Functional reconstitution of a bacterial CO2 concentrating mechanism in Escherichia coli
Figures
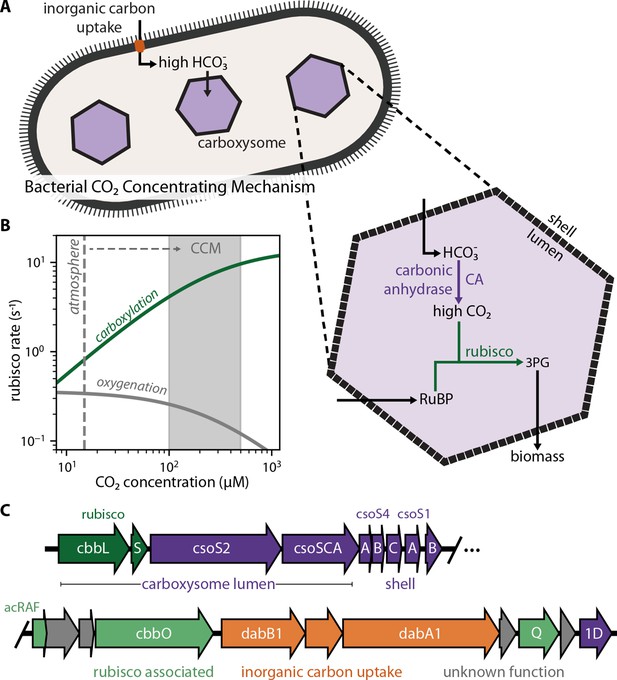
Twenty genes form the basis of a bacterial CCM.
(A) The bacterial CCM consists of at least two essential components - energy-coupled inorganic carbon uptake and carboxysome structures that encapsulate rubisco with a carbonic anhydrase (CA) enzyme (Desmarais et al., 2019; Kaplan et al., 1980; Price and Badger, 1989a, Price and Badger, 1989b; Rae et al., 2013; Shively et al., 1973). Transport generates a large cytosolic HCO3- pool, which is rapidly converted to high carboxysomal CO2 concentration by the carboxysomal CA (Mangan et al., 2016; McGrath and Long, 2014). (B) Elevated CO2 increases the rubisco carboxylation rate (green) and suppresses oxygenation by competitive inhibition (grey). [O2] was set to 270 μM for rate calculations. A more detailed version of this calculation is described in Figure 1—figure supplement 1. (C) H. neapolitanus CCM genes are mostly contained in a 20 gene cluster (Desmarais et al., 2019) expressing rubisco and its associated chaperones (green), carboxysome structural proteins (purple), and an inorganic carbon transporter (orange). Supplementary file 1 gives fuller description of the functions of these 20 genes along with a per-gene bibliography. Figure 1—figure supplement 2 demonstrates that the operon beginning with acRAF indeed encodes a functional inorganic carbon transporter.
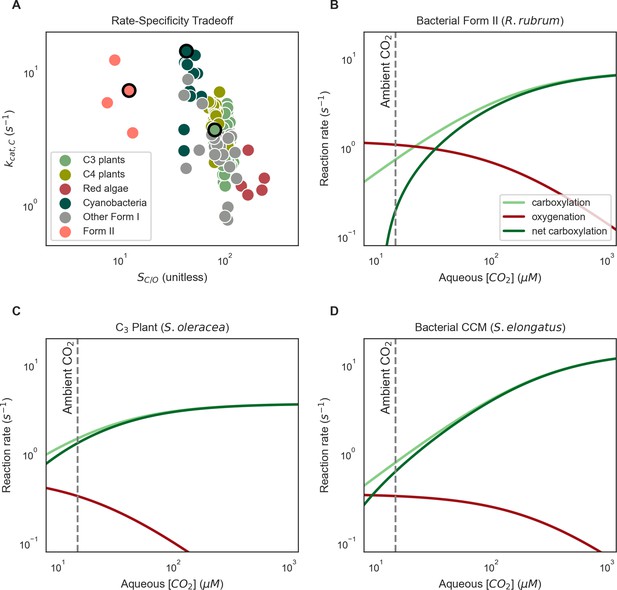
Elevated CO2 overcomes limitations associated with rubisco catalysis.
(A) Kinetic data assembled for ≈300 rubiscos from diverse organisms show that there is limited variation (less than one order of magnitude) in CO2 specificity (SC/O) and maximum carboxylation rate (kcat,C) among the Form I rubiscos found in all photoautotrophs and all bacteria harboring carboxysome CCMs (Flamholz et al., 2019). Moreover, SC/O and kcat,C appear to trade-off with each other. Although this relationship is not strict, rubiscos with high kcat,C values also typically have lower SC/O(Savir et al., 2010; Tcherkez et al., 2006). As carboxylation and oxygenation reactions occur at the same active site, elevated CO2 will both increase the carboxylation rate (until it reaches kcat,C) and also inhibit oxygenation by exclusion of oxygen from the active site. As it relies only on the well-founded assumption that catalysis with CO2 and O2 substrates are mutually exclusive, this mechanism should function for any rubisco. Panels B-D depict this effect for three distinct rubiscos, which are highlighted with black borders in (A). Panels give carboxylation (light green), oxygenation (red) and net carboxylation (dark green) rates as a function of the aqueous CO2 concentration at ambient O2 levels (270 uM at 25 ℃). All curves were calculated using standard kinetic equations for rubisco. Net carboxylation was calculated as the carboxylation rate less ½ the oxygenation rate, which presumes a plant-type photorespiratory pathway that loses one CO2 for every two oxygenation reactions. (B) Bacterial Form II rubiscos are typically found in organisms living in low O2 environments and, accordingly, display low CO2 specificities (SC/O ≈ 10) and relatively high maximum carboxylation rates (kcat,C ≈10–20 s−1, Davidi et al., 2020). As such, Form II rubiscos do not perform well in ambient CO2 and O2 concentrations. (C) C3 plants like spinach do not have CCMs. Furthermore, the CO2 concentration inside the leaf is typically measured to be lower than ambient due to a balance of stomatal conductance and CO2 fixation by rubisco itself (Caemmerer and Evans, 1991). Accordingly, C3 plant rubiscos display high CO2 specificities (SC/O ≈ 100), modest kcat,C ≈ 3 s−1, and perform well at ambient and sub-ambient CO2 levels, displaying relatively little oxygenation and, consequently, net carboxylation rates that are similar to the total carboxylation rate. (D) Rubiscos found in bacteria with a carboxysome CCM typically have relatively low CO2 specificities (SC/O ≈ 50) and fast maximum carboxylation rates relative to other Form I rubiscos (kcat,C ≈ 10 s−1). In general, rubiscos from organisms bearing CCMs (whether bacteria, algae, or plants) tend to have lower CO2 specificities and higher kcat,C than enzymes from related organisms without CCMs (Iñiguez et al., 2020; Savir et al., 2010). The carboxysomal rubisco from S. elongatus PCC 7942 performs worse than a typical C3 plant rubisco in ambient air, but much better in the elevated CO2 environment we presume is maintained by the carboxysome CCM. The aqueous CO2 and O2 concentrations were calculated assuming Henry’s law equilibrium at 25 ℃. Notably, changes in temperature will affect CO2 and O2 solubility (Milo and Phillips, 2015; Sander, 2015) and rubisco kinetics, most notably decreasing CO2-specificity at elevated temperatures (Boyd et al., 2019; Sage et al., 2012).
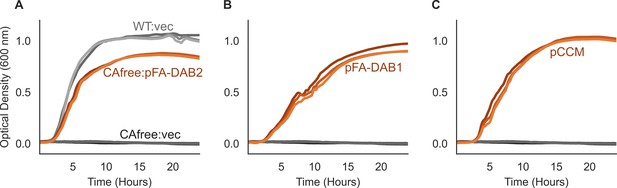
The 20 gene CCM cluster includes a functional DAB-type inorganic carbon transporter.
In previous work, we showed that the H. neapolitanus genome contains two homologous complexes DAB1 and DAB2 that are required for growth in ambient air (Desmarais et al., 2019). We further demonstrated that DAB2 is a two-gene operon whose protein products form a membrane associated complex that is capable of energetically active inorganic carbon uptake, but did not investigate DAB1 in detail. Here, we use DAB1 because it is encoded in the same genomic locus as the carboxysome, in a putative operon that also contains other potentially CCM-relevant genes like rubisco chaperones. See Figure 1C and Supplementary file 1 for further detail on the contents of this operon. As before, we rely on a reporter strain, CAfree, to test inorganic carbon transporters. This strain lacks all endogenous carbonic anhydrases and fails to grow in ambient air as a result (Desmarais et al., 2019). Growth of CAfree is complemented by elevating the CO2 level in the growth chamber, expressing carbonic anhydrases, or by supplying intracellular HCO3- via an inorganic carbon transporter like DAB2. Panel (A) reproduces our previous result - that expression of DAB2 from the pFA backbone complements growth of CAfree in ambient air, such that CAfree:pFA-DAB2 (orange) grows similarly to the wild-type strain (WT:vec, light grey). A negative control (CAfree:vec, dark grey) fails to grow, as expected. (B) DAB1 genes, which are marked in orange in Figure 1C, also rescue growth of CAfree in ambient air. pFA-DAB1 expresses only the DAB1 genes and omits the remaining eight genes in the operon. (C) The pCCM plasmid encodes all 11 genes found in the same putative operon as DAB1. CAfree:pCCM also well grows in ambient air. Cells were grown in LB media with 100 nM aTc induction throughout, with ‘vec’ denoting a vector control of pFA-sfGFP.
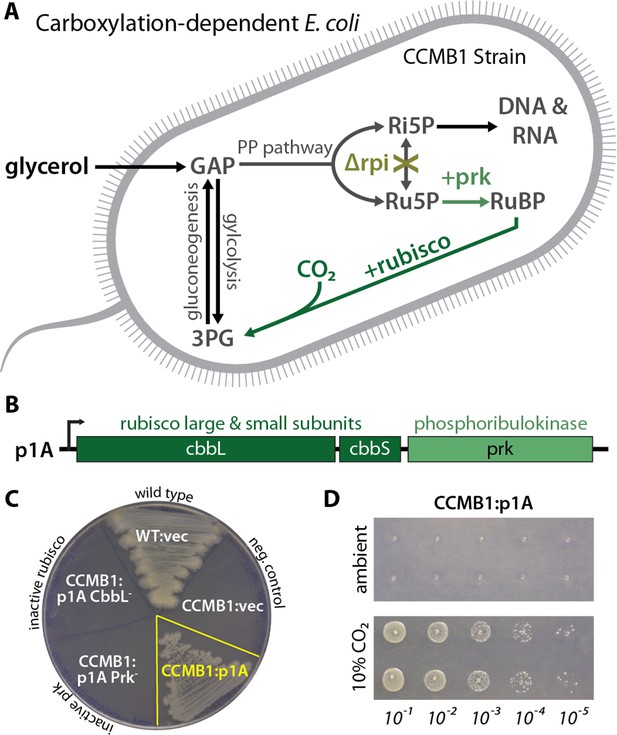
CCMB1 depends on rubisco carboxylation for growth on glycerol.
(A) Ribose-5-phosphate (Ri5P) is required for nucleotide biosynthesis. Deletion of ribose-phosphate isomerase (Δrpi) in CCMB1 blocks ribulose-5-phosphate (Ru5P) metabolism in the pentose phosphate (PP) pathway. Expression of rubisco (H. neapolitanus CbbLS) and phosphoribulokinase (S. elongatus PCC7942 prk) on the p1A plasmid (B) permits Ru5P metabolism, thus enabling growth on M9 glycerol media in 10% CO2 (C). Mutating the rubisco active site (p1A CbbL-) abrogates growth, as does mutating ATP-binding residues of Prk (p1A Prk-). (D) CCMB1:p1A grows well under 10% CO2, but fails to grow in ambient air. Cells were grown on M9 glycerol media throughout. The algorithmic design of CCMB1 is described in Figure 2—figure supplement 4 and Appendix 1. The mechanism of rubisco-dependence is diagrammed in Figure 2—figure supplement 3. Figure supplement 2 demonstrates growth of CCMB1:p1A on various media, Figure 2—figure supplement 5 demonstrates complementation by a variety of bacterial rubiscos and Figure 2—figure supplement 1 demonstrates anaerobic growth of CCMB1:p1A, establishing that oxygenation is not required for growth. Acronyms: ribulose 1, 5-bisphosphate (RuBP), 3-phosphoglycerate (3PG).
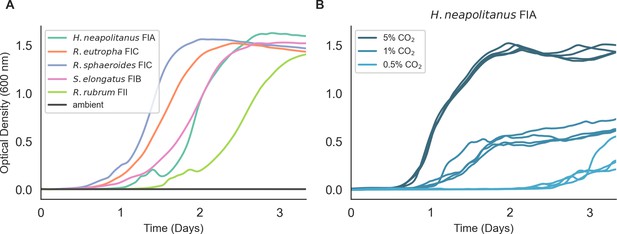
Expression of five kinetically and phylogenetically distinct rubiscos permits CCMB1 growth in glycerol minimal media with 5% CO2.
(A) Expression of five diverse rubiscos in CCMB1 complemented growth in 5% CO2 (colored lines) but not in ambient air (grey). Expressing the carboxysomal Form IA rubisco from H. neapolitanus along with prk on the p1A plasmid (CCMB1:p1A) permits growth in 5% CO2 (teal) but not in ambient air. A catalytically inactive variant (p1A CbbL K194M) failed to grow in both conditions, as expected and shown in Figure 2 and Figure 2—figure supplements 2, 5. The kinetic parameters of this rubisco have not been measured, but it is assumed to be relatively fast (kcat,C ≈ 5–10 s−1) and relatively non-specific towards CO2 (SC/O ≈ 30–50) like other carboxysome-localized Form IA rubiscos. Four additional rubiscos were expressed from an identical plasmid backbone and all of them permitted CCMB1 to grow in 5% CO2 with varying kinetics. The non-carboxysomal Form IC rubisco from Ralstonia eutropha (also known as Cupriavidis necator) is in orange and is the most specific bacterial rubisco known, with kcat,C ≈ 2–3 s−1 and SC/O ≈ 75–85. The Form IC rubisco from Rhodobacter sphaeroides (light blue) and has kcat,C ≈ 1–2 s−1 and SC/O ≈ 55–60. The cyanobacterium S. elongatus PCC 6301 expresses a Form IB rubisco in the same family found in eukaryotic algae and land plants (pink). This enzyme is exceptionally fast for a Form I rubisco, with kcat,C ≈ 10–14 s−1 and SC/O ≈ 40–50. Finally, the model Form II rubisco from R. rubrum (light green) also complemented CCMB1 for growth in 5% CO2. Form II enzymes have relatively high kcat,C ≈ 10 s−1 and low SC/O ≈ 10–20. Biological triplicate measurements were conducted for all rubiscos in panel A and were all consistent. Notably, none of these rubiscos permit growth in ambient air, even though they span a large fraction of the known diversity in maximum carboxylation rate (kcat,C) and CO2-specificity (SC/O). For kinetic measurements of diverse rubiscos, see recent meta-analyses by Flamholz et al., 2019; Iñiguez et al., 2020. For recent measurements of the R. eutropha, R. rubrum, and S. elongatus enzymes, see Davidi et al., 2020; Occhialini et al., 2016; Satagopan and Tabita, 2016. (B) Growth rate and yield of CCMB1:p1A+vec depend on the CO2 concentration, with higher CO2 improving growth (‘vec’ denotes pFA-sfGFP). This result suggests that growth of CCMB1 is indeed coupled to the rate of carboxylation by rubisco, as predicted by the OptSlope algorithm described in Figure 2—figure supplement 4.
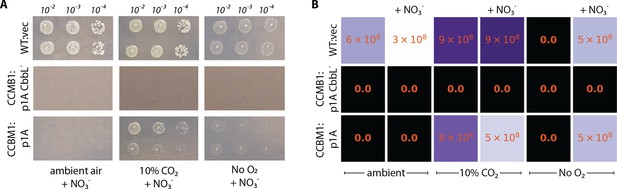
CCMB1 does not require oxygen for growth in minimal media.
(A) Titer plating assays were used to measure the viability of CCMB1:p1A grown on glycerol media under ambient air (≈0.04% CO2, 21% O2), 10% CO2 (balance air), and an anoxic mix of 10% CO2 and 90% N2 (‘No O2’). Since E. coli cannot ferment glycerol, 20 mM nitrate (NO3-) was provided as an alternate electron acceptor as marked. (B) CCMB1:p1A grows on glycerol media in the absence of O2 so long as nitrate is provided. While CCMB1:p1A colonies are noticeably smaller than WT in panel (A), the colony count is indistinguishable, as quantified in panel (B). Experiments were conducted in biological duplicate (i.e. pre-cultures from distinct colonies) with at least two technical replicates (repeated spotting from the same preculture).
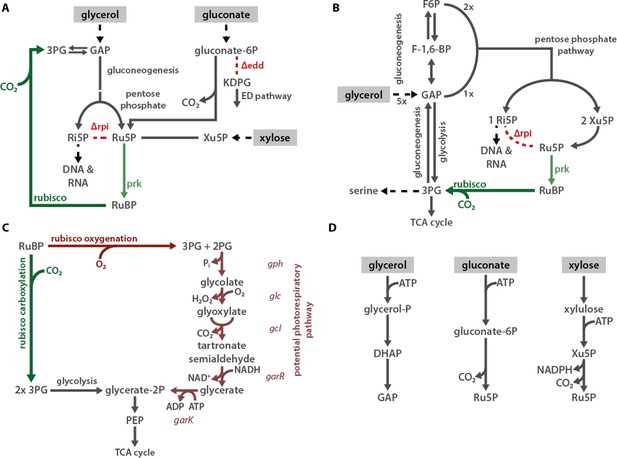
Proposed mechanisms of rubisco-dependent growth in CCMB1.
(A) CCMB1 depends on rubisco and prk for growth in glycerol, gluconate, and xylose minimal media. The common mechanism is an inability to metabolize ribulose-5-phosphate (Ru5P) due to the deletion of both ribose-phosphate isomerase genes (ΔrpiAB). When gluconate or xylose is the growth substrate, Ru5P must be produced in order to metabolize the carbon source. Although wild-type E. coli can metabolize gluconate via the ED pathway, the ED dehydratase knockout (Δedd) in CCMB1 blocks this route and forces 1:1 production of Ru5P from gluconate. Expression of prk and rubisco opens a new route of Ru5P metabolism, thus enabling CCMB1 to grow in gluconate or xylose media. Since extracellular glycerol is converted to glyceraldehyde 3-phosphate (GAP), it can be metabolized through lower glycolysis or through gluconeogenesis. The gluconeogenesis route produces hexoses that enter the pentose phosphate pathway, which is required to synthesize ribose 5-phosphate (Ri5P) for nucleotide and histidine biosynthesis. Depending on the growth rate, products of Ri5P make up 5–25% of E. coli biomass (Bremer and Dennis, 2008; Taymaz-Nikerel et al., 2010). As shown in (B), the pentose phosphate pathway forces co-production of Ri5P, Ru5P and xylulose 5-phosphate (Xu5P). In the absence of rpi activity, there is no pathway for metabolism of Xu5P or Ru5P. This defect is complemented by the expression of rubisco and prk. Notably, rubisco can also oxygenate RuBP, as shown in (C). E. coli can, in principle, recycle the oxygenation product 2-phosphoglycolate (2PG) through an ersatz salvage pathway via tartronate semialdehyde. This pathway is not the dominant mechanism of rubisco complementation because CCMB1:p1A cannot grow in ambient air, where O2 is abundant (Figure 2D). Panel (D) describes the initial metabolism of extracellular glycerol, gluconate, and xylose in E. coli Extracellular carbon sources are marked with a grey background throughout. Abbreviations: 3-phosphoglycerate (3PG), 2-phosphoglycolate (2PG), glyceraldehyde 2-phosphate (GAP), dihydroxyacetone phosphate (DHAP), ribose 5-phosphate (Ri5P), ribulose 5-phosphate (Ru5P), xylulose 5-phosphate (Xu5P), ribulose 1, 5-bisphosphate (RuBP), 2-keto-3-deoxy-6-phosphogluconate (KDGP), fructose 6-phosphate (F6P), fructose 1,6-bisphosphate (F-1,6-BP), phosphoenolpyruvate (PEP).
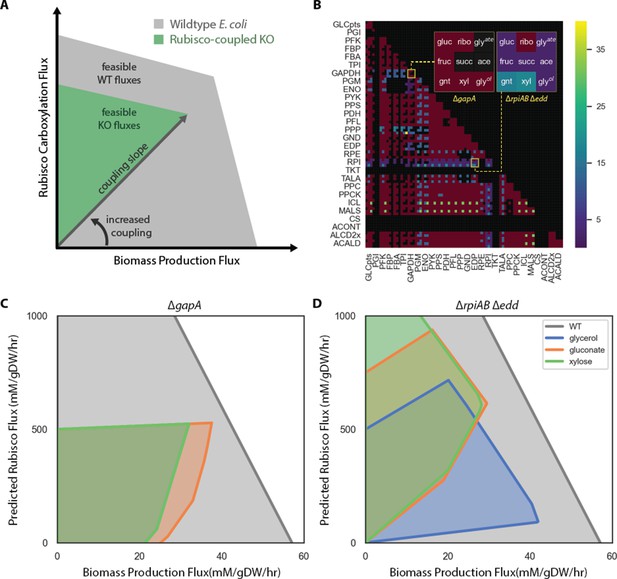
The OptSlope algorithm for designing rubisco-coupled E.colistrains.
Optslope searches for metabolic knockout mutants in which biomass production is coupled to flux through a reaction of choice (e.g. rubisco) at all growth rates. (A) shows the space of feasible biomass production and rubisco fluxes for wildtype (WT, grey) and a knockout mutant (green). For WT, biomass production and, therefore, growth rate, are independent of rubisco at all feasible growth rates (i.e. within the grey polygon). The mutant is ‘rubisco-coupled’ because maximal biomass production requires non-zero rubisco carboxylation flux and increasing biomass production demands increased carboxylation. The slope of this relationship is the ‘coupling slope.’ (B) We computationally generated pairs of E. coli central metabolic knockouts and calculated the coupling slope on nine carbon sources: glucose (gluc), fructose (fruc), gluconate (gnt), ribose (ribo), succinate (succ), xylose (xyl), glycerate (glyate), acetate (ace), and glycerol (glyol). Each double knockout is summarized as a 3 × 3 matrix of coupling slopes. Black denotes a rubisco-independent mutant and maroon a coupling slope of 0. The published mutant ∆gapA (Mueller-Cajar et al., 2007) has a coupling slope of 0 (left), while the ∆rpiAB ∆edd strain is rubisco-coupled on seven of the carbon sources (right). (C) Feasible phase space diagram for the ∆gapA strain shows that biomass production is not coupled to rubisco flux. (D) ∆rpiAB ∆edd has a positive coupling slope in glycerol, gluconate and xylose media.
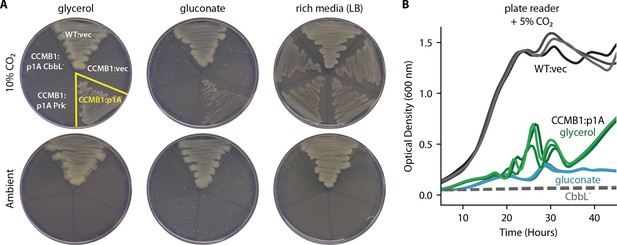
CCMB1 depends on rubisco and prk for growth in minimal media.
(A) Expression of rubisco and prk complements CCMB1 growth on M9 glycerol and gluconate media under 10% CO2, but not in ambient conditions (100 nM aTc induction in M9 plates). Mutations ablating rubisco (CbbL-) or prk (Prk-) activity abrogate growth in selective media but not in LB under 10% CO2. Growth in LB is robust and rubisco-independent in 10% CO2, but CCMB1 does not grow in ambient air even when supplied with rich media because it lacks CA genes (Merlin et al., 2003). Growth curves in (B) show the rubisco-dependence of CCMB1:p1A growth in glycerol (green) and gluconate (blue) media under 5% CO2 in a gas controlled plate reader (Materials and methods). Negative controls (CCMB1:p1A CbbL- in glycerol or gluconate media) and uninduced cultures failed to grow in these conditions (dashed grey lines). Experiments were conducted in technical sextuplicate and replicates were all consistent.
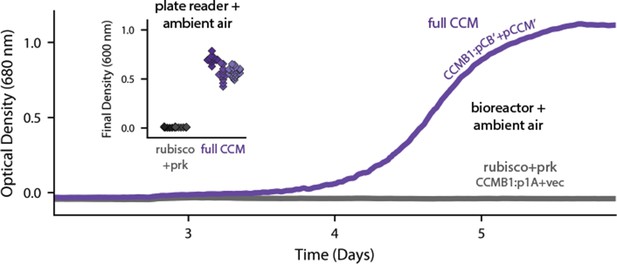
Expression of 20 CCM genes permits growth of CCMB1 in ambient air.
Time course data give representative growth curves from a bioreactor bubbling ambient air. CCMB1:pCB’ + pCCM’ grows well (purple, ‘full CCM’), while rubisco and prk alone are insufficient for growth in ambient air (grey, CCMB1:p1A+vec). Inset: a plate reader experiment in biological triplicate (different shades) gave the same result. Expressing the full complement of CCM genes led to an increase in culture density (optical density at 600 nm) of ≈0.6 units after 80 hr of cultivation. Bootstrapping was used to calculate a 99.9% confidence interval of 0.56–0.64 OD units for the effect of expressing the full CCM during growth in ambient air. Figure 3—figure supplement 1 and Appendix 2 describe the selection procedures in detail while Figure 3—figure supplement 2 shows triplicate growth curves and evaluates statistical significance.
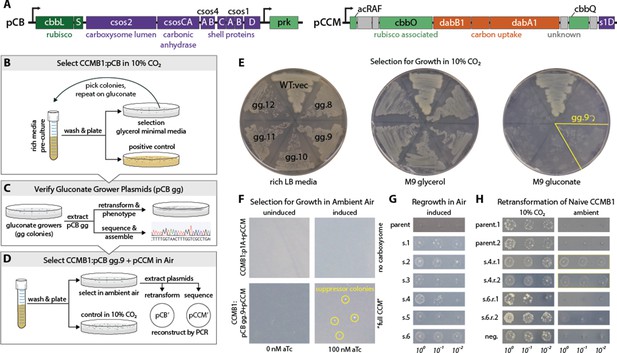
A series of selection experiments produced mutant plasmids that permit rubisco-dependent growth in ambient air.
(A) pCB and pCCM plasmids together encode 20 H. neapolitanus genes including 12 confirmed CCM components. pCB carries kanamycin resistance and has two transcriptional units both expressed under an aTc-inducible PLtetO-1 promoter (Lutz and Bujard, 1997). The first derives from pHnCB10 (Bonacci et al., 2012) and expresses 10 carboxysome proteins. The second expresses phosphoribulokinase (prk). pCCM carries chloramphenicol resistance and expresses an 11 gene operon from H. neapolitanus that contains both putative and confirmed CCM genes (Desmarais et al., 2019). Although pCB expresses both rubisco and prk, CCMB1:pCB did not initially grow in M9 media under 10% CO2 (not shown) and so we undertook a series of selections, described in panels (B–D) that ultimately led to isolation of pCB’ and pCCM’ plasmids that together enable CCMB1 to grow in ambient air. (B) We first selected CCMB1:pCB for growth on minimal media by screening for mutants able to grow on M9 glycerol and then M9 gluconate media. Gluconate growing mutant #9 (gg.9) was used for subsequent experiments as this mutant was found to grow best on gluconate (as shown in E). (C) Plasmid extracted from gg.9 was deep sequenced and electroporated into naive CCMB1 to test for plasmid linkage of growth on minimal. (D) Selection for rubisco-dependent growth in ambient air. A turbid pre-culture of the CCMB1:pCB gg.9+pCCM double transformant was washed and plated on M9 glycerol media under ambient air. Colonies formed after ≈20 days (as shown in F). Forty colonies (s.1–40) were picked into rich media, grown to saturation, washed and plated on M9 glycerol media to verify growth under ambient air. Roughly ¼ of chosen colonies regrew under ambient air to varying degrees (s.1–6 are shown in G). Plasmid extracted from several strains was deep-sequenced and electroporated into naive CCMB1 to test plasmid-linkage of growth on glycerol minimal media in ambient air. Pooled plasmid extracted from s.4 was found to confer replicable growth in ambient air (as shown in H). PCR and Gibson cloning were used to reconstruct the individual pCB and pCCM plasmids from this pool. We termed these reconstructed plasmids pCB’ and pCCM’. (E) Restreaking of gluconate-growing mutants gg.8–12 described in panel B shows that gg.9 grew best on gluconate. (F) CCMB1:pCB gg.9+pCCM double transformants were plated for mutants on M9 glycerol media under ambient air. A negative control lacking carboxysome genes (CCMB1:p1A+pCCM) was plated at the same time. Colonies formed after 20 days (bottom right) only on induced plates (100 nM) and only when all CCM genes were provided (i.e. pCB gg.9 and pCCM). (G) Several of the chosen colonies regrew in ambient air. Growth characteristics varied from colony to colony, suggesting genetic variation. (H) Pooled plasmid extracted from s.4 was found to permit naive CCMB1 to grow in ambient air. For comparison, plasmid from s.6 produced less reproducible growth in ambient air.
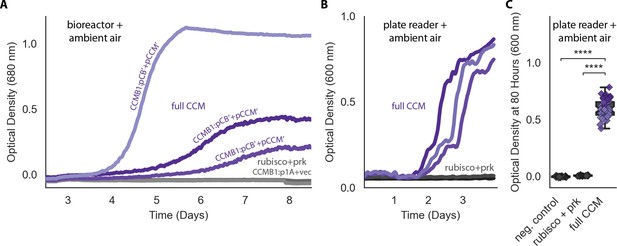
pCB’ and pCCM’ permit CCMB1 to grow in ambient air.
(A) Biological triplicate growth curves from a bioreactor bubbling ambient air. CCMB1 co-transformed with post-selection plasmids pCB’ and pCCM’ (CCMB1:pCB’ + pCCM’) grows well (purple, ‘full CCM’), while rubisco and prk alone are insufficient for growth in air (grey, ‘rubisco+prk’). Maximal growth rates for the 'full CCM' cultures ranged from 0.03 to 0.06 hr−1, corresponding to doubling times of 12–25 hr. As these are biological replicate cultures, heterogeneity in growth kinetics could be due to genetic effects (e.g. point mutations in founding colonies) or non-genetic differences (e.g. varying degree of carboxysome production during pre-culturing). (B) Data for the same strains grown in a 96-well plate in ambient air in a shaking plate reader. Different shades mark biological replicates (pre-cultures deriving from three distinct colonies). Additionally, each preculture was used to inoculate at least 12 technical replicates. (C) Quantification of the experiment in panel (B) using endpoint data at 80 hr for biological and technical replicates. Panel (C) uses the same colors as (A) and (B) with the addition of a rubisco active site mutant as a negative control (grey, CCMB1:p1A- + vec). ‘****’ indicates p<10−10. p-Values were calculated with a Bonferroni-corrected two-sided Mann-Whitney-Wilcoxon test. 104-fold bootstrapping was used to compare ‘full CCM’ data to ‘rubisco + prk’ and estimate a confidence interval for the effect of expressing a full CCM on growth in ambient air, which gave a 99.9% confidence interval of 0.56–0.64 OD units.
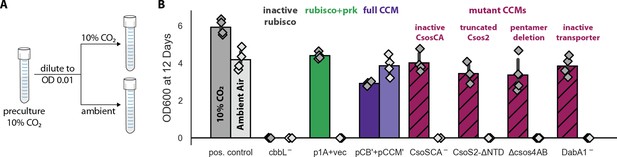
Growth in ambient air depends on the known components of the bacterial CCM.
We generated plasmid variants carrying inactivating mutations to known components of the CCM. (A) Pre-cultures were grown in 10% CO2 and diluted into pairs of tubes, one of which was cultured in 10% CO2 and the other in ambient air (Materials and methods). Strains were tested in biological quadruplicate and culture density was measured after 12 days to ensure an endpoint measurement of capacity to grow. (B) Targeted mutations to CCM components ablated growth in ambient air while permitting growth in 10% CO2, as expected. The left bar (darker color) gives the mean endpoint density in 10% CO2 for each strain. The right bar (lighter color) gives the mean in ambient air. Error bars give a 95% confidence interval for the mean. From left to right, in pairs: a positive control for growth (a complemented carbonic anhydrase knockout in grey, see Materials and methods) grew in 10% CO2 and ambient air, while a negative control CCMB1 strain carrying catalytically inactive rubisco (CCMB1:pCB’ CbbL-+pCCM’) failed to grow in either condition; CCMB1 expressing rubisco and prk but no CCM genes (green, CCMB1:p1A+vec) grew only in 10% CO2; CCMB1:pCB’+pCCM’ grew in 10% CO2 and ambient air, recapitulating results presented in Figure 3. The following four pairs of maroon bars give growth data for strains carrying targeted mutations to CCM genes: an inactivating mutation to carboxysomal carbonic anhydrase (CCMB1:pCB’ CsoSCA-+pCCM’), deletion of the CsoS2 N-terminus responsible for recruiting rubisco to the carboxysome (CCMB1:pCB’ CsoS2 ΔNTD+pCCM’), deletion of pentameric vertex proteins (CCMB1:pCB’ ΔcsoS4AB + pCCM’), and inactivating mutations to the DAB carbon uptake system (CCMB1:pCB’ DabA1- + pCCM’). All four CCM mutations abrogated growth in air while permitting growth in 10% CO2. The positive control is the CAfree strain expressing human carbonic anhydrase II (Materials and methods). Figure 4—figure supplement 1 describes statistical analyses, a 4-day replicate experiment, and additional mutants testing the contribution of rubisco chaperones to the CCM. Figure 4—figure supplement 2 gives measurements of media pH after growth in 10% CO2 and ambient air. Detailed description of all plasmid and mutation abbreviations is given in Supplementary file 1.
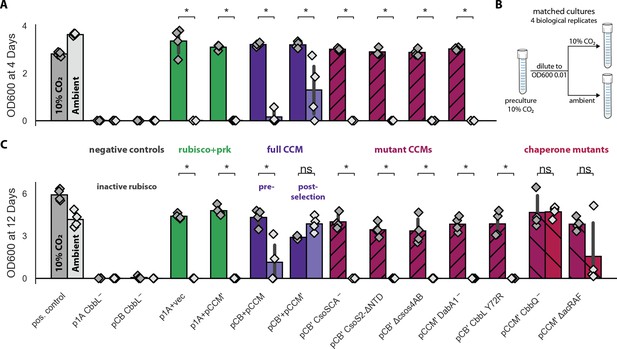
Targeted mutations to the CCM eliminate growth in ambient air.
Pre-cultures were grown to saturation in 10% CO2 and then diluted to an optical density of 0.01 (600 nm) into two tubes (Materials and methods). One tube was grown in 10% CO2 and the other in ambient air, as diagrammed in (B). Cells were incubated for 4 days before measuring optical density in (A) and 12 days in (C). The left bar (darker color) gives the mean endpoint density of biological quadruplicate cultures in 10% CO2 and the right bar (lighter color) gives the mean in ambient air. Error bars give a 95% confidence interval of measurements. (A) and (C) share the leftmost 11 strains. From left to right: a positive control (grey, grows in both conditions), two negative controls carrying active site mutants of rubisco (CCMB1:p1A-+vec and CCMB1:pCB’ CbbL-+pCCM’), CCMB1 expressing rubisco and prk but no CCM genes (green, CCMB1:p1A+vec) or an incomplete set of CCM genes (green, CCMB1:p1A+pCCM’), CCMB1:pCB+pCCM which carries the pre-selection CCM plasmids (purple), and CCMB1:pCB’+pCCM’ which carries the post-selection plasmids. ‘vec’ denotes an appropriate vector control (pFA-sfGFP). The following pairs of maroon bars describe strains carrying plasmids with targeted CCM mutations: CCMB1:pCB’ CsoSCA-+pCCM’ which carries an inactivating mutation to carboxysomal carbonic anhydrase, CCMB1:pCB’ CsoS2 ΔNTD +pCCM’ harboring a deletion of the N-terminal domain of CsoS2 responsible for recruiting rubisco to the carboxysome, CCMB1:pCB’ ΔcsoS4AB + pCCM’ lacking both genes pentameric vertex proteins, and CCMB1:pCB’ DabA1- + pCCM’ carrying an inactivated DAB carbon uptake system. (A) CCMB1 grows well in ambient air only when given a full complement of CCM genes on the post-selection plasmids. All mutations to the CCM abrogate growth in air (maroon). Panel (C) shows consistent results over a 12-day time period. (C) describes three additional mutants: CCMB1:pCB’ CbbL Y72R + pCCM’ carrying a mutation to the rubisco large subunit that eliminates rubisco-CsoS2 binding, CCMB1:pCB’ + pCCM’ CbbQ- harboring inactivating mutation to the CbbQ subunit of the rubisco activase complex, and CCMB1:pCB’ + pCCM’ ΔacRAF lacking the putative rubisco chaperone acRAF (CCMB1:pCB’ + pCCM’ ΔacRAF). Ablation of rubisco-CsoS2 interaction should eliminate recruitment of rubisco to the carboxyome (Oltrogge et al., 2020). Accordingly, the Y72R mutation eliminated growth in air. Chaperone mutants (CbbQ or acRAF) were both viable in air, although removal of acRAF produced a substantial growth defect (2.5 fold in mean and 8.5 fold in median final density). The positive control strain is the CAfree strain expressing human carbonic anhydrase II (Materials and methods). p-Values calculated by a one-sided Mann-Whitney-Wilcoxon test. ‘*’ denotes a p<0.05. Detailed description of all plasmid abbreviations is given in Supplementary file 1.
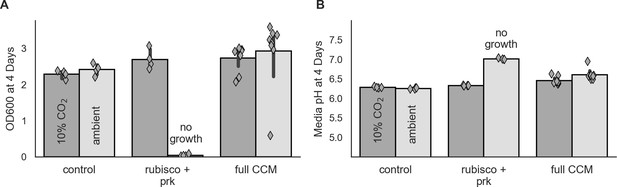
Tandem endpoint measurements of growth and culture pH.
Pre-cultures were grown to saturation in 10% CO2 and then diluted to an optical density of ≈0.01 (600 nm) as in Figure 4 (Materials and methods). One tube was grown in 10% CO2 and the other in ambient air. The media pH was measured in technical triplicate prior to the experiment and was found to be 7.02 ± 0.01. Cells were incubated for 4 days before measuring optical density and pH of culture supernatants. In both panels, the left bar (darker color) gives the mean endpoint density or pH of biological replicate cultures in 10% CO2 and the right bar (lighter color) gives the mean in ambient air. Error bars give a 95% confidence interval of measurements. Consistent with Figure 4, panel (A) shows that the positive control (leftmost, CAfree:vec+pFA-HCAII) and CCMB1:pCB’+pCCM’ grow (rightmost) in both conditions, while CCMB1:p1A+vec fails to grow in ambient air (center). Panel (B) gives endpoint pH measurements for the same cultures. In all cases, growth led to a relative acidification of the media, consistent with chemiosmotic H+ pumping from the cytoplasm to the media. For the control, the endpoint pH was ≈6.3 in both conditions. For CCMB1:pCB’+pCCM’ (‘full CCM’) the endpoint pH was ≈6.5 in 10% CO2 and ≈6.6 in ambient. These differences between conditions and between control and experimental samples are consistent with inward-directed H+ pumping mechanism proposed for the DAB-type transport systems (Desmarais et al., 2019), but could also be due to large differences in the growth rate and intracellular metabolic flux distributions due to the multiple central metabolic knockouts in the CCMB1 strain.
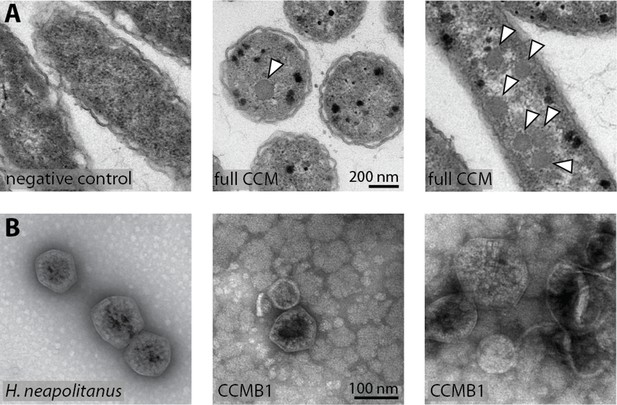
CCMB1:pCB’+pCCM’ produces carboxysomes when grown in air.
(A) Polyhedral bodies resembling carboxysomes are evident in electron micrographs of CCMB1:pCB’+pCCM’ cells grown in air (full CCM, both images on the right) but were not observed in a negative control lacking pCB and pCCM plasmids (left, Methods). All panels have equal scale. (B) Carboxysome structures purified from CCMB1:pCB’+pCCM’ grown in ambient air (Materials and methods, right) resemble structures isolated from the native host (left) in size and morphology. Figure 5—figure supplement 2 gives full size and additional images clearly showing rubisco inside isolated carboxysomes. SDS-PAGE gels in Figure 5—figure supplement 1 demonstrate co-migration of rubisco large and small subunits with carboxysomes structures through the purification procedure.
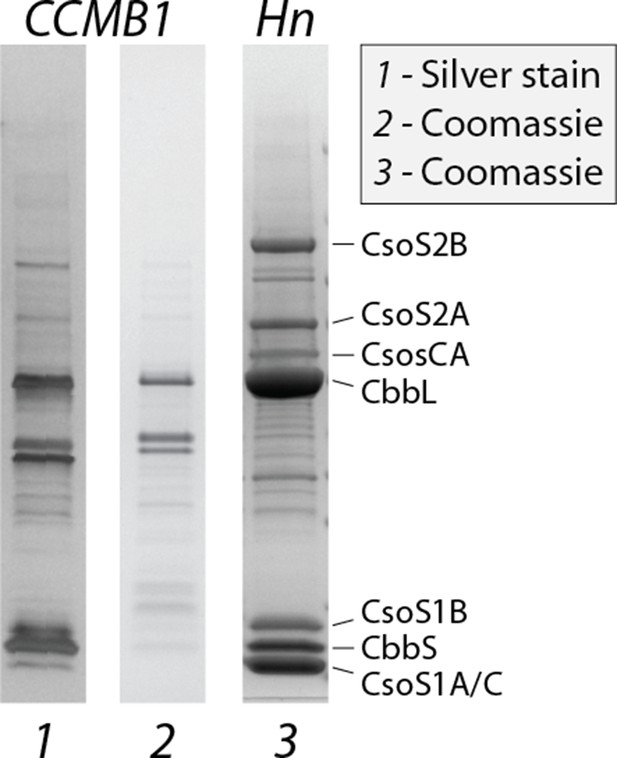
Carboxysomes purified from CCMB1:pCB’ + pCCM’ contain rubisco and other known carboxysome components.
Lanes 1 and 2 give purifications from CCMB1:pCB’+pCCM’ grown in ambient air stained with silver and coomassie stains, respectively. Lane 3 is a purification from wild-type H. neapolitanus (Materials and methods). The legend on the right marks the carboxysome components that are typically visible on preparations from the native host: the shell proteins CsoS1CAB, two forms of the disordered protein CsoS2, the carbonic anhydrase CsoSCA, and the rubisco large and small subunits CbbLS. CbbLS bands are evident in all lanes, and all three purifications were found to contain carboxysome-like structures when imaged by transmission electron microscopy, as shown in Figure 5 and Figure 5—figure supplement 2.
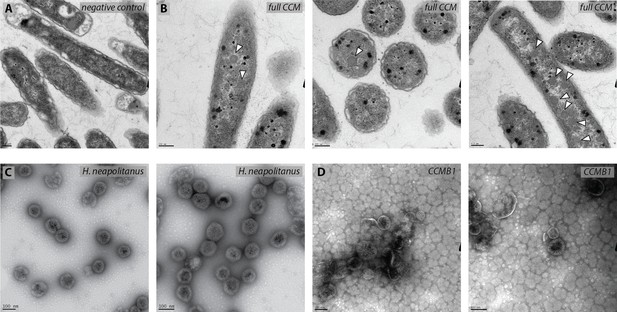
CCMB1:pCB’ + pCCM’ produces polyhedral bodies resembling carboxysomes when grown in ambient air.
Thin section transmission electron micrographs of a negative control strain (A) and experimental cells (B) show that air-grown CCMB1:pCB’+pCCM’ cells (‘full CCM’ in panel B) contain morphological carboxysomes (white arrows). The negative control for carboxysome expression is CAfree:pFE-sfGFP + pFA-HCAII (Materials and methods). Expression of CCM was associated with production of black-staining stress granules, which were not observed in images of the negative control. (C) Carboxysomes isolated from wild-type H. neapolitanus displayed regular pseudo-icosahedral structures with ≈100 nm diameter, as expected (Shively et al., 1973). (D) Carboxysomes isolated from CCMB1 were less regular, but clearly resemble native structures. Preparations also contained ‘rosette’ structures we often observe when isolating carboxysomes from E. coli (far right panel). Purification yields from CCMB1:pCB’+pCCM’ were much lower than is typical for preparations from wild-type E. coli which may explain the relative abundance of rosette structures.
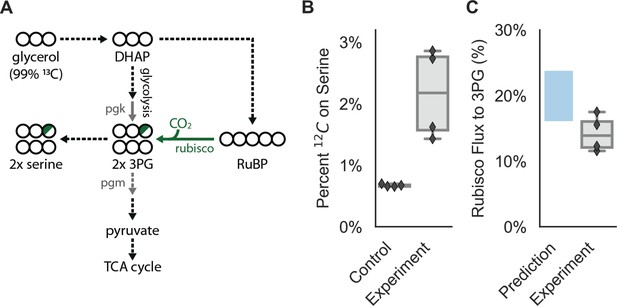
CCMB1:pCB’+pCCM’ fixes CO2 from ambient air into biomass.
Biological replicate cultures were grown in ambient air in M9 media containing 99% 13C labeled glycerol such that 12CO2 from air is the dominant source of 12C. In (A) 13C is depicted as open circles and partial 12C incorporation is indicated in green. As serine is a direct metabolic product of 3PG, we expect 12C enrichment on serine when rubisco is active in CCMB1 cells. 3PG also derives from glycolytic metabolism of glycerol, so complete 12C labeling of serine was not expected. (B) The 12C composition of serine from CCMB1:pCB’ + pCCM’ (‘Experiment’) is roughly threefold above the control strain (CAfree:vec+pFA-HCAII), which grows in a rubisco-independent manner (Materials and methods). Figure 6—figure supplement 1 gives 12C composition of all measured amino acids. (C) The fraction of 3PG production due to rubisco was predicted via Flux Balance Analysis and estimated from isotopic labeling data (Materials and methods, Appendix 3). Estimates of the rubisco flux fraction exceeded 10% for all four biological replicates and the mean estimate of ≈14% accords reasonably with predictions ranging from 16 to 24%. Appendix 3 and Figure 6—figure supplements 2–3 detail the flux inference procedure and give additional evidence for in vivo carboxylation from the fragmentation of serine.
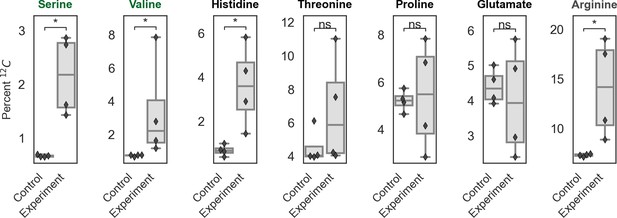
Isotopic composition of amino acids from total biomass hydrolysate.
Cells were grown under ambient air in M9 media containing 99% 13C-labeled glycerol (0.4% v/v) so that nearly all 12C in biomass must derive from inorganic carbon. The isotopic composition of amino acids in total biomass hydrolysate of CCMB1:pCB’ + pCCM’ and an appropriate rubisco-independent control were measured via LC-MS (Materials and methods). The control strain is CAfree complemented with the human carbonic anhydrase II, which does not express rubisco (Materials and methods). Serine and valine, which are marked in green, are downstream of the rubisco product 3PG in E. coli central metabolism and, accordingly, show significantly greater 12C incorporation in CCMB1:pCB’ + pCCM’ than the control. Most of the carbon atoms in histidine derive from ribose, which might contain rubisco-derived carbon atoms if the addition of rubisco enables cycling of carbon deriving from CO2 through the pentose phosphate pathway and gluconeogenesis. This could explain the significant increase in histidine labeling in the experimental samples relative to the control. Threonine, proline, and glutamate are synthesized from precursors deriving from the TCA cycle and thus their carbon atoms are not expected to derive primarily from 3PG (Szyperski, 1995). Arginine is synthesized via a rubisco-independent carboxylation of glutamate by the addition of carboxyphosphate (Gleizer et al., 2019), and so the difference between arginine and glutamate labeling is used to calculate the isotopic composition of intracellular inorganic carbon (Ci, Appendix 3). Notably, intracellular Ci derives both from extracellular Ci (predominantly 12C) and intracellular decarboxylation of the 99% 13C glycerol carbon source. As such, the composition will depend on Ci uptake as well as the rate of glycerol metabolism. Control cells also grew faster than CCMB1:pCB’+pCCM, which might explain why arginine from these cells contains significantly less 12C than control samples (i.e. due to rapid decarboxylation of glycerol).
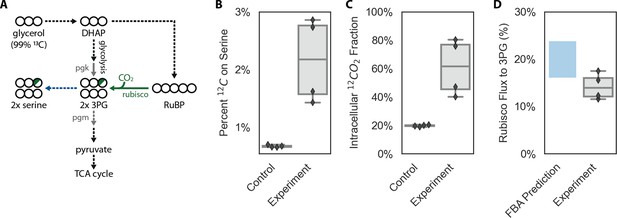
12C enrichment on serine is consistent with intracellular CO2 fixation.
Cells were grown under ambient air in M9 media containing 99% 13C-labeled glycerol (0.4% v/v) so that nearly all 12C in biomass must derive from inorganic carbon. In (A) 13C atoms are depicted as open circles and fractional 12C labeling by a partial green fill color. In CCMB1, 3-phosphoglycerate (3PG) can be produced either through glycolytic metabolism of glycerol (via dihydroxyacetone-phosphate, DHAP) or through rubisco-catalyzed carboxylation of RuBP. At most ⅙ of the carbon atoms on 3PG will be 12C when rubisco is active in vivo. In practice, this fraction will be less than ⅙ because some of the intracellular inorganic carbon pool (Ci) derives from decarboxylation of 13C-labeled glycerol and also because a large fraction of intracellular 3PG is produced through glycolysis (Appendix 3). Serine is a direct metabolic product of 3PG and so reports on the labeling of 3PG. As such, we measured the 12C composition of amino acids in total protein hydrolysate via LC-MS (Materials and methods). (B) Serine from CCM-expressing CCMB1 cells (‘Experiment’) displayed roughly threefold higher 12C labeling than controls, which grow in a rubisco-independent manner (Materials and methods). (C) Rubisco carboxylation draws from the intracellular inorganic carbon pool, whose 12C composition can be inferred for each sample by comparing the labeling of L-arginine and L-glutamate (Materials and methods). The mean 12C fraction of intracellular Ci was estimated to be 20% ± 1% and 61% ± 20% for the control and experiment, respectively. (D) These values were integrated to estimate the percent of 3PG production flux that is due to carboxylation by rubisco (Appendix 3), which was inferred to be 14% ± 3%. These values compare favorably with predictions made via Flux Balance Analysis (16-24%, Appendix 3). A sampling approach, described in Appendix 3, was used to estimate the uncertainty in these rubisco flux inferences. 99% confidence intervals on the rubisco flux fraction were strictly positive for each biological replicate, with 99% of all posterior estimates between 5% and 20.3% across all four replicates.
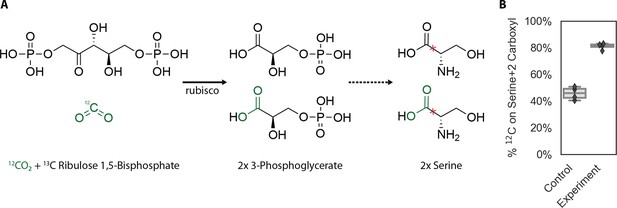
Fragmentation of serine M+2 isotopologues confirms rubisco-catalyzed CO2 addition in growing cells.
In (A) CO2 and carboxyl groups deriving from it are marked in green. When rubisco carboxylates ribulose 1, 5-bisphosphate, it produces two molecules of 3-phosphoglycerate only one of which has a CO2-derived carboxylic acid. Serine biosynthesis does not alter this position, and so we expect one CO2-derived carboxylic acid for every two serines that ultimately derive from rubisco-catalyzed carboxylation of ribulose 1, 5-bisphosphate. If fully 13C-labeled ribulose 1, 5-bisphosphate undergoes carboxylation with 12CO2, it would result in one fully 13C-labeled 3-phosphoglycerate and one with 12C at the carboxyl position as diagrammed. In panel (B), we summarize data from multiple reaction monitoring of the fragmentation of the serine M+2 isotopologue. This molecule has two additional mass units due to two 13C atoms. Fragmentation cleaves the bond between the carboxyl carbon and the ɑ-carbon on L-serine, marked by a red ‘x’ (Piraud et al., 2003), enabling us to ask: what fraction of the carboxyl carbons are 12C? If the 12C was incorporated at random, it would appear in the carboxyl position 33% of the time. However, if the carboxyl position derives appreciably from carboxylation by rubisco, we expect 12C enrichment at the carboxyl position as described in panel (A). We find that the control strain contains ≈45% 12C at this position, consistent with some background incorporation of CO2 into carboxyl groups in central metabolism, but that the experimental strain (CCMB1:pCB’+pCCM’ grown in ambient air) incorporates nearly twofold more 12C at the same position (≈80%), as would be expected if rubisco produces a substantial fraction of intracellular 3-phosphoglycerate.
Additional files
-
Supplementary file 1
This file comprises five supplementary tables.
Table 1 describes the strains used in this study; Table 2 details all plasmids used; Table 3 gives primer sequences used in genotyping assays; Table 4 describes mutations observed during selection experiments; Table 5 gives a detailed description of all 20 genes expressed in this study with a detailed bibliography describing the evidence underpinning our current understanding of the molecular funciton of each gene.
- https://cdn.elifesciences.org/articles/59882/elife-59882-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/59882/elife-59882-transrepform-v2.pdf





