wit mutants (witA12/witB11), reporter activity was reduced by 88.5% to 24 ± 23 nuclei (n = 9). In the pMad-binding site mutant (CM2Δmad), reporter activity was reduced by 41% to 131 ± 25 nuclei (n = 9) compared to controls that had reporter expression in 221 ± 36 nuclei (n = 6). CM3 (CM3,witA12/+ controls) reporter was expressed in 212 ± 50 nuclei, of which 68 ± 19 (32%) nuclei are pMad-positive (n = 9). In wit mutants, reporter activity was reduced by 64% to 77 ± 15 nuclei (n = 9). In the pMad-binding site mutant (CM3Δmad), reporter activity was reduced by 67% to 76 ± 20 nuclei (n = 9) compared to controls that had reporter expression in 228 ± 52 nuclei (n = 9). CM4 reporter was expressed in 339 ± 62 nuclei, of which 134 ± 25 (34%) nuclei were pMad-positive (n = 16). In wit mutants, reporter activity was reduced by 47% to 181 ± 22 nuclei (n = 17). CM5 reporter was expressed in 805 ± 58 nuclei in the VNC, of which 217 ± 10 (27%) nuclei were pMad-positive (n = 7). In wit mutants, reporter activity was reduced by 30% to 592 ± 94 nuclei (n = 6). CM6 was expressed in 61 ± 13 nuclei (n = 5). In wit mutants, reporter activity was reduced by 50% to 30 ± 10 nuclei (n = 5). Finally, CM7 (CM7,witA12/+ controls) reporter was expressed in 49 ± 11 nuclei, of which 9 ± 2 (18%) nuclei are pMad-positive (n = 5). In wit mutants, reporter activity was reduced by 75% to 12 ± 3 nuclei (n = 5). In the pMad-binding site mutant (CM7Δmad), reporter activity was reduced by 50% to 23 ± 9 nuclei (n = 7) compared to controls that had reporter expression in 46 ± 12 nuclei (n = 6). Significance was calculated with One-way ANOVA with a Tukey post hoc multiple comparisons test for the following genotypes: CM1 control versus wit mutants p<0.0001 and control versus pMad-binding site mutants p=0.0143; CM2 control versus wit mutant and control versus pMad-binding site mutants p<0.0001; CM7 control versus wit mutants p<0.0001 and control versus pMad-binding site mutants p=0.0021. Student`s t-test was used for: CM4 p<0.0001; CM5 p=0.0004; CM6 p=0.0028. For CM3, as the wit control samples were non-normally distributed (Shapiro Wilk test), significance was calculated with the Mann-Whitney U-test for control versus wit mutants p<0.0001; Student's t-test was used for control versus pMad-binding site mutants p<0.0001. Each point represents the total number of reporter-expressing cells in the VNC of a single animal, n indicates the number of VNCs analyzed and data is reported as mean ± SD. Genotypes: All control and pMad-binding site mutant lines examined here were heterozygous (w;;CM#/+); wit mutants (w;;CM#,witA12/witB11).
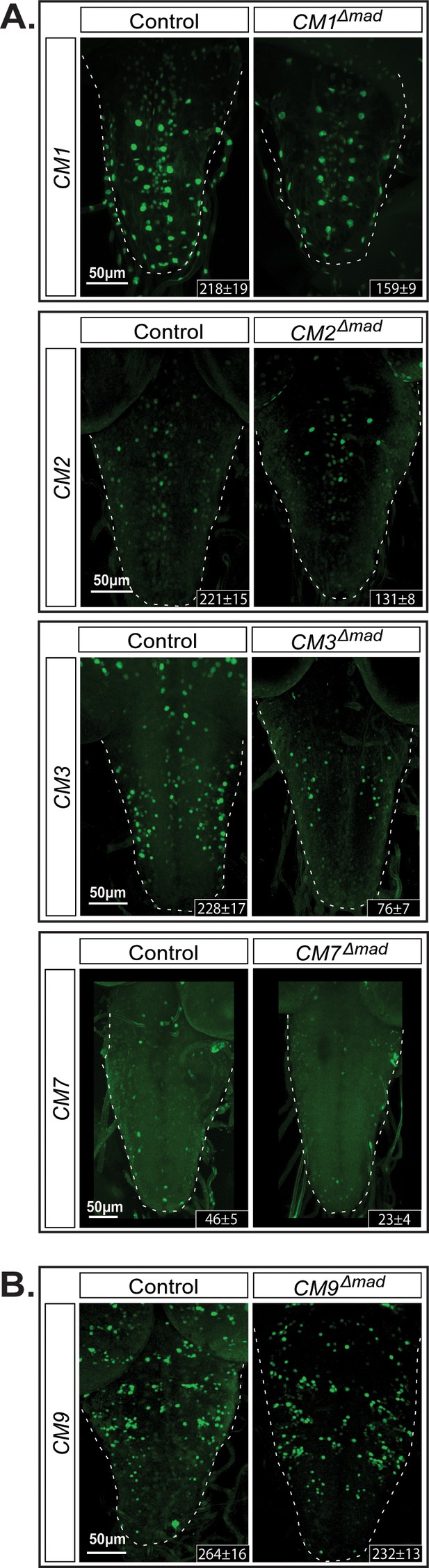
The GGCGCC pMad-binding site is necessary for reporter expression in vivo.
We introduced specific mutations into the pMad-binding site of the BMP-LA motif (GGCGCC >TGATGA) of four wit-responsive fragments (CM1, CM2, CM3, and CM7) and one non-wit responsive fragment (CM9) to verify whether reporter activity was dependent on pMad binding. (A) CM9 showed no significant loss of reporter expression in the CM9Δmad mutant compared to the control. (B) All four wit-responsive fragments exhibited a significant loss of reporter expressing cells; however, this loss was less pronounced than the loss in wit mutants, apart from CM3. Full z-projections though the whole VNC are shown. Genotypes: All control and pMad-binding site mutant lines examined here were heterozygous (w;;CM#/+).
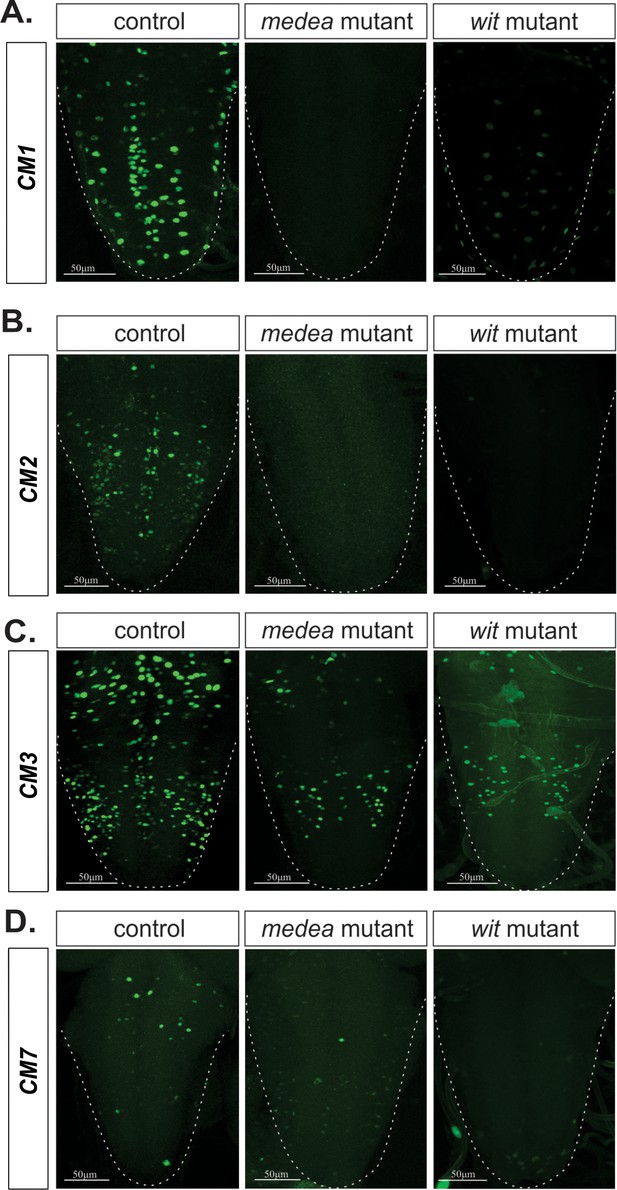
Medea is necessary for BMP-dependent activity of four wit-responsive BMP-LAs.
(A–D) Representative images of CM1, CM2, CM3, and CM7 control reporter expression compared to the same lines in a Medea and wit mutant backgrounds. In all cases, reporter expression loss in both mutants exhibit the same pattern. Full z-projections though the whole VNC are shown.Genotypes: All control lines examined here were heterozygous (w;;CM#/+); wit nulls (w;;CM#, witA12/witB11); Medea mutants (w;;CM#, Medc246/Med13).
Tables
Summary table of expression pattern and wit-responsiveness for BMP-LA containing DNA fragments tested in vivo.
The first column indicates the name for each of the cloned BMP-LA containing DNA fragments; these were sorted based on intensity and pattern of reporter expression. The second column provides information on reporter expression in the VNC, (more plus signs indicate higher intensity), and the final column provides details on expression pattern. The third and fourth columns indicate fragments that exhibited pMad and reporter co-expression, as well as the ones that were shown to be wit-responsive. Bolded letters indicate the enhancer fragments that were further tested for wit-responsiveness. The expression pattern was assessed in wandering third instar larvae.
| DNA fragment | VNC expression | Reporter/pMad stain overlap | Wit responsive | VNC expression details |
|---|---|---|---|---|
| CM5 | +++ | √ | √ | neurons and glia |
| CM1 | +++ | √ | √ | medial and lateral neurons |
| CM4 | +++ | √ | √ | medial and lateral neurons |
| CM3 | +++ | √ | √ | medial and lateral neurons |
| CM2 | ++ | √ | √ | medial and lateral neurons |
| CM7 | + | √ | √ | sparse |
| CM6 | + | √ | √ | sparse |
| CM8 | +++ | √ | - | medial neurons |
| CM9 | ++ | √ | - | lateral neurons |
| CM10 | + | √ | - | sparse |
| CM11 | ++ | - | - | neurons and glia |
| CM12 | + | - | - | sparse |
| CM13 | + | - | - | sparse |
| CM14 | + | - | - | sparse |
| CM15 | + | - | - | sparse |
| CM16 | + | - | - | sparse |
| CM17 | + | - | - | low intensity |
| CM18 | + | - | - | low intensity |
| CM19 | + | - | - | low intensity |
| CM20 | - | - | - | none |
| CM21 | - | - | - | none |
| CM22 | - | - | - | none |
| CM23 | - | - | - | none |
| CM24 | - | - | - | none |
Additional files
-
Supplementary file 1
Summary of wildtype and mutant Tv4-enhancer sequences tested for in vivo reporter activity.
(A) The full length Tv4-enhancer was isolated from Oregon R and the sequence is shown here. (B-D) We introduced substitution mutants into the full length Tv4-enhancer to generate three mutant enhancers that were tested in Figures 1 and 7. (E,F) The Homeodomain Response Element (HD-RE) and BMP-Response cis-Element (BMP-RCE) were identified previously within the Tv4-enhancer. The sequences shown in E, F are a 6× concatemer of the HD-RE (E) and 4× concatemer of the BMP-RCE (F) enhancers used to drive EYFP in the TvHD-RE-EYFP and TvBMP-RE-EYFP transgenes, respectively in Figure 2. Bolded black letters are the Mad and Medea binding sites of the BMP-LA. Bolded red letters are the substitution mutations introduced. Green letters are the Restriction enzyme sites or Restriction scar sites in the concatemeric sequences of E,F. (b) List of 128 BMP-LA motifs in the Drosophila genome with an average PhastCons score over 0.55. The BMP-LA motifs are ranked according to their average PhastCONS score. The location of each motif is indicated according to the August 2014 (BDGP Release 6 + ISO1 MT/dm6) Assembly. The first column indicates the assigned name for the BMP-LA as used throughout the manuscript or its location based on the nearest gene. Out of these 128 motifs, we found that 68% (87/128) is located within an intron or untranslated region (UTR), while the remaining 32% (41/128) are intergenic relative to the nearest gene. Blue BMP-LAs indicate those tested but not found to be BMP-dependent. Bolded red letters indicate the BMP-LA motifs in genomic fragments found to be BMP-dependent in third instar larva VNC. *Average evolutionary conservation score of a motif calculated based on the base-by-base conservation of each position in the motif. (c) Summary of BMP-LA containing genomic fragments tested for in vivo reporter activity. We prioritized 24 enhancers to test in vivo based on their proximity to genes expressed (4th column) in the third instar larvae VNC. Reporters were sorted based on the distance of the BMP-LA enhancer to the nearest CNS-expressed gene transcription start site (TSS). Bolded letters indicate the BMP-LA containing genomic fragments found to be wit-dependent in the VNC of third instar larvae. *Average evolutionary conservation score of a motif calculated on the base-by-base conservation of each position in the motif.
- https://cdn.elifesciences.org/articles/59650/elife-59650-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59650/elife-59650-transrepform-v2.pdf





