Feedback control of Wnt signaling based on ultrastable histidine cluster co-aggregation between Naked/NKD and Axin
Figures
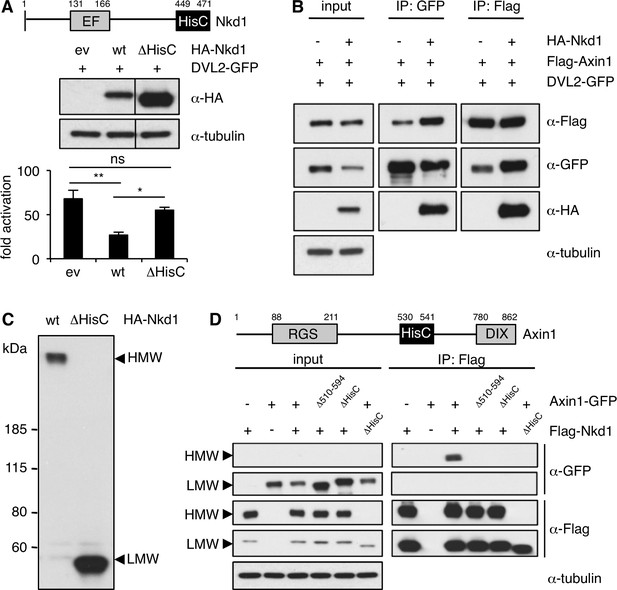
Nkd1 co-aggregates with Axin1 via highly conserved HisCs.
(A) SuperTOP assays, monitoring the signaling activity of overexpressed DVL2-GFP upon co-expression with wt or ΔHisC HA-Nkd1 in transfected HEK293T cells (used in this and all subsequent figures unless otherwise stated); expression levels were monitored by Western blot (above); error bars, SEM of > 3 independent experiments; one-way ANOVA with multiple comparisons; *p < 0.05, **p < 0.01. (B) CoIP assays between co-expressed proteins, as indicated above panels. (C) Western blot of wt and ΔHisC HA-Nkd1, revealing HisC-dependent HMW aggregates; left, positions of molecular weight markers (LMW, corresponding to monomeric Nkd1); in subsequent figures, only HMW and LMW regions are shown (see also supplementary information, for full-length gels). (D) CoIP assays between co-expressed proteins, as indicated above panels, monitoring ΔHisC-dependent co-aggregation of Flag-Nkd1 and Axin1-GFP (note that the amounts of transfected DNA has been adjusted in this experiment, to obtain comparable expression levels of different constructs).
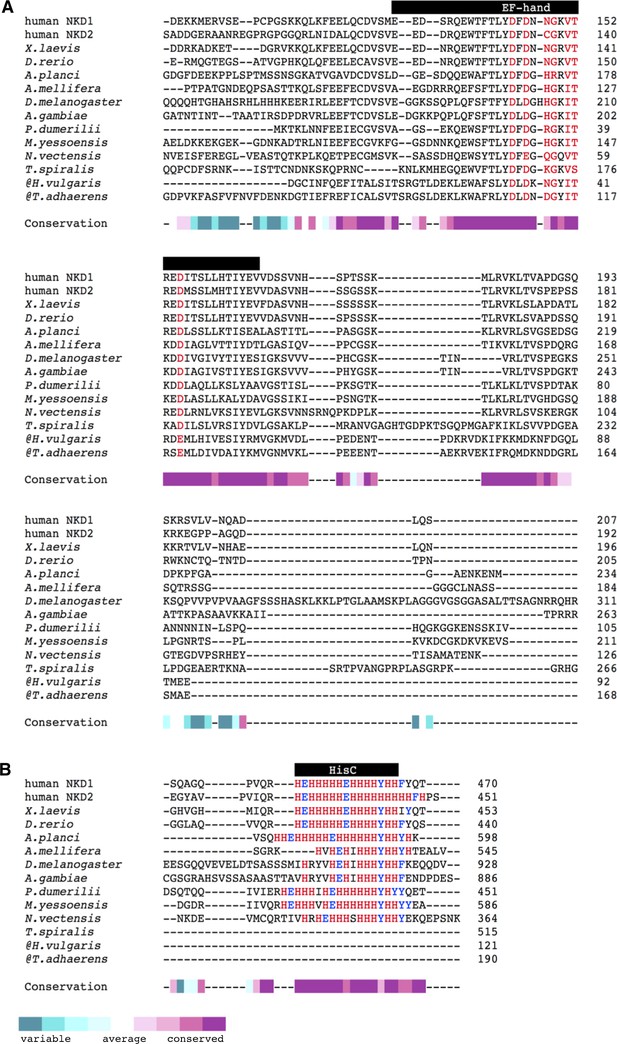
Sequence conservation of Naked/NKD orthologs.
Sequence alignments of the (A) EF-hand and the (B) C-terminal HisC cluster across diverse animal species. @, C-terminal sequences not available. Conservation score was calculated with the ConSurf webserver.
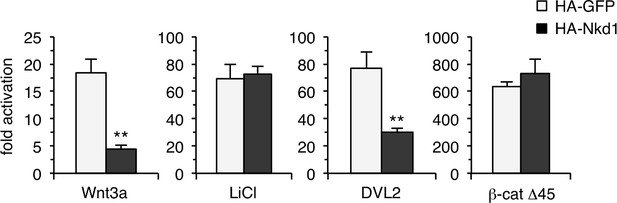
Effects of overexpressed NKD1 on β-catenin signaling.
SuperTOP assays in transfected HEK293T cells, monitoring the effects of overexpressed HA-Nkd1 on β-catenin-dependent transcription in response to stimulation for 6 hr with Wnt3a-conditioned media or 20 mM LiCl (control 20 mM NaCl), or to overexpressed DVL2-GFP or activated β-catenin (β-cat Δ45) in transfected HEK293T cells; error bars, SEM of > 3 independent experiments; Student’s t-test; **p < 0.01.
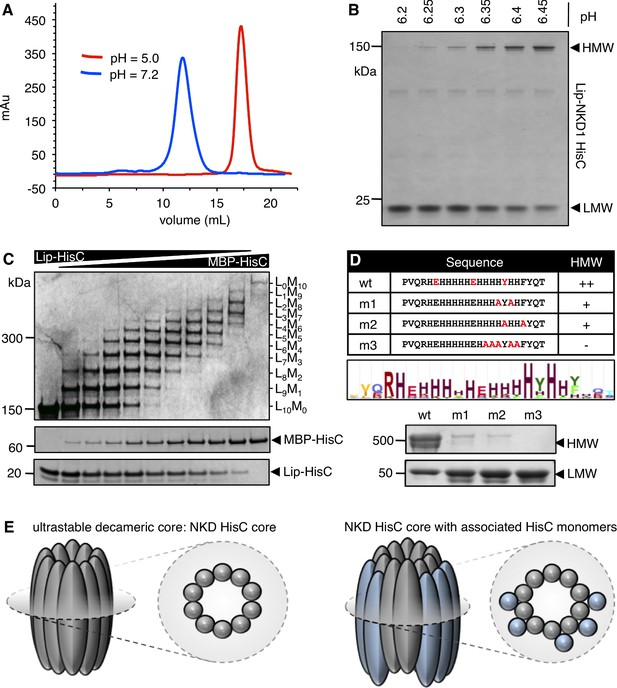
NKD1 HisC forms ultra-stable decameric core aggregates.
(A, B) pH-dependent aggregation of purified recombinant Lip-NKD-HisC in vitro, monitored by (A) SEC or (B) PAGE. (C) Co-aggregation assays after mixing Lip-NKD-HisC and MBP-NKD-HisC at different ratios, revealing a decameric core (note that the self-aggregation of MBP-NKD-HisC is considerably less efficient than that of Lip-NKD-HisC, presumably owing to the large size of the MBP tag). (D) Mutational analysis, revealing that in vitro aggregation depends on conserved aromatic and charged residues within HisC. (E) Cartoon of NKD-HisC core and associated HisC monomers.
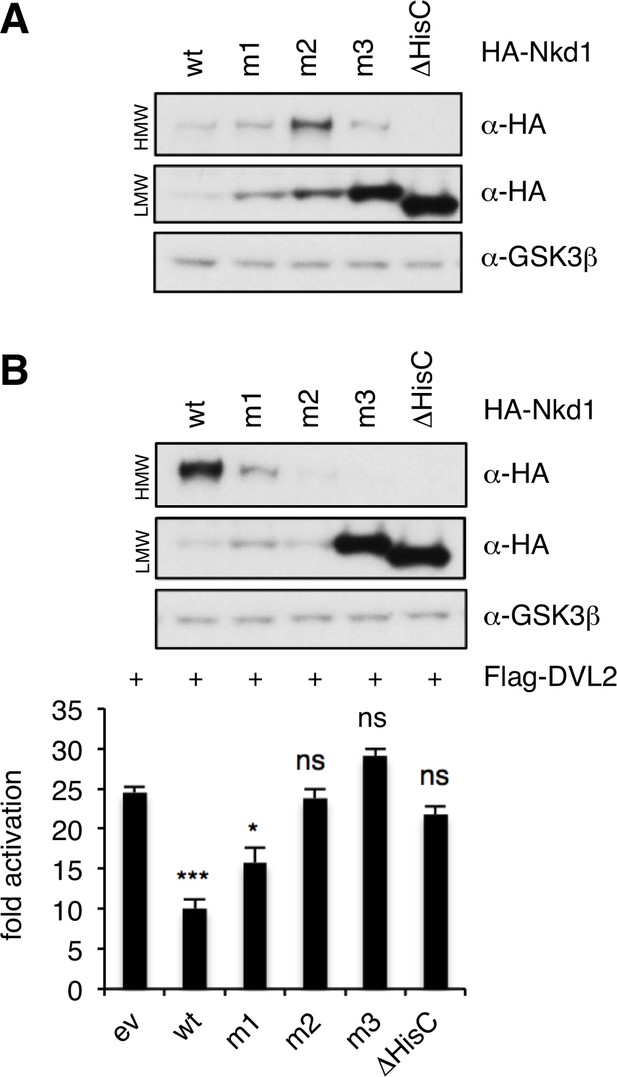
Effects of overexpressed Nkd1 HisC mutants on β-catenin signaling.
Western blots monitoring expression of HA-Nkd1 and specific HisC point-mutants thereof (see Figure 2D) alone (A) and in the presence of Flag-DVL2 (B). SuperTOP assays in transfected HEK293T cells, monitoring the effects of overexpressed HA-Nkd1 on β-catenin-dependent transcription owing to overexpressed Flag-DVL2; error bars, SEM of > 3 independent experiments; one-way ANOVA with multiple comparisons; *p < 0.05, ***p < 0.001, ns = not significant.
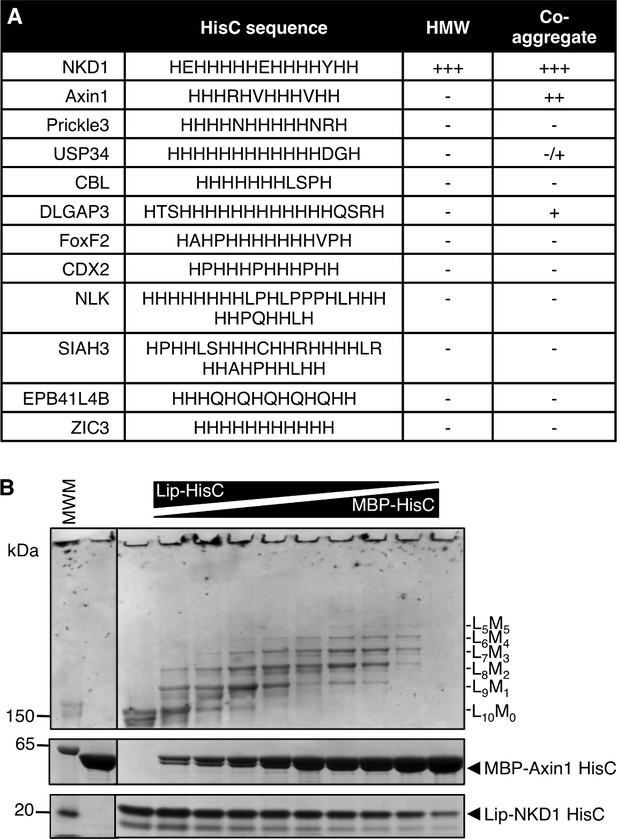
NKD1 HisC co-aggregates with Axin1 HisC.
(A) Selected HisC-containing proteins in the human genome (see also text) and their ability to co-aggregate with NKD1 HisC. (B) Co-aggregation assays after mixing Lip-NKD-HisC and MBP-Axin1-HisC at different ratios (MWM, molecular weight markers).
-
Figure 3—source data 1
HisC proteins in the human genome and their subcellular location.
- https://cdn.elifesciences.org/articles/59879/elife-59879-fig3-data1-v2.docx
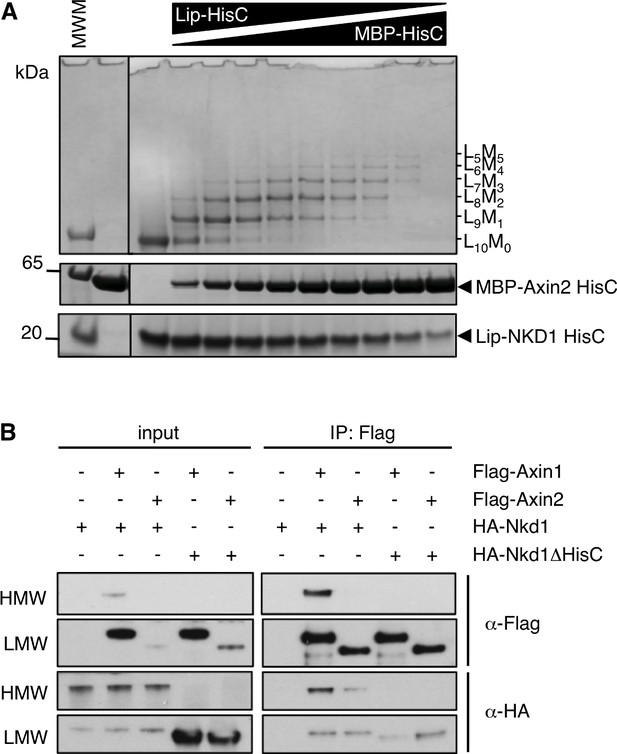
Co-aggregation between NKD1 and Axin2.
(A) Co-aggregation assays after mixing purified recombinant Lip-NKD1-HisC and MBP-Axin2-HisC (KHVHHHYIHHHA) at different ratios. (B) CoIP assays between proteins co-expressed in transfected HEK293T cells, as indicated above panels, revealing formation of HMW co-aggregates between HA-Nkd1 and Flag-Axin1 but not Flag-Axin2.
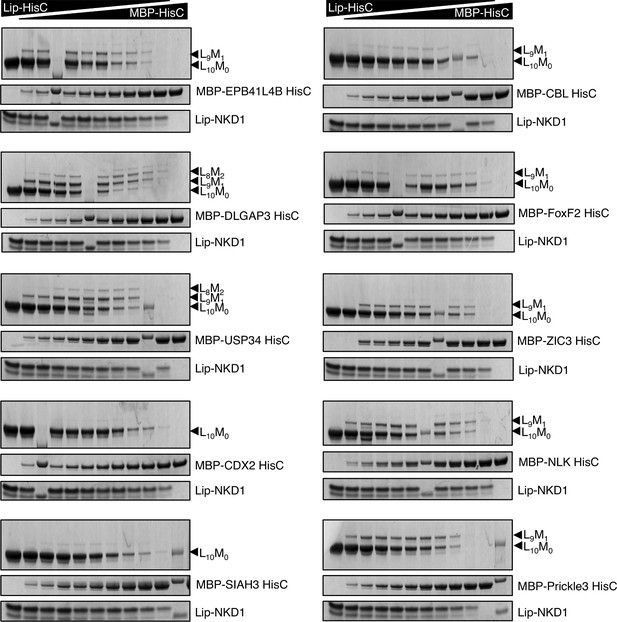
Systematic co-aggregation tests with NKD1-HisC.
Co-aggregation assays after mixing purified recombinant Lip-NKD1-HisC and MBP-HisC from various non-nuclear proteins, as indicated on the right (see main Figure 3A, for HisC sequences).
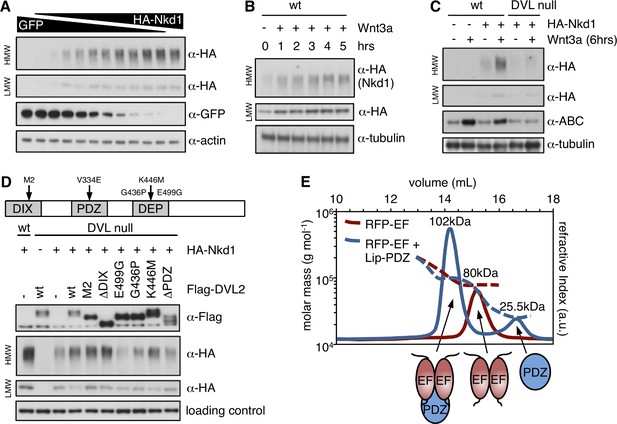
HisC aggregation is promoted by DVL2.
(A–D) Western blots monitoring the formation of HisC aggregates in wt or mutant HEK293T cells dependent on (A) Nkd1 concentration, (B) duration of Wnt stimulation, (C) DVL or (D) complementation of DVL null cells by re-expression of wt or mutant DVL2 (E499G, G436P block DVL2 dimerization; K446M blocks Frizzled receptor binding) (Gammons et al., 2016a). (E) SEC-MALS, monitoring complex formation between purified recombinant RFP-NKD1-EF and Lip-DVL2-PDZ (as indicated by cartoons); numbers correspond to Mr values determined by MALS (expected: RFP-NKD1-EF dimer, 71.8 kDa; Lip-DVL2-PDZ, 22.0 kDa; complex, 93.8 kDa).
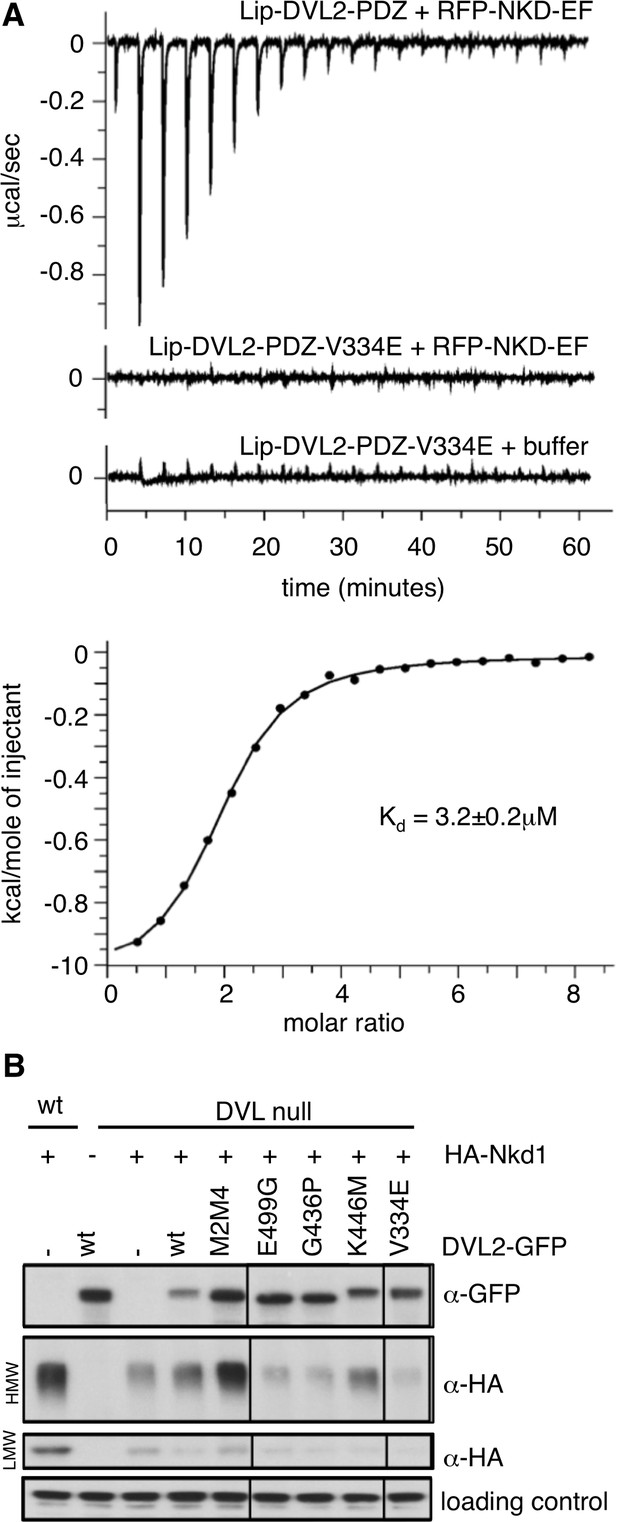
Binding of the NKD1 EF-hand to the DVL2 PDZ cleft.
(A) ITC profiles for NKD1 EF-hand binding to the wt DVL2 PDZ domain or its V334E cleft mutant; Kd value obtained with wt DVL2 PDZ domain. (B) Western blots monitoring the formation of Nkd1 HisC aggregates dependent on complementation of DVL null cells by re-expression of wt or mutant DVL2 (see also main Figure 4D).
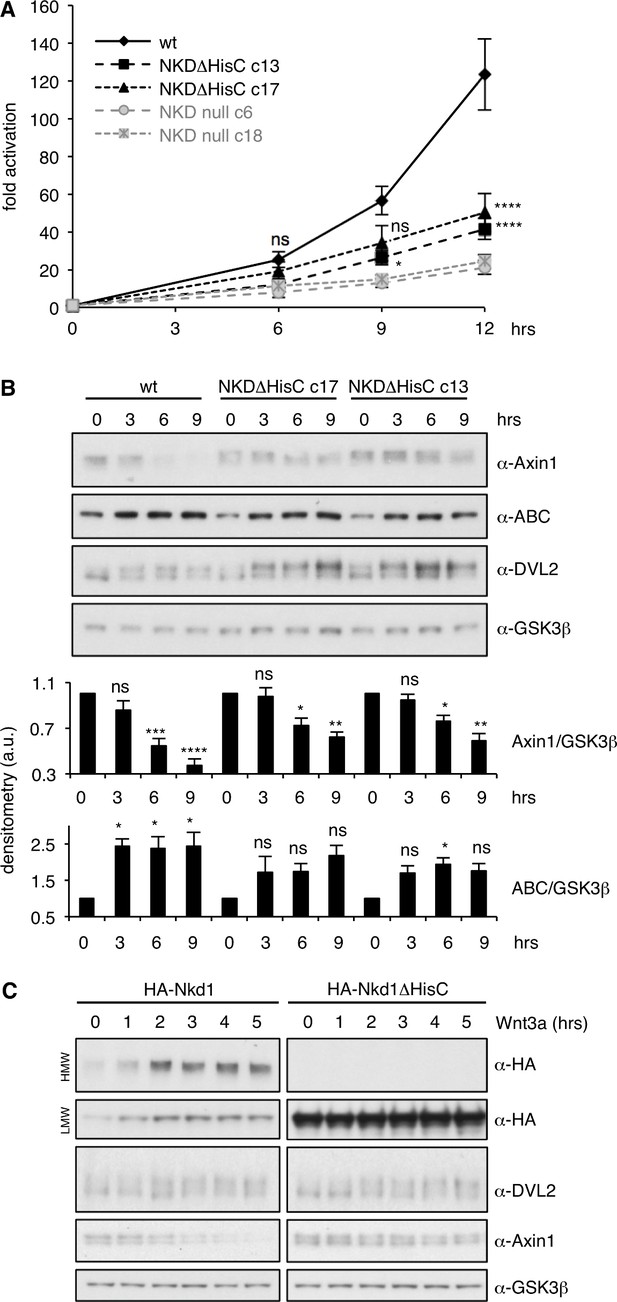
NKD is required for sustained Wnt signaling.
(A) SuperTOP assays in HEK293T cells, monitoring the signaling response of NKDΔHisC, NKD null-mutant or control cells after 6 hr of Wnt3a stimulation (two independently isolated lines each, see also Figure 5—figure supplement 1); error bars, SEM of > 3 independent experiments; two-way ANOVA with multiple comparisons (for c13 and c17; see Figure 5—figure supplement 2 for null lines); *p < 0.05, ****p < 0.0001. (B) Representative western blots monitoring expression levels of endogenous proteins in two independent NKDΔHisC lines following 0–9 hr of Wnt stimulation (as indicated), quantified by densitometry of >3 independent experiments relative to GSK3β as internal control (a.u., arbitrary units); error bars, SEM of > 3 independent experiments; two-way ANOVA with multiple comparisons; ∗p < 0.05, ∗∗p < 0.01,∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, ns = not significant. For the corresponding analysis of NKD null cell lines, see Figure 5—figure supplement 2. (C) Western blots monitoring the effects of overexpressed wt or ΔHisC HA-Nkd1 on the levels of endogenous proteins (indicated on the right; GSK3β, internal control) after 0–5 hr of Wnt stimulation.
-
Figure 5—source data 1
Oligonucleotides used for CRISPR engineering of HEK293T cells and confirmation of lesions.
- https://cdn.elifesciences.org/articles/59879/elife-59879-fig5-data1-v2.docx
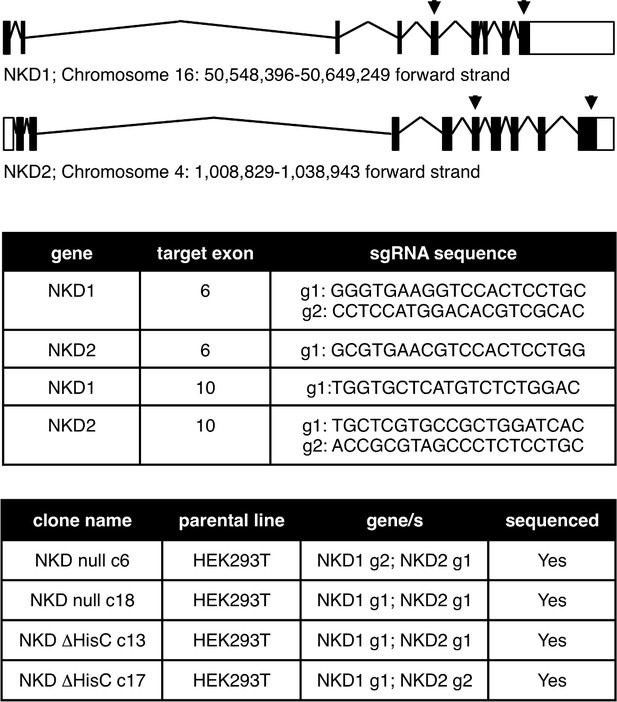
CRISPR engineering of HEK293T cells.
Top, genomic locations and organizations of the human NKD1 and NKD2 loci, with positions of CRISPR gRNAs indicated; underneath, sequences of gRNAs used for generating truncations in two different NKD null (c6 and c18) or ΔHisC (c13 and c17) lines, each verified by sequencing of both genes.
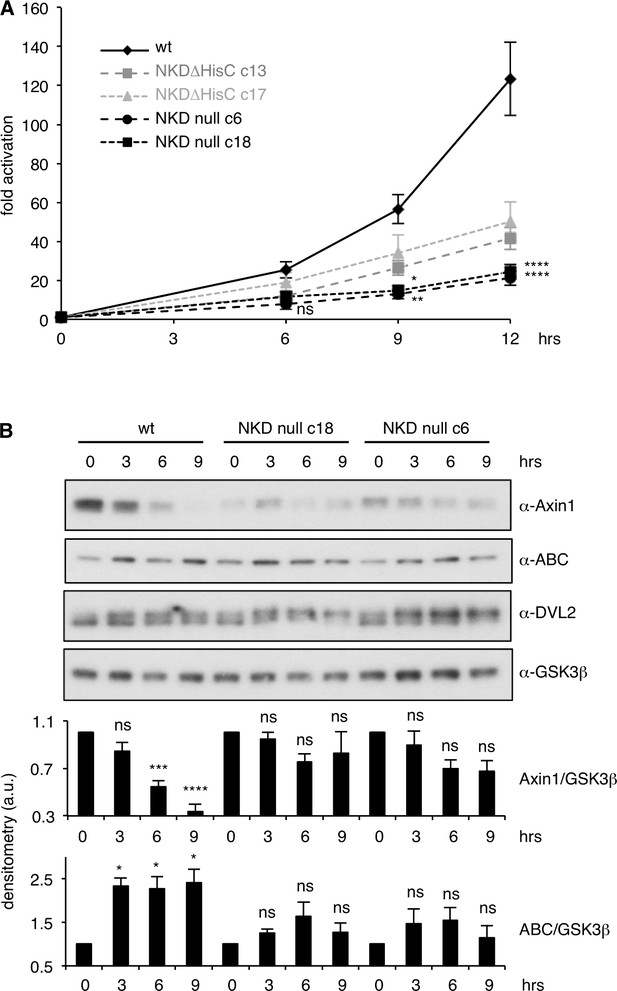
NKD is required for sustained Wnt signaling.
(A) Western blots corresponding to SuperTOP assays shown in main Figure 5A, monitoring expression levels of endogenous proteins in NKD null cells, quantified by densitometry underneath, normalized to 0 hr time point for each cell line (A.U., arbitrary units); SEM of >3 independent experiments; two-way ANOVA with multiple comparisons; *p < 0.05, ***p < 0.001, ****p < 0.0001; ns = not significant. We note that the levels of endogenous Axin1 are consistently reduced in unstimulated NKD null cells compared to their parental controls, perhaps reflecting a compensatory mechanism of these null cells to maximize their tonic Wnt signaling levels (see also Gammons et al., 2016b).
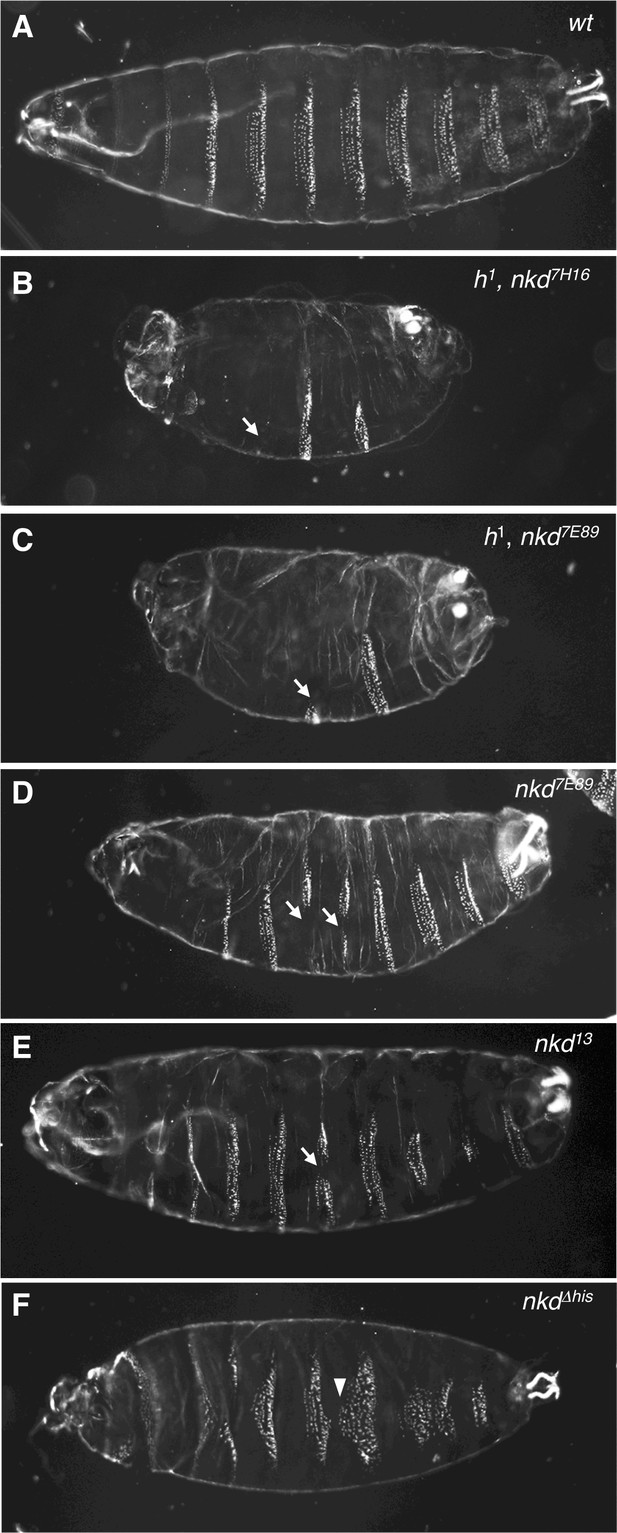
Allele-dependent Wg signaling defects in nkd mutant Drosophila embryos.
Dark-field images of ventral cuticles of (A) wt and (B–F) homozygous mutant embryos, as indicated in panels (nkd7H16, nkd7E98, pre-existing alleles; nkd13, nkdΔhis CRISPR-engineered alleles), showing regions of excess naked cuticle (arrows) signifying ectopic Wg signaling. Note that the ‘near-naked’ phenotype is only seen in embryos bearing h1 (B, C); excess naked cuticle regions are seen infrequently in null-mutant embryos without h1 (D, E), or not at all in nkdΔhis embryos bearing a HisC deletion (F) whose most penetrant phenotype are excess small denticles (arrowheads) signifying reduced Wg signaling.
-
Figure 6—source data 1
Oligonucleotides used for Drosophila CRISPR engineering and confirmation of lesions.
- https://cdn.elifesciences.org/articles/59879/elife-59879-fig6-data1-v2.docx
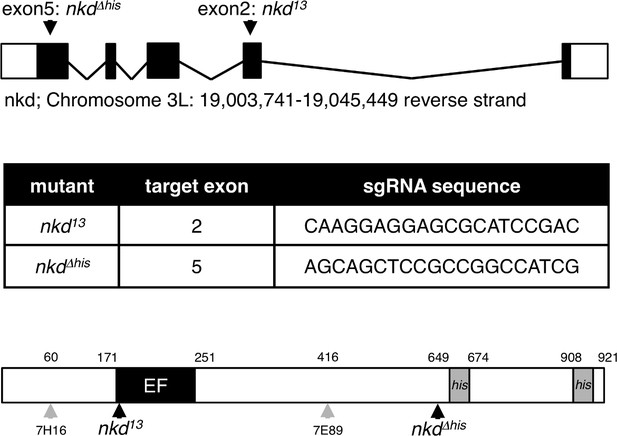
CRISPR-engineered Drosophila nkd alleles.
Top, genomic location and organization of the Drosophila nkd locus, with positions of CRISPR gRNAs indicated; underneath, sequences of gRNAs used for generating two different nkd truncations, each verified by sequencing; bottom, cartoon of Naked, with domains and positions of various alleles indicated.
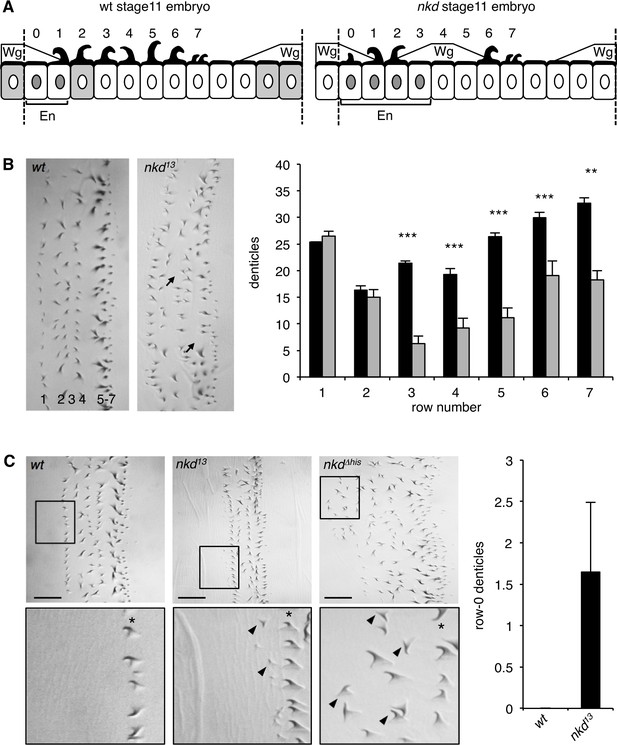
Quantification of context-dependent Wg signaling defects in embryos bearing CRISPR alleles.
(A) Cartoons of row-specific ventral denticles relative to expression domains of Wg, En, and Nkd in stage 11 wt (left) and nkd13 null-mutant (right) embryos; numbers indicate denticle rows; see also text. (B) Left, examples of abdominal denticle belts in wt and nkd13 homozygous embryos, with arrows pointing to missing denticles; right, quantitation of numbers of denticles per row in wt (black bars) or nkd13 null-mutant (white bars) embryos (n = 10 near-normal denticle belts); Student’s unpaired t-test; **p < 0.01, ***p < 0.001. (C) Left, nkd13 and nkdΔhis homozygous but not wt embryos exhibit excess row-0 denticles (arrowheads; asterisks mark row-1); right, quantitation of excess row-0 denticles in nkd13 mutants (n = 20 near-normal denticle belts).
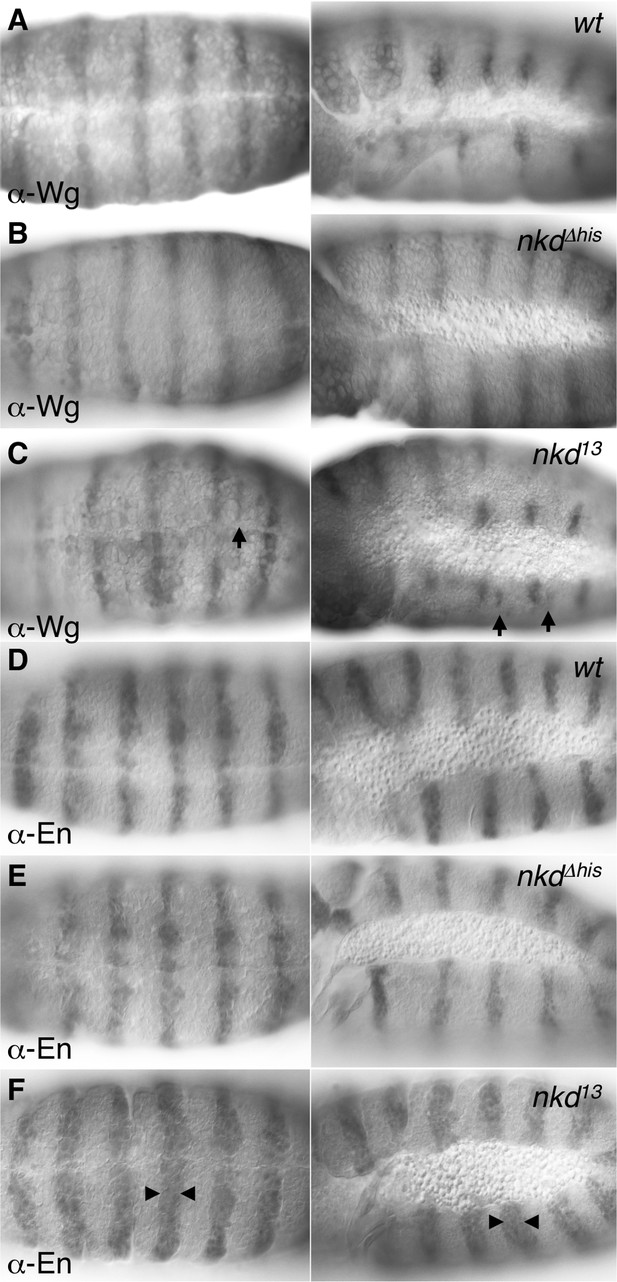
Ectopic Engrailed and Wingless expression in nkd mutant Drosophila embryos.
5–6 hr old wt and nkd mutant embryos, as indicated in panels, stained for Wingless (Wg, A–C) or engrailed (En, D–F) expression; left, ventral view; right, side view. nkd null (nkd13) but not ΔHisC mutants (nkdΔhis) exhibit an expansion of En expression (arrowheads) in every segment, and sporadic patches of ectopic Wg expression (arrows). The latter appear to correlate with the sporadic regions of excess Naked cuticle shown in main Figure 6D,E, and thus likely accounts for the partially ‘Naked’ phenotype of these embryos.
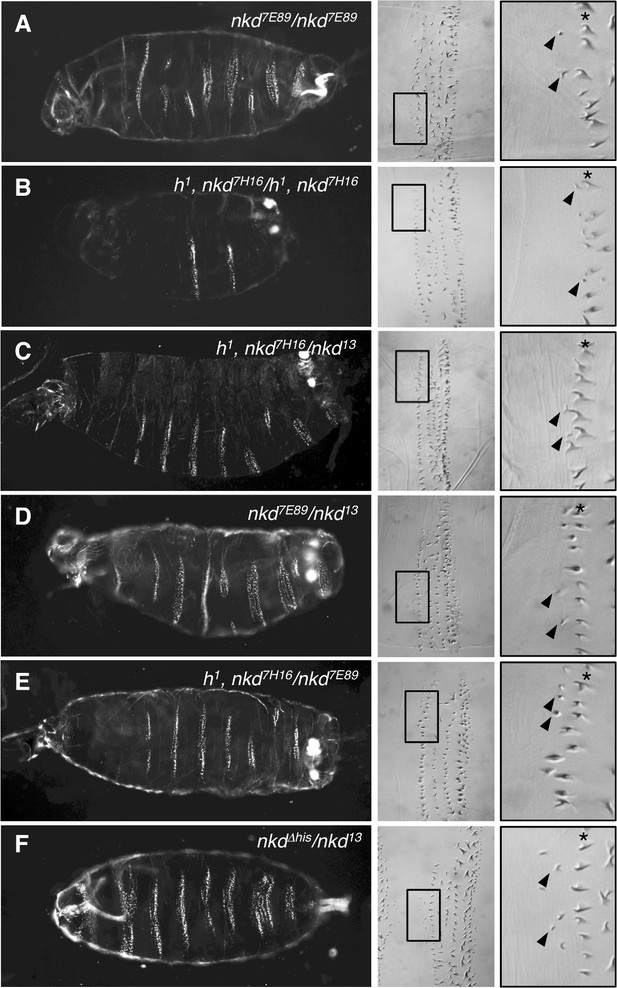
Excess anterior denticles in transheterozygous embryos bearing different combinations of nkd alleles.
Images of ventral cuticles of embryos bearing different combinations of nkd and h1 alleles as indicated in panels, exhibiting excess anterior denticles. Left, dark-field; right, DIC.
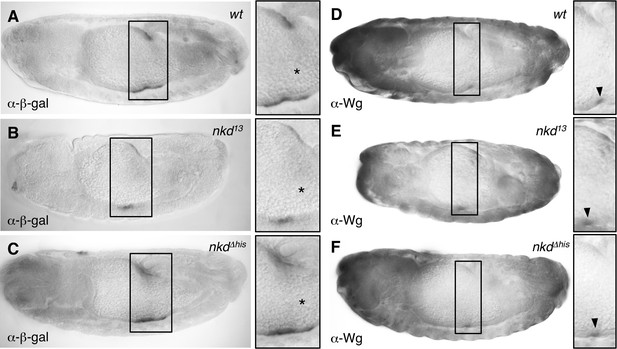
Reduced expression of Wg-responsive Ubx midgut reporter gene in nkd null-mutant embryos.
(A–C) Side views of 12–14 hr old embryos bearing UbxB.lacZ, fixed and stained with α-β-galactosidase (α-β-gal) antibody, showing reduced expression of UbxB.lacZ in nkd13 but not nkdΔhis homozygous embryos (* in high-magnification views mark positions of Wg signaling sources in the middle midgut). (D–F) Side views of ∼12-hr old wt or nkd mutant embryos, fixed and stained with α-Wg antibody, revealing apparently normal Wg expression (marked by arrowheads in high-magnification views) in the middle midgut of nkd mutants.
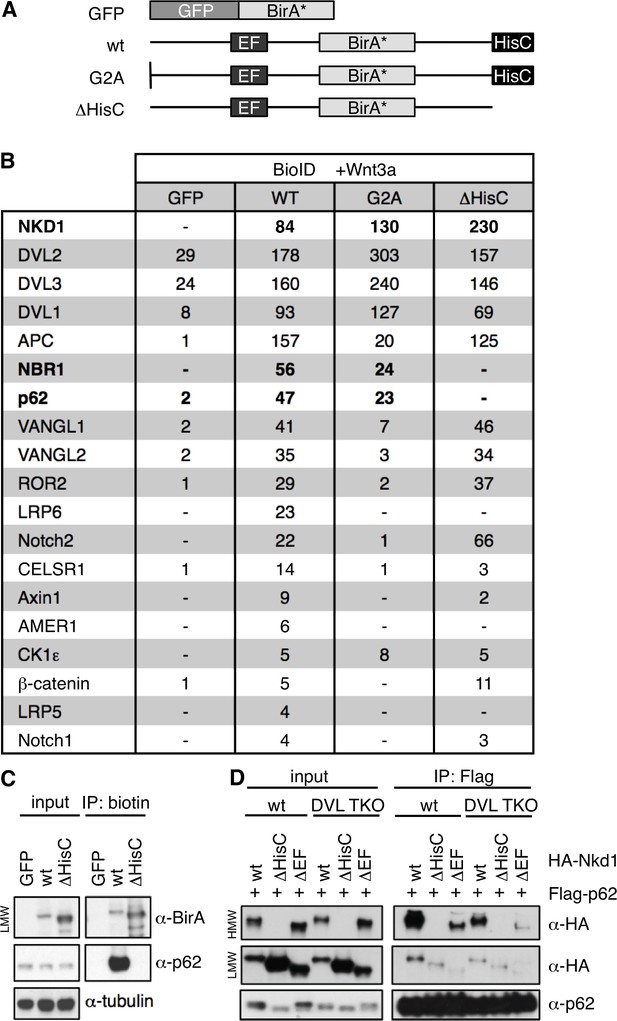
p62 is a HisC-dependent interactor of Nkd1.
(A) Cartoons of BirA* baits used for BioID proximity labeling. (B) Selected BioID hits obtained with wt or mutant Nkd1-BirA* stably integrated in HEK293 cells (see also text); numbers correspond to unweighted spectral counts > 95% probability (for full list, see Figure 8—source data 1); note that APC is large (~311 kDa) and relatively abundant, explaining the high numbers of spectral counts. (C) BioID proximity-labeling assays in HEK293 cells with stably integrated baits, monitoring association between endogenous p62 and Nkd1-BirA* or Nkd1ΔHisC-BirA* (only LMW Nkd1-BirA* is shown since HMW Nkd1-BirA* is too large to be detectable on blots). (D) CoIP assays between proteins co-expressed in HEK293T cells, as indicated above panels, monitoring association between HA-p62 and wt or mutant HA-Nkd1.
-
Figure 8—source data 1
Full list of BioID hits obtained with wt or mutant Nkd1-BirA*.
- https://cdn.elifesciences.org/articles/59879/elife-59879-fig8-data1-v2.docx
Additional files
-
Supplementary file 1
Uncropped Western blots.
- https://cdn.elifesciences.org/articles/59879/elife-59879-supp1-v2.pdf
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59879/elife-59879-transrepform-v2.docx





