Activation of a neural stem cell transcriptional program in parenchymal astrocytes
Figures
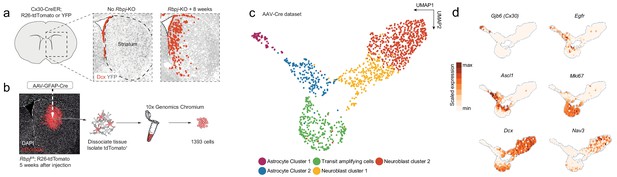
Neurogenesis by striatal astrocytes can be reconstructed using single-cell RNA sequencing.
(a) Deletion of the gene encoding the Notch-mediating transcription factor Rbpj activates a latent neurogenic program in striatal astrocytes (Magnusson et al., 2014). Nuclei of Dcx+ neuroblasts are indicated by red dots. Not all striatal astrocytes undergo neurogenesis, shown by the restricted distribution of Dcx+ neuroblasts and the fact that many recombined astrocytes (gray) remain even 2 months after Rbpj deletion. (b) We performed single-cell RNA sequencing using two protocols. For the AAV-Cre dataset, we deleted Rbpj exclusively in striatal astrocytes using a Cre-expressing AAV and sequenced the transcriptomes of recombined cells five weeks later. (c) Dimensionality reduction using UMAP captures the progression from astrocytes, through proliferating transit-amplifying cells, to neuroblasts. Panel (d) shows markers for the different maturation stages.
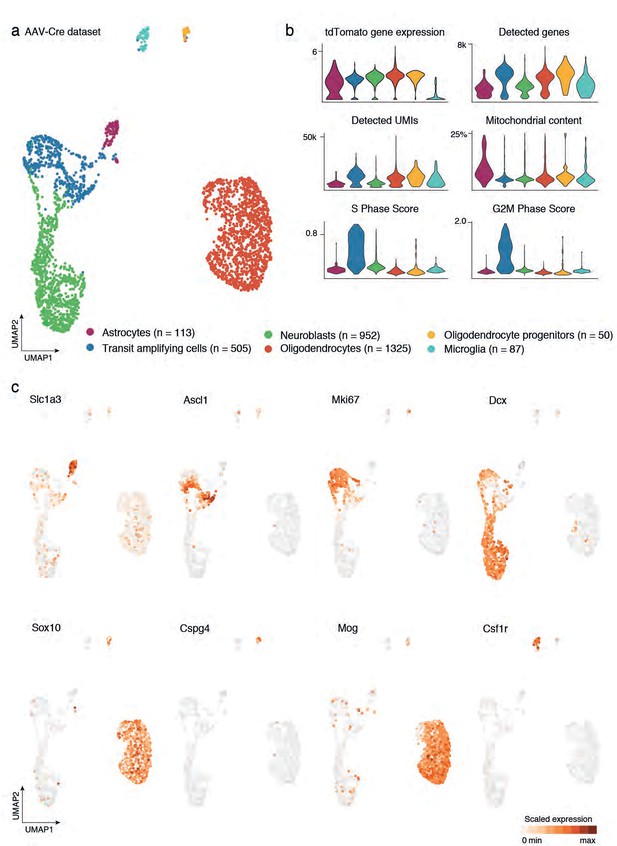
Cell census from AAV-Cre dataset after local recombination of striatal cells.
(a) UMAP visualization indicates the cell types composing the full AAV-Cre dataset. This sample encompasses astrocytes and their neurogenic progeny, along with cells from the oligodendroglial lineage, and microglia. Total number of cells detected for each cluster is reported in the legend. (b) Violin plots display the gene expression level of tdTomato, which was used as a means to select cells recombined after viral injection. In addition, we report per-cluster numbers of detected genes and UMIs, as well as the percentage of mitochondrial genes, as a measure of quality of the cells. Finally, we report the cell cycle score (S and G2M phase score) to indicate cells, such as transit amplifying cells that are actively dividing. (c) Expression of classical cell type markers are displayed on the UMAP plot to discriminate between populations indicated in (a).
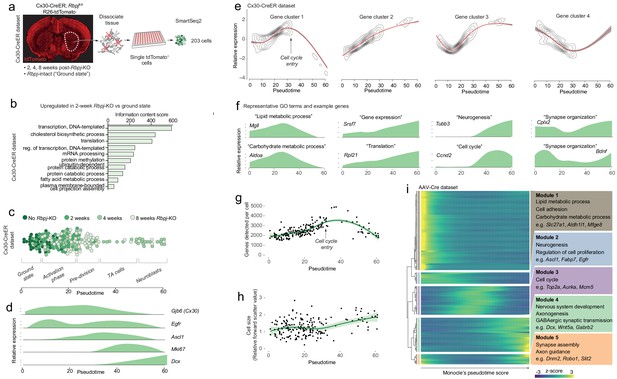
Neurogenesis by astrocytes is dominated by early transcriptional changes associated with metabolism and gene expression.
(a) We prepared a second single-cell RNA sequencing dataset using SmartSeq2. This dataset consists of cells isolated from Cx30-CreER; Rbpjfl/fl mice at 2, 4 and 8 weeks after Rbpj deletion, and ground-state astrocytes with intact Rbpj. (b) Two weeks after Rbpj deletion, astrocytes have upregulated genes mainly associated with metabolism and gene expression. (c) Dimensionality reduction using Monocle organizes astrocytes and their progeny along pseudotime (see Figure 2—figure supplement 2 for cell state definitions). (d) Pseudotime captures the neurogenic trajectory of astrocytes, as shown by plotting the expression of classical neurogenesis genes along this axis. (e) Gene clustering and GO analysis shows distinctive dynamics of functional gene groups with example genes shown in (f). Genes involved in lipid and carbohydrate metabolism are upregulated initially but downregulated as cells enter transit-amplifying divisions. Genes involved in transcription and translation are expressed at steadily increasing levels. Accordingly, the number of genes detected per cell increases initially (g) – an increase not explained by a mere increase in cell size (h). A gene cluster with remarkable dynamics contains genes involved in synapse organization (e), presumably important for both astrocytes and neurons. (i) All these findings are supported by a pseudotemporal analysis of the AAV-Cre dataset. The AAV-Cre dataset, however, only contains cells from the 5-week time point and thus does not show changes that happen early in the neurogenic program. (TA cells, transit-amplifying cells).
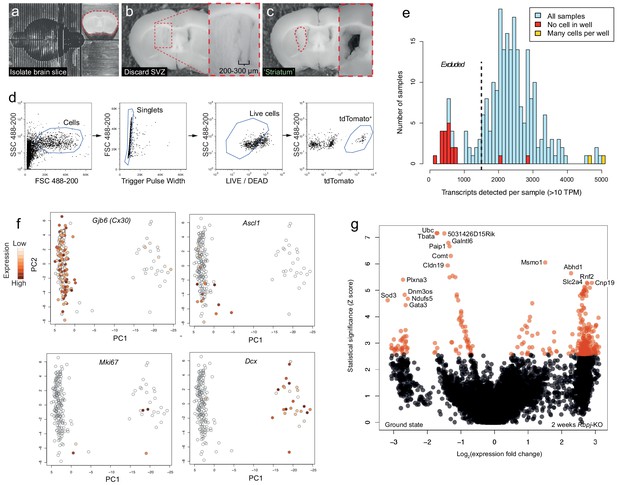
Isolation of tdTomato+ cells for single-cell RNA sequencing and initial analyses.
(a–c) The dissection procedure for isolation of brain regions is shown. (a) First, a ~ 2 mm thick slice was isolated from the mouse brain. (b) The SVZ, including 200–300 µm of the medial striatum, was carefully removed and discarded before the striatum was isolated (c; inset shows region after removal of striatum piece). (d) Striatal cells were sorted based on LIVE/DEAD marker and tdTomato expression. (e) All wells with no or multiple cells were excluded from further analysis. In addition, all samples with <1500 unique transcripts detected were excluded, a conservative exclusion criterion based on the number of transcripts detected in wells into which no cell was sorted. (f) Principal component analysis of striatal cells shows the existence of astrocytes (Gjb6+), activated astrocytes and transit-amplifying cells (Ascl1+, Mki67+) and neuroblasts (Dcx+). Note the relatively small number of transit-amplifying cells, probably a result of transit-amplifying cells only existing for a short time, and at only one of our experimental time points. (g) A volcano plot illustrates genes differentially expressed two weeks after Rbpj deletion. Red data points lie above the statistical significance threshold 2.576 (a = 0.005).
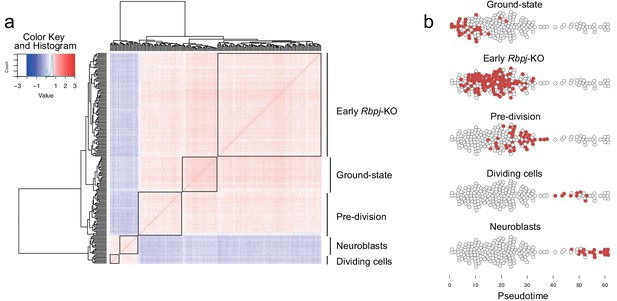
Hierarchical clustering of striatal astrocytes undergoing neurogenesis.
(a) A correlation heatmap of all 203 striatal cells revealed five cell groups. (b) These groups were distributed sequentially along pseudotime and thus likely represent distinct stages along neurogenesis by astrocytes. These groups were used to generate Monocle’s gene clustering algorithm (see Figure 2c): We performed differential expression analysis of each cell group versus the rest, and used the resulting five gene lists as input to the gene clustering algorithm (see Materials and methods).
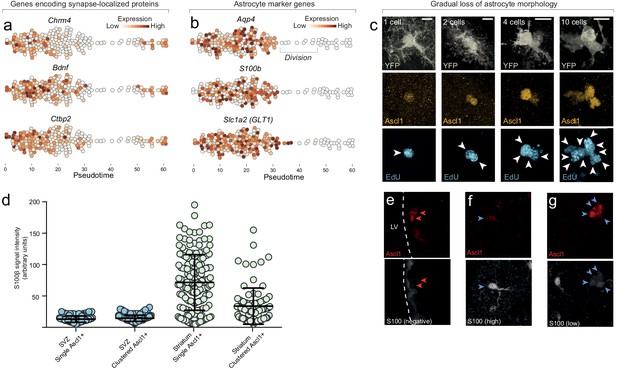
Astrocytes gradually lose astrocyte features as they enter the neurogenic program.
(a) Genes encoding proteins localized to synapses are downregulated early upon entry into the neurogenic program, suggesting that some normal astrocyte functions are immediately compromised as neurogenesis is initiated. Interestingly, these same synapse-associated genes are upregulated again as cells differentiate into neuroblasts, suggesting that they are important for synapse maintenance in both astrocytes and neurons. (b) Common astrocyte marker genes such as Aqp4, S100b and Slc1a2, are maintained in astrocytes approximately until they enter transit-amplifying divisions. (c) Neurogenic astrocytes maintain normal astrocyte morphology even as they have upregulated Ascl1 and initiated DNA synthesis in preparation for transit-amplifying divisions (one-cell stage. Note that EdU, which was administered for the last 2 weeks prior to sacrifice, has been incorporated into the DNA of this single astrocyte. This indicates that astrocytes initiate S phase while still retaining astrocyte features.). As transit-amplifying divisions occur, astrocytic processes are gradually lost by the dividing cells (‘2–10 cells’. See also Video 1). (d, e) The astrocyte marker S100β is not expressed by neurogenic-lineage cells in the SVZ, neither in single Ascl1+ cells or clustered transit-amplifying cells ([red arrowheads]; each data point in (d) is one cell). In contrast, many striatal astrocytes express high levels of S100β (d, f [blue arrowheads]). Consequently, many (but not all) striatal transit-amplifying cells retain low amounts of residual S100β protein (d, g [blue arrowheads]) as remnants of their origin as parenchymal astrocytes. Thus, lingering S100β protein can be used as a short-term lineage-tracing marker for transit-amplifying cells derived from striatal astrocytes. (LV, lateral ventricle). Scale bar (c): 10 μm.
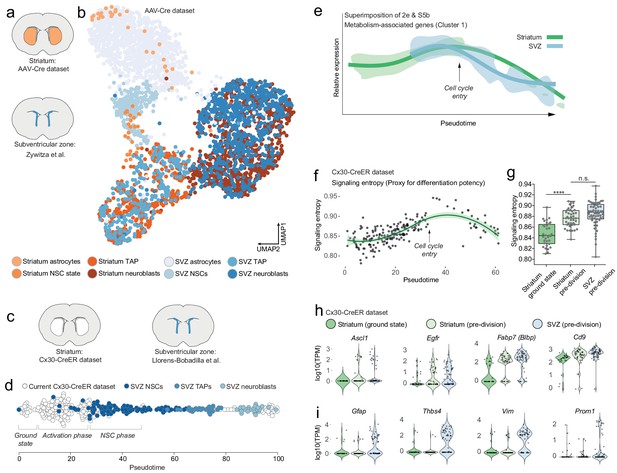
Striatal astrocytes are upstream of SVZ neural stem cells in the neurogenic lineage.
(a) We analyzed our AAV-Cre dataset together with a previously published dataset of SVZ cells. (b) All maturational stages along the striatal neurogenic trajectory cluster together with the corresponding stages in the SVZ. The AAV-Cre dataset has few ground-state astrocytes because all cells were isolated five weeks after Rbpj deletion. (c) We therefore compared our Cx30-CreER dataset with a previously published dataset of SVZ cells using Monocle’s pseudotime reconstruction (d). This analysis suggests that striatal astrocytes are located upstream of SVZ stem cells in the neurogenic lineage, as if representing highly quiescent neural stem cells that, upon Rbpj deletion, initiate a neurogenic program very similar to that of the SVZ stem cells. (e) Overlaying the expression pattern of metabolism-associated genes (compare Figure 2e, S5b) shows that striatal astrocytes initiate neurogenesis with a phase of metabolic gene upregulation, whereas SVZ stem cells permanently reside on this metabolic peak. (f) Signaling entropy, a reliable proxy for differentiation potency, increases in Rbpj-deficient astrocytes to reach the same levels as in SVZ stem cells (g). (h) Prior to entering transit-amplifying divisions, striatal astrocytes are in their most stem cell-like state, having upregulated many markers of neural stem cells (each dot is one cell. Note the bursting expression pattern of many of these genes). (i) Some classical markers, however, are not upregulated.
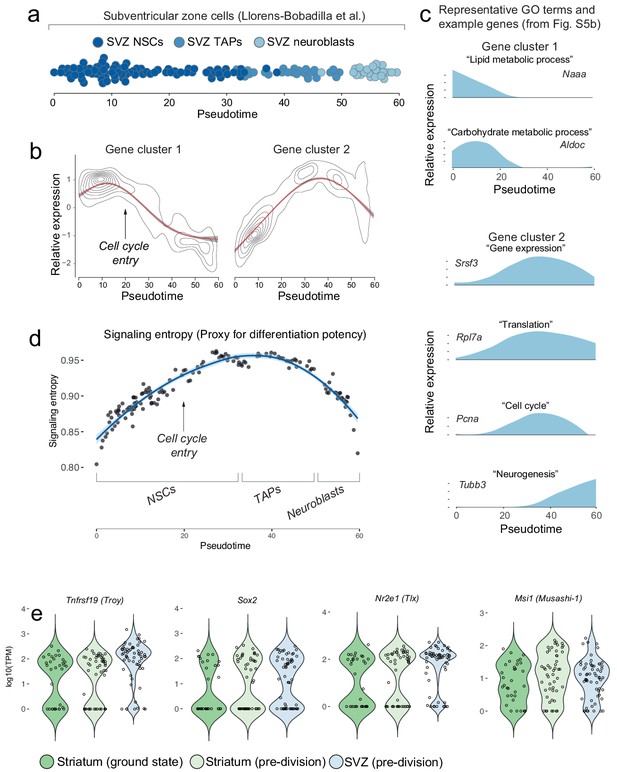
Gene clustering analysis of SVZ neural stem cells and their progeny from the dataset published by Llorens-Bobadilla et al.
We analyzed a previously published dataset of SVZ neural stem cells and their progeny. (a) Monocle reconstructs SVZ neurogenesis along pseudotime (cells are colored based on cell groups defined in Llorens-Bobadilla et al.). (b–c) An analysis using Monocle’s gene clustering algorithm shows that, as SVZ stem cells enter transit-amplifying divisions, genes associated with lipid and carbohydrate metabolism are downregulated, whereas genes associated with gene expression are upregulated. (d) Signaling entropy, an approximation of a cell’s differentiation potency, increases as SVZ stem cells are activated and initiate transit-amplifying divisions. (e) Some classical markers of neural stem cells are already expressed by ground-state striatal astrocytes and their RNA levels do not change after Rbpj deletion.
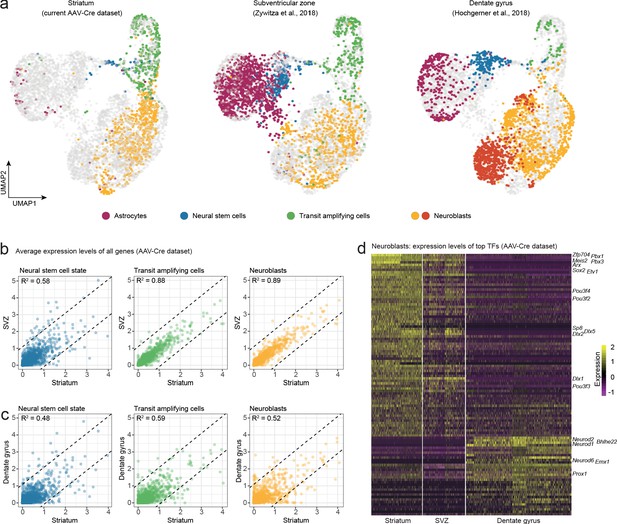
Neuroblasts generated by striatal astrocytes are more similar to SVZ neuroblasts than to DG neuroblasts.
We analyzed our AAV-Cre dataset together with two previously published datasets from the SVZ and DG. (a) Cells from each maturational stage cluster together regardless of origin, highlighting that neurogenic cells transition through the same general stages in all three regions. (b) We performed pairwise comparisons between brain regions by plotting the average expression levels of all genes. Striatal astrocytes in the stem cell-like pre-division state are only moderately similar to SVZ neural stem cells (b; R2 = 0.58) but become more similar as they develop into transit-amplifying cells (b; R2 = 0.88) and neuroblasts (b; R2 = 0.89; b), indicating that striatal astrocytes and SVZ stem cells converge on nearly identical neurogenic programs. (c) In contrast, stem cells in the DG and their progeny never acquire more than moderate similarity to the striatal cells. (d) A heat map shows some of the top transcription factors that define the similarities and differences among neuroblasts. This suggests that striatal astrocytes are primed to generate very similar neuronal subtypes as the SVZ stem cells.
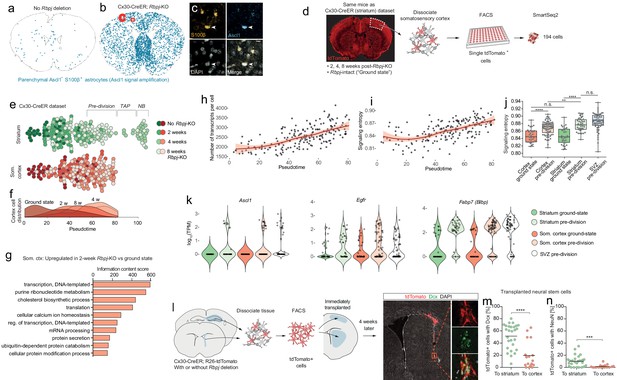
Astrocytes outside the striatum initiate an incomplete neurogenic program in response to Rbpj deletion.
(a–c) Four weeks after Rbpj deletion, astrocytes throughout the brain have upregulated low levels of Ascl1 protein, only detectable by using fluorescence-signal amplification (SVZ cells not analyzed or visualized for clarity). (d) We sequenced RNA from somatosensory cortical astrocytes, isolated from the same mice as the striatal cells. (e) We reconstructed the neuronal differentiation trajectory of striatal and somatosensory cortical cells together using Monocle. This analysis reveals that the cortical astrocytes also progress along this trajectory but all stall prior to transit-amplifying divisions. (f) This lineage progression is more clearly visible in a plot showing cell distribution along pseudotime. (g) Rbpj-deficient cortical astrocytes upregulate genes primarily related to transcription, translation and lipid metabolism, just like striatal astrocytes. (h) The number of transcripts detected per cell also increases. (i–j) At the same time, their signaling entropy increases to nearly the same levels as in Rbpj-deficient striatal astrocytes or SVZ stem cells, indicating an increase in differentiation potency. (k) When cortical astrocytes halt their neurogenic development, they are in their most neural stem cell-like state, having upregulated classical markers of neural stem cells to similar levels as in SVZ stem cells, at least at the RNA level. (l) We isolated neural stem cells from Cx30-CreER; tdTomato mice and immediately transplanted them to litter mates. Four weeks later, a lower proportion of transplanted cells had matured into neuroblasts (m) and neurons (n) in the cortex than in the striatum (each data point represents one recipient mouse). This indicates that the cortical environment is less permissive to neurogenesis than that of the striatum, possibly explaining the failure of astrocytes to complete the neurogenic program there.
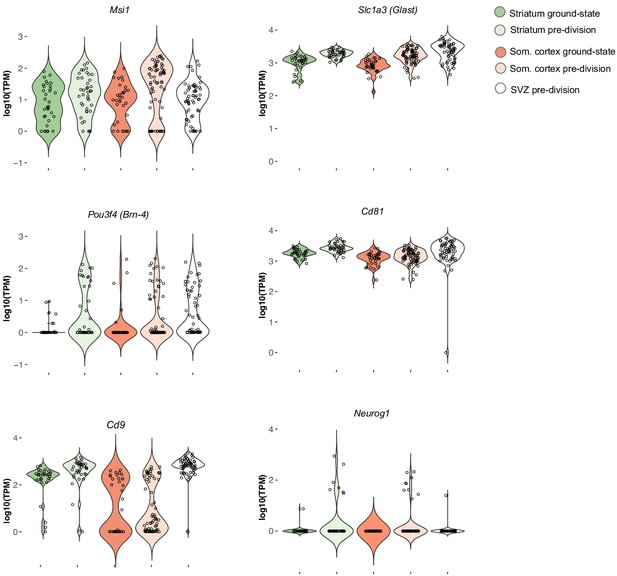
Expression levels of neural stem cell markers in striatum, somatosensory cortex and SVZ.
In the pre-division stage, cortical astrocytes have upregulated many neurogenesis-associated genes to the same levels as in the striatum and SVZ. One neural stem cell marker, Cd9, was conspicuously lower in the cortical astrocytes than in the striatum and SVZ. Another gene, Neurog1, was uniquely upregulated by the parenchymal astrocytes and was not expressed at any differentiation stage in the SVZ.
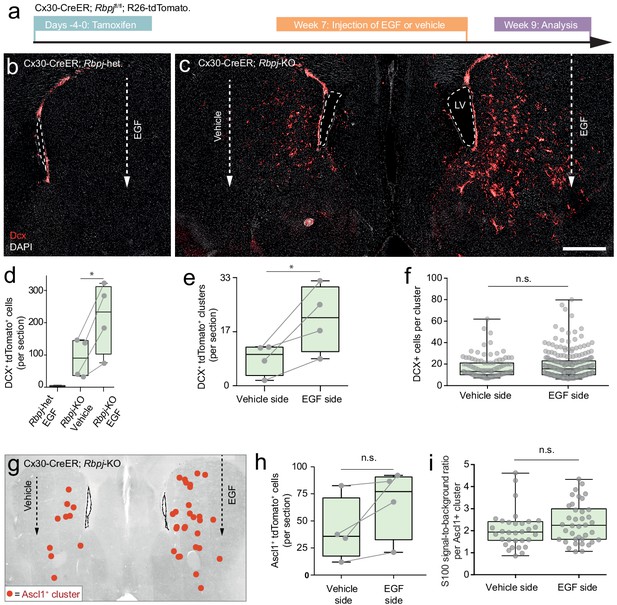
Stalled neurogenic astrocytes in the striatum can resume neurogenesis if exposed to EGF.
(a) Striatal astrocytes were exposed to EGF delivered through a single injection into the lateral striatum. (b, d) In Rbpj-heterozygous control mice, EGF neither causes ectopic migration of SVZ neuroblasts nor neurogenesis by striatal astrocytes. (c, d) However, in Rbpj knockout mice, EGF injection leads to a doubling of the number of striatal neuroblasts compared to the vehicle-injected contralateral striatum of the same mice (each data point represents one mouse; lines connect left and right striata from the same mice). (e) EGF injection leads to more striatal neuroblast clusters, but each cluster does not contain more cells (f), indicating that EGF recruits more halted astrocytes into neurogenesis rather than causing increased proliferation by the same number of astrocytes. (g–h) Some mice also show a dramatic increase in the number of transit-amplifying cells; however, this increase was not as pronounced as that of neuroblast clusters. (i) We used residual S100β protein as a short-term lineage tracing marker of striatal astrocyte-derived cells (see Figure 2—figure supplement 3d–g). EGF injection does not lead to any influx of SVZ-derived S100β-negative transit-amplifying cells (each dot is one cluster; plot shows sum of three brain sections [n = 3 mice]). Error bars in (d–f, h–i) represent maximum and minimum values. Scale bar (c): 500 μm; LV: lateral ventricle.
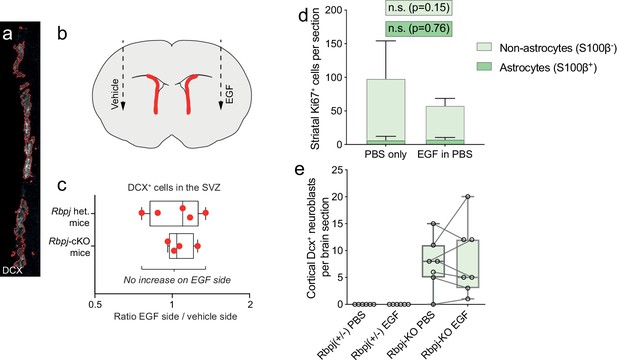
EGF injection into the lateral striatum does not change the amount of SVZ neuroblasts.
(a) Dcx signal intensity was used as a proxy for Dcx cell numbers in the SVZ. Dcx signal intensity was quantified using CellProfiler, which delineates Dcx+ cells and measures their signal intensity (red outlines in a). (b) In mice injected with EGF into one striatum and a vehicle solution in the other striatum, we could not detect any change in the amount of SVZ neuroblasts in the EGF-injected side, neither in mice heterozygous or homozygous for the Rbpj mutation. This is shown in (c); here, each red dot represents one animal, and the position of each dot indicates whether that mouse had more SVZ neuroblasts on the vehicle side or the EGF side. On average, there is no significant difference between the vehicle side and the EGF side, neither in Rbpj-heterozygous or Rbpj–KO mice. This indicates that EGF injection in the lateral striatum does not significantly affect the number of SVZ neuroblasts. (d) EGF injection into the striatum of wild-type mice did not lead to increased proliferation of striatal astrocytes or non-astrocytes 48 hr after injection (n = 5 mice per experimental group). (e) EGF injection into the somatosensory cortex did not trigger stalled Rbpj-KO astrocytes to re-initiate neurogenesis there. However, the small injury caused by inserting a thin syringe needle was enough of a stimulus to do so in some stalled astrocytes. Each data point in (e) represents one mouse; lines connect hemispheres of the same mice.
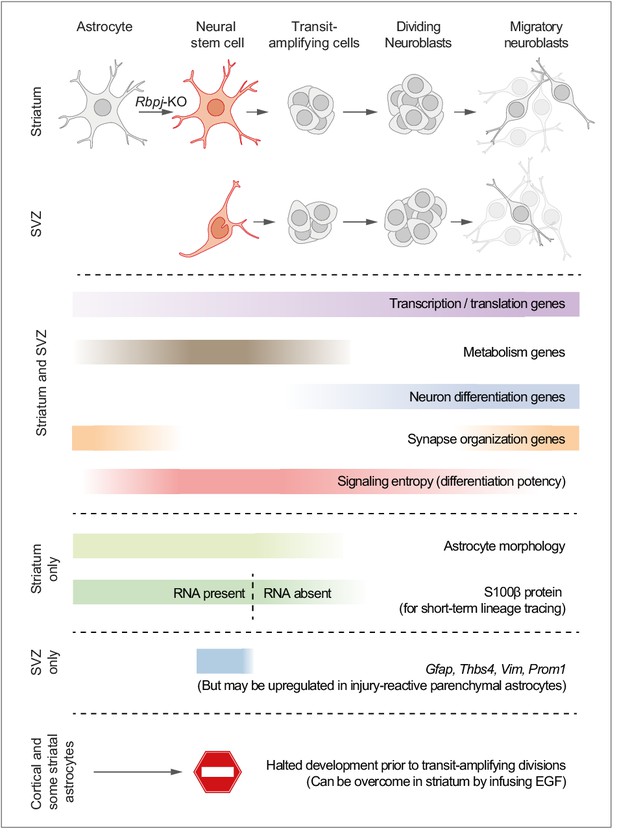
Proposed model summarizing the main findings of this study.
We propose a model in which striatal astrocytes can be viewed as highly quiescent neural stem cells that, upon Rbpj deletion, activate a neurogenic program very similar to that of SVZ stem cells. Some features of the neurogenic program are unique to these parenchymal astrocytes: Their distinct astrocyte morphology is partly retained even after initiation of transit-amplifying divisions (see Figure 2—figure supplement 3c and Video 1). Furthermore, S100β protein is found in many parenchymal astrocytes, but not in SVZ stem cells, and lingers at low levels in astrocyte-derived transit-amplifying cells even when the S100b gene is no longer expressed (dashed line), acting as a short-term lineage tracing marker. Other features are instead unique to the SVZ stem cells, such as expression of Gfap, Thbs4, Vim and Prom1, although markers of reactive astrogliosis, such as Gfap, are upregulated in neurogenic striatal astrocytes after stroke (Magnusson et al., 2014). Astrocytes all over the brain initiate the neurogenic program in response to Rbpj deletion, but all astrocytes in the somatosensory cortex, and many in the striatum, halt their development prior to entering transit-amplifying divisions. In the striatum, stalled astrocytes can resume neurogenesis if exposed to EGF.
Videos
Gradual loss of astrocyte morphology in astrocyte-derived transit-amplifying cells.
Striatal astrocytes retain their branched astrocyte morphology (see YFP signal) even after they have initiated transit-amplifying divisions in vivo. Shown are several examples of different Ascl1+ astrocytes and astrocyte-derived transit-amplifying cells at various stages of transit-amplifying divisions. EdU indicates that these cells had divided within the 2 weeks leading up to the analysis (Materials and methods). Note that astrocyte processes gradually become less branched and more retracted the longer transit-amplifying divisions proceed.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Mus musculus, C57BL/6; M, F) | Cx30-CreER | PMID:17823970 | ||
| Strain, strain background (Mus musculus, C57BL/6; M, F) | R26-tdTomato (Ai14) | PMID:25741722 | ||
| Strain, strain background (Mus musculus, C57BL/6; M, F) | R26-YFP | PMID:11299042 | ||
| Strain, strain background (Mus musculus, C57BL/6; M, F) | Rbpj(fl/fl) | PMID:11967543 | ||
| Antibody | anti-Ascl1 (Rabbit polyclonal) | Cosmo Bio | Product No: CAC-SK-T01-003 (RRID:AB_10709354) | Used at 1:2000 |
| Antibody | anti-Dcx (Goat polyclonal) | Santa Cruz Biotechnology | Product No: sc-8066 (RRID:AB_2088494) | Used at 1:500 |
| Antibody | anti-GFP (Chicken polyclonal) | Aves Labs | RRID:AB_2307313 | Used at 1:2000 |
| Antibody | anti-Ki67 (Rat monoclonal) | eBioscience | Catalog No: SolA15 (RRID:AB_10853185) | Used at 1:2000 |
| Antibody | anti-S100 (Rabbit polyclonal) | DAKO | Catalog No: GA50461-2 (RRID:AB_2811056) | Used at 1:200 (Directly conjugated to A647) |
| Peptide, recombinant protein | Epidermal growth factor | Sigma-Aldrich | Catalog No: E4127 | |
| Recombinant DNA reagent | AAV8-GFAP(0.7)-iCre-WPRE | Vector Biolabs | Catalog No: VB4887 | |
| Commercial assay or kit | Papain Dissociation System | Worthington | Catalog No: LK003160 | |
| Commercial assay or kit | Adult Brain Dissociation Kit | Miltenyi Biotec | Catalog No: 130-107-677 | |
| Commercial assay or kit | Magnetic MACS Multistand | Miltenyi Biotec | Catalog No: 130-042-303 | |
| Commercial assay or kit | MACS Large Cell Column | Miltenyi Biotec | Catalog No: 130-042-202 | |
| Commercial assay or kit | Myelin Removal Beads II | Miltenyi Biotec | Catalog No: 130-096-731 | |
| Commercial assay or kit | AMPure XT beads | Beckman Coulter | Catalog No: A63881 | |
| Commercial assay or kit | Nextera XT dual indexes | Illumina | ||
| Commercial assay or kit | Chromium Single Cell 3’ Library and Gel Bead Kit (v3) | 10X Genomics | Catalog No: CG000183 | |
| Chemical compound, drug | Percoll | GE Healthcare | Catalog No: 17-0891-01 | |
| Chemical compound, drug | EdU | Thermo Fisher | Catalog No: C10424 | |
| Chemical compound, drug | LIVE/DEAD Blue Dead Cell Stain Kit | Thermo Fisher | Catalog No: L23105 | |
| Software, algorithm | R | R Foundation For Statistical Computing | ||
| Other | Agilent Bioanalyzer | Agilent | ||
| Other | BD Influx FACS machine | BD | ||
| Other | 36-gauge beveled needle | World Precision Instruments | Catalog No: NF36BV-2 | |
| Other | NovaSeq 6000 | Illumina | ||
| Other | Illumina HiSeq 2500 | Illumina |
Additional files
-
Source data 1
Raw data for plots.
- https://cdn.elifesciences.org/articles/59733/elife-59733-data1-v2.xlsx
-
Supplementary file 1
Genes differentially expressed between ground-state and Rbpj-deficient astrocytes.
Data is shown for both striatum and somatosensory cortex. Astrocytes at the pre-division stage are in their most stem cell-like state. Therefore, genes differentially expressed at this time point represent potential marker genes for the astrocyte stem cell state.
- https://cdn.elifesciences.org/articles/59733/elife-59733-supp1-v2.xlsx
-
Supplementary file 2
Lists of genes characteristic for the four gene clusters in Figure 2e.
Results from a GO analysis (DAVID) is provided for each gene cluster.
- https://cdn.elifesciences.org/articles/59733/elife-59733-supp2-v2.xlsx
-
Supplementary file 3
Details about mice transplanted with neural stem cells (Figure 6l–n).
This list provides information about the mice that were used as transplantation donors (see also Methods).
- https://cdn.elifesciences.org/articles/59733/elife-59733-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59733/elife-59733-transrepform-v2.docx





