A simple and effective F0 knockout method for rapid screening of behaviour and other complex phenotypes
Figures
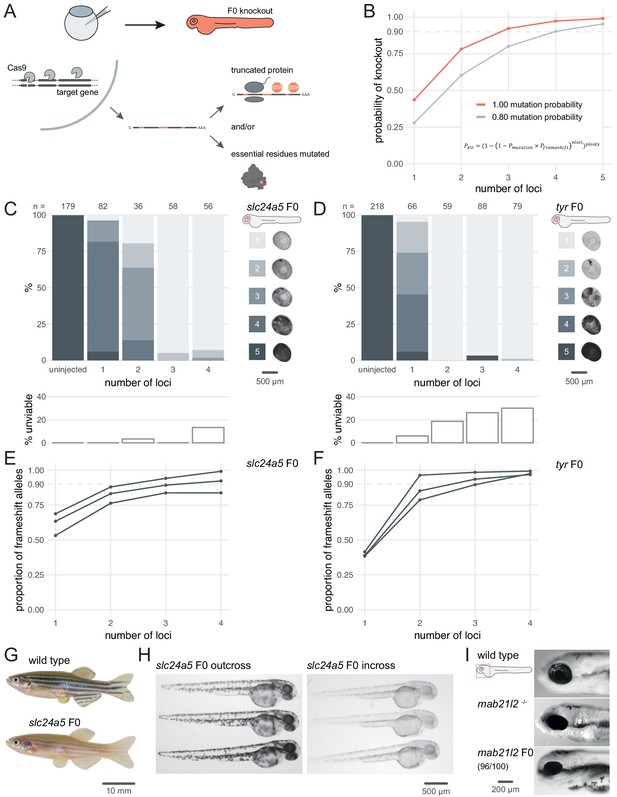
Three synthetic gRNAs per gene achieve over 90% biallelic knockouts in F0.
(A) Schematic of the F0 knockout strategy. Introduction of indels at multiple loci within the target gene leads to frameshift and premature stop codons and/or mutation of essential residues. (B) Simplified theoretical model of biallelic knockout probability as a function of number of targeted loci, assuming frameshift is the sole knockout mechanism. PKO, probability of biallelic knockout; Pmutation, mutation probability (here, 1.00 or 0.80); Pframeshift, probability of frameshift after mutation (0.66); nloci, number of targeted loci. (C–D) (top) Phenotypic penetrance as additional loci in the same gene are targeted. Pictures of the eye at 2 dpf are examples of the scoring method. (bottom) Unviability as percentage of 1-dpf embryos. (E–F) Proportion of alleles harbouring a frameshift mutation if 1, 2, 3, or 4 loci in the same gene were targeted, based on deep sequencing of each targeted locus. Each line corresponds to an individual animal. (G) 2.5-month wild-type and slc24a5 F0 knockout adult fish (n = 41). (H) 2-dpf progeny from slc24a5 F0 adults outcrossed to wild types (n = 283) or incrossed (n = 313). (I) Example of 3-dpf wild type, mab21l2u517 mutant, and mab21l2 F0 embryos (n = 96/100 injected). See also Figure 1—figure supplement 1.

Cas9 and gRNA achieve highest phenotypic penetrance at 1-to-1 ratio.
(A–B) (top) Phenotypic penetrance as gradually more Cas9 was injected; 1:6: 4.75 fmol Cas9, 1:3: 9.5 fmol Cas9, 1:2: 14.25 fmol Cas9, 1:1: 28.5 fmol Cas9. gRNA was kept constant at 28.5 fmol. Pictures of the eye at 2 dpf are examples of the scoring method (reproduced from Figure 1C,D). (bottom) Unviability as percentage of 1-dpf embryos.
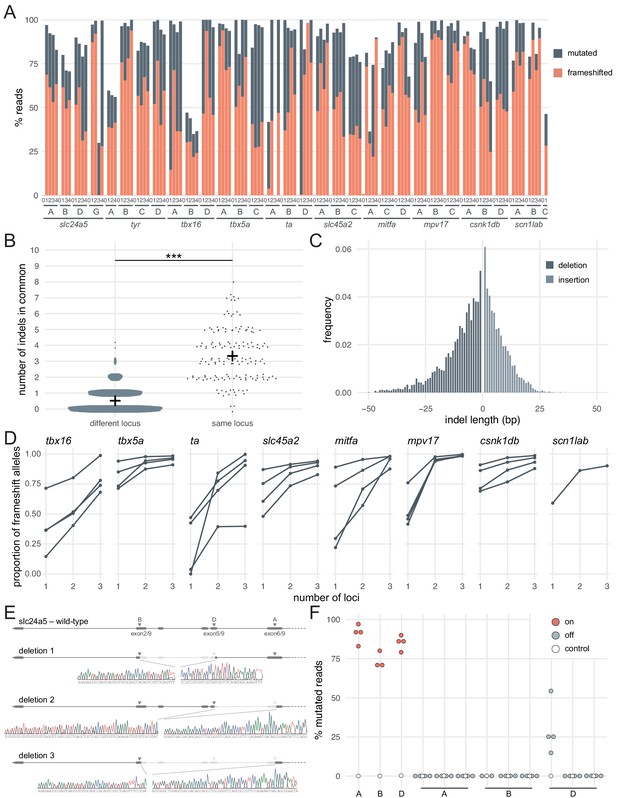
Deep sequencing of loci targeted in F0 embryos.
(A) Percentage of reads mutated (height of each bar, grey) and percentage of reads with a frameshift mutation (orange) at each gene, locus (capital letters refer to IDT’s database), larva (within each gene, the same number refers to the same individual animal; 0 is uninjected control). (B) Number of indels in common when intersecting the top 10 most frequent indels of two samples from different loci or from the same locus but different animals. Black crosses mark the means. *** p < 0.001; Welch’s t-test. (C) Frequency of each indel length (bp). Negative lengths: deletions, positive lengths: insertions. (D) Proportion of alleles harbouring a frameshift mutation if 1, 2, or three3 loci in the same gene were targeted, based on deep sequencing of each targeted locus. Each line corresponds to an individual animal. (E) Sanger sequencing of amplicons spanning multiple targeted loci of slc24a5. Arrowheads indicate each protospacer adjacent motif (PAM), capital letters refer to the crRNA/locus name. (F) Deep sequencing of the top 3 predicted off-targets of each slc24a5 gRNA (A, B, D). Each data point corresponds to one locus in one animal. Percentage of mutated reads at on-targets is the same data as in A (slc24a5 loci A, B, D). See also Figure 2—figure supplement 1.

Frequency at which each nucleotide around the targeted site was deleted.
Nucleotide positions are given in relation to the protospacer adjacent motif (PAM), whose position is set at 0 (0 = PAM nucleotide adjacent to the gRNA binding site). The nucleotide 4 bp before the PAM (−4) was the most frequently deleted (67% of all deletions removed it). n = 4016 unique deletions.
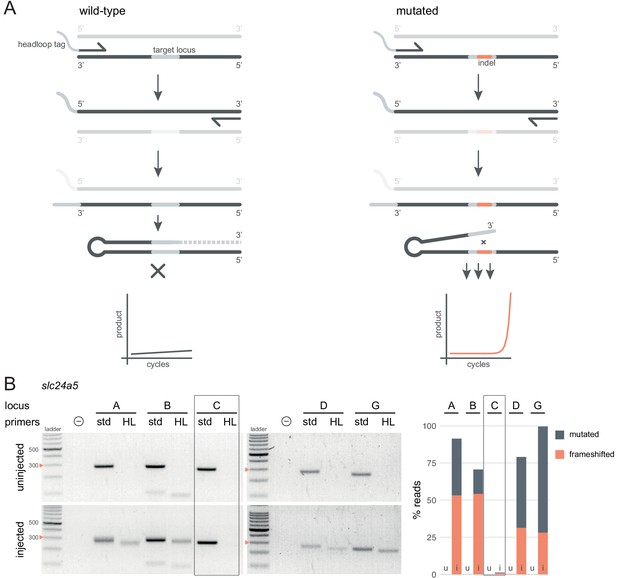
Headloop PCR is a rapid, sequencing-free method to confirm design of gRNAs.
(A) Principle of headloop PCR. A headloop tag complementary to the target locus is added to one PCR primer (here, to the forward primer). During PCR, the first elongation incorporates the primer and its overhang; the second elongation synthesises the headloop tag. (left) If the template is wild-type, the complementary tag base-pairs with the target locus and directs elongation (hatched sequence). The amplicon forms a hairpin secondary structure, which prevents its subsequent use as template. (right) If the targeted locus is mutated, the tag is no longer complementary to the locus. The amplicon remains accessible as a template, leading to exponential PCR amplification. (B) (left) Target loci (A, B, C, D, G) of slc24a5 amplified with the PCR primers used for sequencing (std, standard) or when one is replaced by a headloop primer (HL). Orange arrowheads mark the 300 bp ladder band. (right) Deep sequencing results of the same samples as comparison (reproduced from Figure 2A, except locus C); u, uninjected control, i, injected embryo. Framed are results for gRNA C, which repeatedly failed to generate many mutations. See also Figure 3—figure supplement 1, 2, 3.
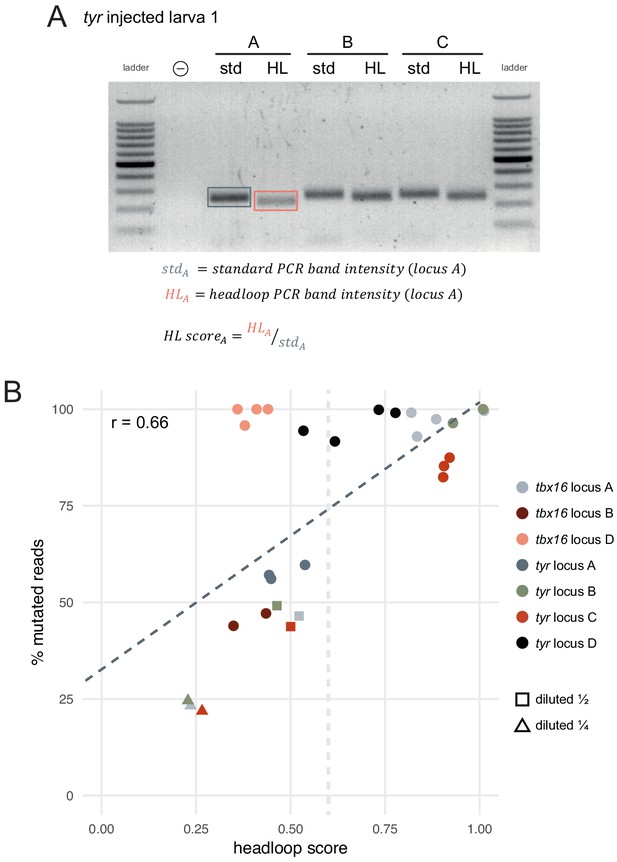
Headloop PCR score can predict the proportion of mutated alleles.
(A) The headloop score (HL score) for a given sample (here, tyr locus A larva 1) was calculated as the ratio between the headloop PCR band intensity (HL) and the standard PCR band intensity (std). As the standard PCR band intensity represents the sum of wild-type and mutated alleles, and the headloop PCR band intensity represents the mutated alleles only, the ratio approximates the proportion of mutated alleles in the sample. (B) Percentage of mutated reads (deep sequencing) as a function of headloop score. Each data point corresponds to one targeted locus in one F0 knockout animal. Some samples were artificially created to simulate mediocre gRNAs by mixing genomic DNA from an injected embryo with genomic DNA from the uninjected control in a 1:1 (diluted ½, squares) or 1:3 ratio (diluted ¼, triangles). For these, the percentage of mutated reads was not measured by deep sequencing but estimated by dividing the percentage of mutated reads of the original sample by 2 (diluted ½) or 4 (diluted ¼). The dark grey dashed line is the line of best fit by linear regression: proportion of mutated reads = 0.33 + 0.69 × headloop score; R2 = 0.44. Headloop score is a significant predictor of the proportion of mutated reads, p < 0.001 (linear regression); r = 0.66 (Pearson). The light grey dashed vertical line indicates a tentative threshold at headloop score 0.6 to discriminate mediocre gRNAs. Two tbx16 locus B samples were excluded because they appeared degraded on the agarose gel. n = 29 samples; each data point is the mean of at least three technical replicates.

Headloop PCR is sensitive to small deletions.
(A) 5’–3’ genomic region of the wild-type and mutated alleles of the stable mutant lines we used to test the detection of small deletions by headloop PCR. apoea: 1 bp deletion followed by a T > A transversion. cd2ap: 2 bp deletion. Framed: headloop tag; grey underlay: headloop tag binding site. (B) For each mutant line (apoea, cd2ap), 32 embryos issued from a heterozygous to wild-type outcross were genotyped (around 50% of embryos expected to be heterozygous and the rest wild-type). Bands on agarose gels alternate between standard PCR (std) and headloop PCR (HL). The genotype (wild-type or heterozygous) was called based on the presence or absence of the headloop PCR band. If the embryo was wild-type, headloop PCR did not generate a product. If the embryo was heterozygous, headloop PCR generated a product, as half of the template DNA was mutated. (C) All genotype calls by headloop PCR were then verified by Sanger sequencing. Samples traces are included here. For the standard PCR heterozygous samples, two Sanger traces run alongside each other past the mutation site, as templates from both the wild-type and the heterozygous alleles were sequenced. Conversely, the Sanger trace for the headloop PCR heterozygous samples remains unique past the mutation as only the mutated allele was sequenced, which confirms that the wild-type allele was not amplified. (D) Summary of genotyping results by headloop PCR and Sanger sequencing for the two clutches of 32 embryos.
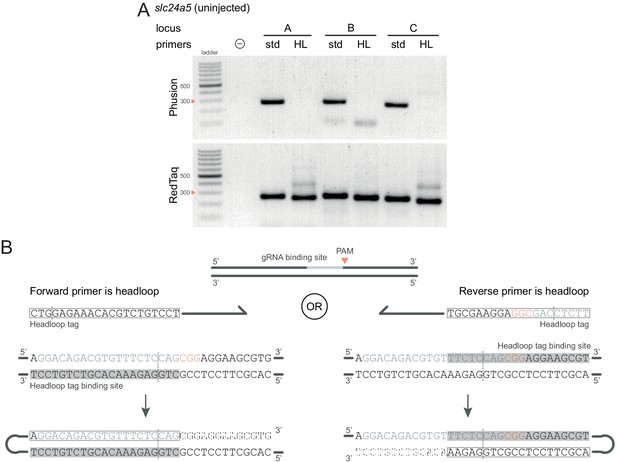
Technical considerations for headloop PCR.
(A) Comparison between results obtained with a proofreading (Phusion Hot Start II) or a non-proofreading (REDTaq) DNA polymerase for three target loci (A, B, C) of slc24a5 amplified with the PCR primers used for sequencing (std, standard) or when one is replaced by a headloop primer (HL). Samples were uninjected controls. Orange arrowheads mark the 300 bp ladder band. (B) Headloop primer designs, using slc24a5 locus G as an example. To perform headloop PCR, the forward or reverse primer from a previously verified primer pair is modified with a 5’ tag sequence and used in conjunction with its unmodified partner. The sequence of the headloop tag is selected so that the predicted Cas9 cleavage site (dashed line) is located towards the 5’-end of the tag. (left) If the modified primer and the gRNA binding site are in the same direction (headloop tag is added to the forward primer and gRNA binding site is on the 5’–3’ genomic strand), the reverse-complement of the gRNA binding site is sufficient (grey underlay). (right) If the modified primer and the gRNA binding site are in opposite directions (headloop tag is added to the reverse primer while gRNA binding site is on the 5’–3’ genomic strand), a sequence that includes the protospacer adjacent motif (PAM, orange font) and shifted from the gRNA binding site is sufficient. In both cases, after second strand elongation, the tag is able to bind the target sequence and direct elongation (hatched sequences) to form a hairpin, suppressing exponential amplification of the wild-type haplotype. Framed: headloop tag; grey font: gRNA binding site; grey underlay: headloop tag binding site.
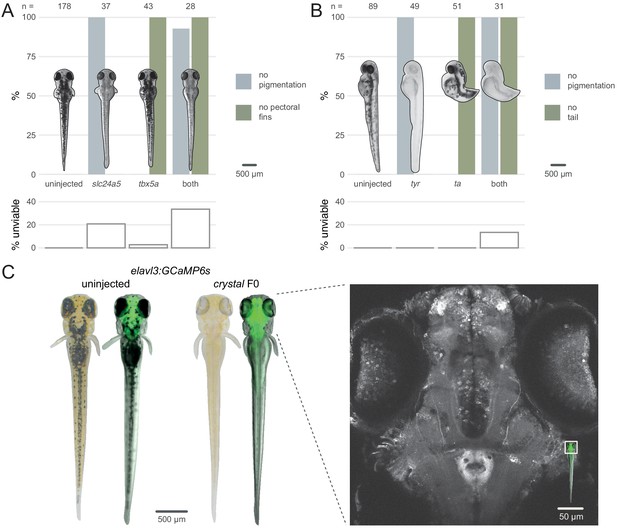
Multiple gene knockout in F0.
(A) Penetrance of single (slc24a5, tbx5a) and combined (both) biallelic knockout phenotype(s) in F0. Pictures of example larvae were taken at 3 dpf. No pigmentation refers to embryos clear of eye pigmentation at 2 dpf, as in Figure 1C. Pectoral fins were inspected at 3 dpf. (bottom) Unviability as percentage of 1-dpf embryos. (B) Penetrance of single (tyr, ta) and combined (both) biallelic knockout phenotype(s) in F0. Pictures of example larvae were taken at 2 dpf. (bottom) Unviability as percentage of 1-dpf embryos. (C) (left) Pictures of example elavl3:GCaMP6s larvae at 4 dpf. Left was uninjected; right was injected and displays the crystal phenotype. Pictures without fluorescence were taken with illumination from above to show the iridophores, or lack thereof. (right) Two-photon GCaMP imaging (z-projection) of the elavl3:GCaMP6s, crystal F0 larva shown on the left. (inset) Area of image (white box). See also Figure 4—video 1, Figure 4—video 2.
Two-photon, live imaging of the brain of an elavl3:GCaMP6s, crystal F0 4-dpf larva.
(inset) Area of image (white box).
Two-photon, live imaging of the right eye of an elavl3:GCaMP6s, crystal F0 4-dpf larva.
(inset) Area of image (white box).
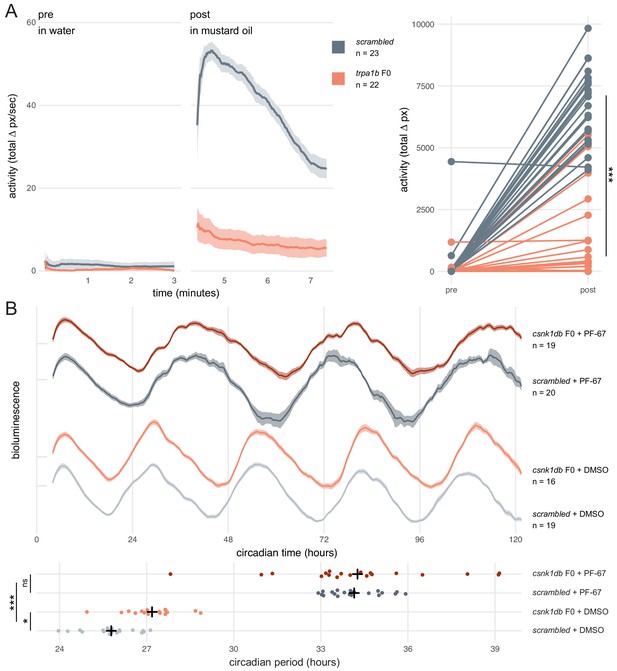
Dynamic, continuous traits are accurately assessed in F0 knockouts.
(A) Escape response to mustard oil in trpa1b F0 knockouts. (left) Activity (total Δ pixel/second) of scrambled controls and trpa1b F0 knockout larvae at 4 dpf. Pre: 3-min window before transfer to 1 µM mustard oil. Post: 3-min window immediately after. Traces are mean ± standard error of the mean (SEM). (right) Total activity (sum of Δ pixel/frame over the 3-min window) of individual larvae before and after transfer to 1 µM mustard oil. *** p < 0.001 (Δ total activity scrambled vs trpa1b F0); Welch’s t-test. (B) Circadian rhythm quantification in csnk1db F0 knockout larvae. (top) Timeseries (detrended and normalised) of bioluminescence from per3:luciferase larvae over five subjective day/night cycles (constant dark). Circadian time is the number of hours after the last Zeitgeber (circadian time 0 9 am, morning of 4 dpf). DMSO: 0.001% dimethyl sulfoxide; PF-67: 1 µM PF-670462. Traces are mean ± SEM. (bottom) Circadian period of each larva calculated from its timeseries. Black crosses mark the population means. ns, p = 0.825; * p = 0.024; *** p < 0.001; pairwise Welch’s t-tests with Holm’s p-value adjustment. See also Figure 4—video 1.
Mustard oil assay on 4-dpf trpa1b F0 knockout larvae.
For illustration, 1 µM mustard oil was applied to Petri dishes with n = 10 trpa1b F0 knockout larvae and n = 10 scrambled-injected control larvae (final concentration 0.66 µM mustard oil). Data in Figure 5A were collected during a separate experiment in which larvae were tracked individually.
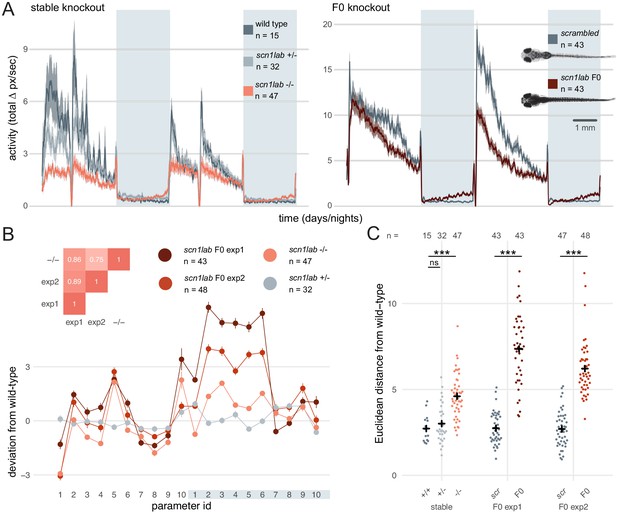
Multi-parameter behavioural phenotypes are closely replicated in F0 knockouts.
(A) Activity (total Δ pixel/second) of larvae across 2 days (14 hr each, white background) and two nights (10 hr each, grey background). Traces are mean ± SEM. (left) Stable scn1labΔ44 mutant line, from 5 to 7 dpf. The drops in activity in the middle of each day is an artefact caused by topping-up the water. (right) scn1lab F0 knockout, from 6 to 8 dpf. This replicate is called scn1lab F0 experiment 1 in B and C. (inset) Pictures of example scrambled-injected control and scn1lab F0 larvae at 6 dpf. (B) Behavioural fingerprints, represented as deviation from the paired wild-type mean (Z-score, mean ± SEM). 10 parameters describe bout structure during the day and night (grey underlay). Parameters 1–6 describe the swimming (active) bouts, 7–9 the activity during each day/night, and 10 is pause (inactive bout) length. (inset) Pairwise correlations (Pearson) between mean fingerprints. (C) Euclidean distance of individual larvae from the paired wild-type or scrambled-injected (scr) mean. Black crosses mark the population means. ns, p > 0.999; *** p < 0.001; pairwise Welch’s t-tests with Holm’s p-value adjustment. See also Figure 6—figure supplement 1.
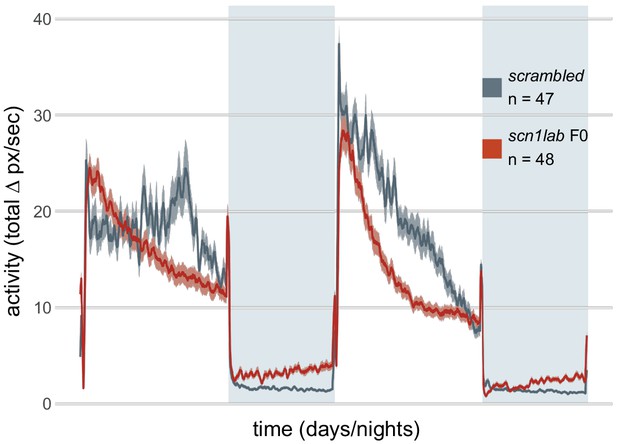
Activity (total Δ pixel/second) of scn1lab F0 knockout larvae across 2 days (14 hr each, white background) and two nights (10 hr each, grey background), from 6 to 8 dpf.
This replicate is called scn1lab F0 experiment 2 in Figure 6C,D.
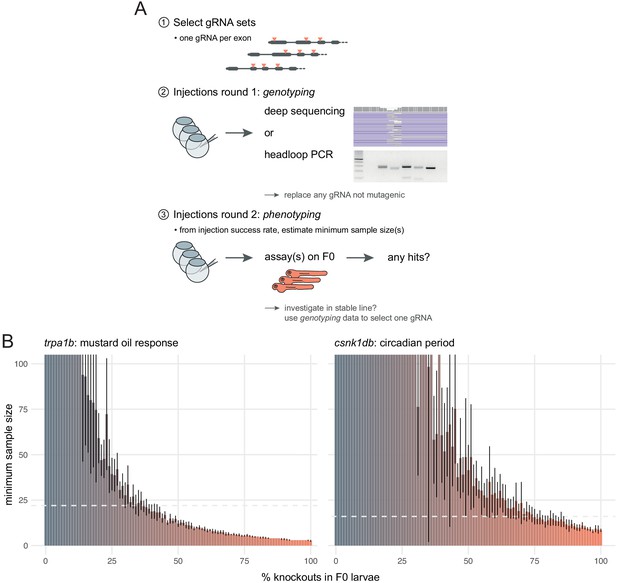
Recommendations for F0 knockout screens.
(A) Suggestion for the design of an F0 screen, based on a three-step process: (1) selection of gRNAs; (2) verification that all gRNAs are mutagenic; (3) phenotyping. During round 1 (step 2), we recommend targeting a pigmentation gene such as slc24a5 to quantify success rate at injections before estimating minimum samples sizes and commencing the screen. (B) Minimum sample size to detect a phenotype at 0.8 statistical power and 0.05 significance level as more knockouts are present in the population of F0 larvae. Based on 10 simulations with 100 animals in each group (scrambled control, F0 knockout). Mean ± standard deviation across the 10 simulations. (left) Minimum sample sizes to detect the lack of response to mustard oil of the trpa1b knockouts. Dashed line indicates the real sample size of the experiment (n = 22, Figure 5A). (right) Minimum sample sizes to detect the lengthened circadian period of csnk1db knockouts. Dashed line indicates the real sample size of the experiment (n = 16, Figure 5B).

Most mutations are found at low frequencies in individual animals.
Frequency of each mutation in individual F0 knockout embryos (samples). Mutations are ranked from the most frequent in the embryo (a 3-bp deletion in gene ta, locus D, embryo 1 was present in 602/605 of the reads) to the least frequent (a 11-bp insertion in gene csnk1db, locus B, embryo 4 was present in 1/3189 of the reads). n = 7015 unique mutations.

scn1lab F0 knockouts do not have consistently more severe behavioural phenotypes than stable scn1lab mutant lines.
Total time active, represented as deviation from the paired wild-type (+/+) or scrambled-injected (scr) mean (Z-score), during the day (left) or the night (right). The tracking spanned two days and two nights (as in Figure 6A), so each animal contributed two data points to each plot. F0 exp1: scn1lab F0 experiment 1, same experiment as in Figure 6A right; F0 exp2: scn1lab F0 experiment 2, same experiment as in Figure 6—figure supplement 1; Δ44: stable scn1labΔ44 mutant line, same experiment as in Figure 6A left; didy: stable scn1lab didy mutant line. Black crosses mark the population means. (day, in order displayed) F0 exp1: *** p < 0.001; F0 exp2: *** p < 0.001; Δ44: ns p > 0.999, *** p < 0.001; didy: * p = 0.021, ** p = 0.002. (night, in order displayed) F0 exp1: ns p > 0.999; F0 exp2: ** p < 0.004; Δ44: ns p = 0.186, ns p > 0.999; didy: ns p > 0.999, *** p < 0.001. Statistics by pairwise Welch’s t-tests with Holm’s p-value adjustment.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Danio rerio) | slc24a5 | Ensembl | ENSDARG00000024771 | |
| Gene (D. rerio) | tyr | Ensembl | ENSDARG00000039077 | |
| Gene (D. rerio) | mab21l2 | Ensembl | ENSDARG00000015266 | |
| Gene (D. rerio) | tbx16 | Ensembl | ENSDARG00000007329 | |
| Gene (D. rerio) | tbx5a | Ensembl | ENSDARG00000024894 | |
| Gene (D. rerio) | ta (tbxta) | Ensembl | ENSDARG00000101576 | |
| Gene (D. rerio) | slc45a2 | Ensembl | ENSDARG00000002593 | |
| Gene (D. rerio) | mitfa | Ensembl | ENSDARG00000003732 | |
| Gene (D. rerio) | mpv17 | Ensembl | ENSDARG00000032431 | |
| Gene (D. rerio) | trpa1b | Ensembl | ENSDARG00000031875 | |
| Gene (D. rerio) | csnk1db | Ensembl | ENSDARG00000006125 | |
| Gene (D. rerio) | scn1lab | Ensembl | ENSDARG00000062744 | |
| Genetic reagent (D. rerio) | Tg(elavl3:GCaMP6s)a13203 | PMID:28892088 | ZFIN ID: ZDB-ALT-180502–2 | Kim et al., 2017 |
| Genetic reagent (D. rerio) | mitfaw2 (nacre) | PMID:10433906 | ZFIN ID: ZDB-ALT-990423–22 | Lister et al., 1999 |
| Genetic reagent (D. rerio) | Tg(per3:luc)g1 | PMID:15685291 | ZFIN ID: ZDB-ALT-050225–2 | Kaneko and Cahill, 2005 |
| Genetic reagent (D. rerio) | Tg(elavl3:EGFP)knu3 | PMID:11071755 | ZFIN ID: ZDB-ALT-060301–2 | Park et al., 2000 |
| Genetic reagent (D. rerio) | mab21l2u517 | PMID:32930361 | mutant line | Wycliffe et al., 2020 |
| Genetic reagent (D. rerio) | scn1labΔ44 | This study | mutant line | Available from Hoffman lab |
| Sequence-based reagent | Alt-R CRISPR-Cas9 crRNAs | IDT | see Supplementary file 1 | |
| Sequence-based reagent | Alt-R CRISPR-Cas9 Negative Control crRNA #1 | IDT | Catalog #: 1072544 | see Supplementary file 1 |
| Sequence-based reagent | Alt-R CRISPR-Cas9 Negative Control crRNA #2 | IDT | Catalog #: 1072545 | see Supplementary file 1 |
| Sequence-based reagent | Alt-R CRISPR-Cas9 Negative Control crRNA #3 | IDT | Catalog #: 1072546 | see Supplementary file 1 |
| Sequence-based reagent | Alt-R CRISPR-Cas9 tracrRNA | IDT | Catalog #: 1072532 | |
| Sequence-based reagent | PCR primers | Thermo Fisher | see Supplementary file 1 | |
| Peptide, recombinant protein | Alt-R S.p. Cas9 Nuclease V3 | IDT | Catalog #: 1081058 | |
| Chemical compound, drug | Mustard oil (allyl isothiocyanate) | Sigma-Aldrich | Catalog #: W203408 | |
| Chemical compound, drug | Beetle luciferin | Promega | Catalog #: E1601 | |
| Chemical compound, drug | PF-670462 | Sigma-Aldrich | Catalog #: SML0795 | |
| Software, algorithm | ampliCan | PMID:30850374 | bioconductor.org/packages/release/bioc/html/amplican.html | |
| Software, algorithm | BioDare2 | PMID:24809473 | biodare2.ed.ac.uk | |
| Software, algorithm | ZebraLab | ViewPoint Behavior Technology | viewpoint.fr/en/p/software/zebralab-zebrafish-behavior-screening | |
| Software, algorithm | MATLAB scripts for behaviour analysis: Vp_Extract.m and Vp_Analyse.m | PMID:32241874 | Scripts included in the GitHub and Zenodo repositories (see Data/resource sharing) | |
| Software, algorithm | R packages | CRAN | see Supplementary file 2 | |
| Software algorithm | Command line packages | Conda | see Supplementary file 2 | |
| Software algorithm | MATLAB toolboxes | MathWorks | see Supplementary file 2 | |
| Software, algorithm | R v3.6.2 | CRAN | r-project.org | |
| Software, algorithm | MATLAB R2018a | MathWorks | mathworks.com/products/matlab.html |
Additional files
-
Supplementary file 1
Sequences.
1) Details about crRNAs and PCR primers used for MiSeq. 2) Details about off-target loci sequenced and PCR primers used for MiSeq. 3) Headloop PCR: sequences of modified primers (i.e. primer with 5′ headloop tag). 4) Sequences of the standard PCR and headloop PCR primers used to genotype the apoea and cd2ap alleles. 5) scn1labΔ44: ZFNs binding sequences, PCR primers and allele.
- https://cdn.elifesciences.org/articles/59683/elife-59683-supp1-v1.xlsx
-
Supplementary file 2
Packages.
1) Packages used in R scripts. 2) Packages used in command line. 3) Toolboxes used in MatLab scripts.
- https://cdn.elifesciences.org/articles/59683/elife-59683-supp2-v1.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59683/elife-59683-transrepform-v1.docx






