Structures reveal gatekeeping of the mitochondrial Ca2+ uniporter by MICU1-MICU2
Figures
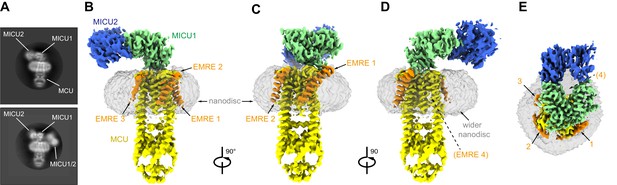
Cryo-EM reconstruction of the holocomplex.
(A) 2D class averages showing side-views of complexes with one or two MICU1-MICU2 heterodimers associated (top and bottom panels, respectively). (B–E) Cryo-EM reconstruction of the holocomplex, shown in orthogonal views. Densities are colored accordingly: MCU (yellow), EMRE (orange), MICU1 (green), MICU2 (blue), and nanodisc (semitransparent gray). Numbers in (E) represent the locations of EMRE subunits. The location of where EMRE 4 would be expected to bind is indicated by parentheses (D–E).
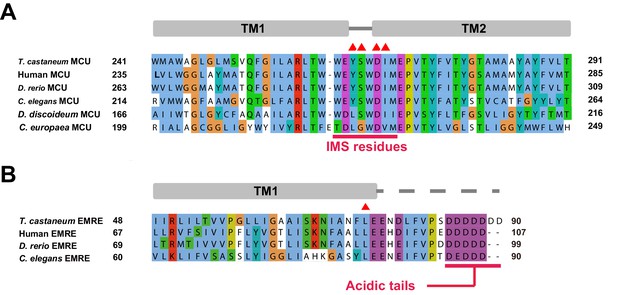
Sequence alignments of MCU and EMRE.
(A) The amino acid sequences of transmembrane regions of T. castaneum, C. europaea, D. discoideum, C. elegans, D. rerio (Zebrafish) and human MCUs are aligned and colored according to the ClustalW convention (NCBI Reference Sequence: XP_008192975.1 (T. castaneum), UniProt accession numbers: W2SDE2, Q54LT0, Q21121, Q08BI9 and Q8NE86, respectively). Secondary structures are indicated with cylinders representing α-helices, and residues facing the intermembrane space are indicated with a red line. Residues involved in MICU1-binding are highlighted with red triangles. (B) The amino acid sequences of T. castaneum, C. elegans, D. rerio (Zebrafish) and human EMREs are aligned and colored in the same manner (UniProt accession numbers: D6X268_TRICA, Q9U3I4, A0A0J9YJ98 and Q9H4I9, respectively). Secondary structures are indicated with bars representing α-helices, solid lines representing structured loop regions, and dashed lines representing disordered regions. The acidic C-terminal tails of EMRE proteins are highlighted with a red line. Leu 92 is noted with a red triangle.
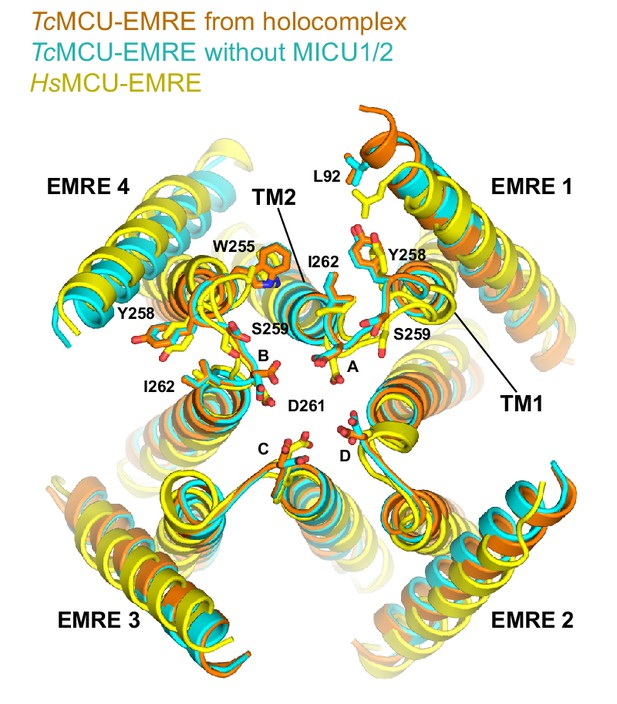
Comparison of the MICU1 receptor sites in human and beetle MCU-EMRE complexes.
Superposition of TcMCU-EMRE from the holocomplex (gold), TcMCU-EMRE without MICU1-MICU2 (cyan), and human MCU-EMRE (yellow, PDB: 6O58) is shown from the perspective of the IMS. MCU and EMRE are shown as cartoons. Residues involved in MICU1 binding are labeled and depicted as sticks (RMSD between these for human and beetle MCU/EMRE is 1.5 Å for Cα atoms).
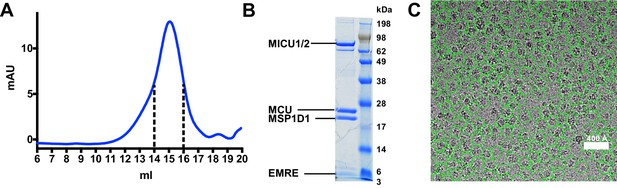
Purification of the MCU-EMRE-MICU1-MICU2 holocomplex under resting ([Ca2+]free <1 nM) conditions and cryo-EM micrograph.
(A) Size exclusion chromatography (SEC) profile of the purified holocomplex. Peak fractions between the dashed lines were collected for cryo-EM analysis. (B) SDS-PAGE analysis of the SEC fractions (Coomassie stained). Protein components are labeled. (C) Portion of a representative micrograph (defocus of −2.5 μm). Automatically picked particles are circled in green.
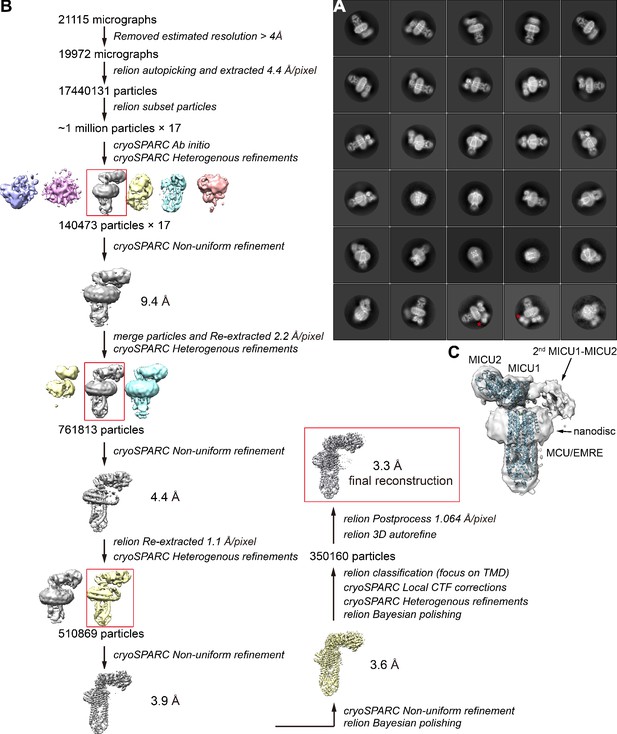
Flowchart of cryo-EM data processing of holocomplex.
(A) Representative 2D class averages ranked in order of class distribution (highest to lowest occupancy from upper left to lower right). Asterisks denote a second MICU1-MICU2 complex in certain classes. (B) Flowchart of cryo-EM data processing and 3D reconstruction. (C) Cryo-EM map (semitransparent representation, ~10 Å resolution) from the ~2% of particles that contain two MICU1-MICU2 heterodimers per channel assembly. The atomic structure of the holocomplex containing one MICU1-MICU2 heterodimer is shown in cartoon representation for comparison.

Cryo-EM analysis of the holocomplex.
(A) FSC curves of the final 3D reconstruction. The half-map (black) and map-to-model (orange) FSC curves are shown (dotted lines indicate 0.143 and 0.5 levels and corresponding resolutions). (B) Euler angle distribution plot of the final 3D reconstruction. (C) Local resolution estimation (calculated with Resmap). The unsharpened map is colored as indicated. Middle and right panels are slices through the map. (D–E). EM densities for indicated regions of the atomic model.
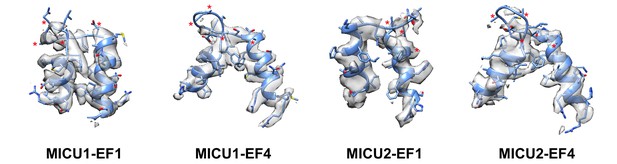
The EF-hands of MICU1 and MICU2 are in apo conformations in the holocomplex structure.
Local densities (transparent surface representations) of the four EF-hand domains are shown in the context of the atomic model (cartoon and sticks). Densities for Ca2+ ions are absent and the Ca2+-binding residues (asterisks) have poorly defined density.
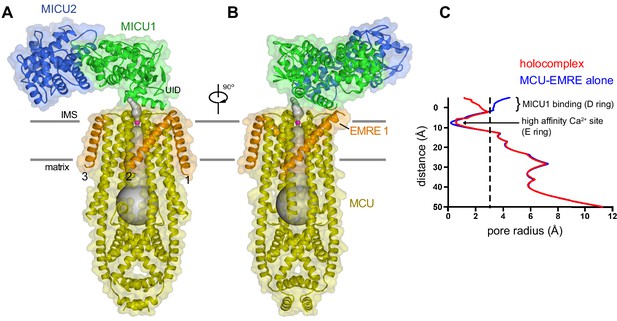
Architecture of the holocomplex.
(A and B) Cartoon representations of the holocomplex, colored as in Figure 1, and shown with semitransparent surfaces. Gray bars represent approximate boundaries of the hydrophobic core of the membrane. A Ca2+ion in the E ring is drawn as a purple sphere. The pore (semitransparent gray surface) is depicted as the minimal radial distance from its center to the nearest van der Waals protein contact. (C) Pore dimensions in structures of TcMCU-EMRE with and without MICU1-MICU2 (red and blue, respectively). A dashed line indicates the radius of a hydrated Ca2+ ion.
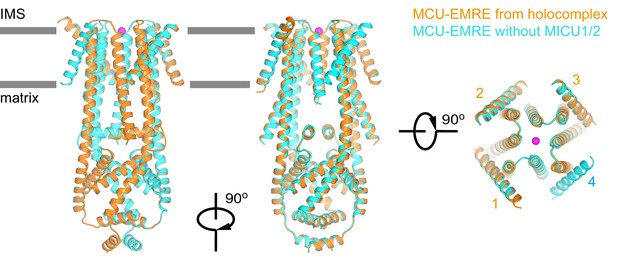
Comparison of the structures of TcMCU-EMRE with and without MICU1-MICU2.
Superposition of the structures of TcMCU-EMRE from the holocomplex (gold) and TcMCU-EMRE without MICU1-MICU2 (cyan) are shown in three orthogonal views. Ca2+ is depicted as a purple sphere. EMRE subunits are labeled 1 to 4. The RMSD for Cα atoms is 0.24 Å between the structures.
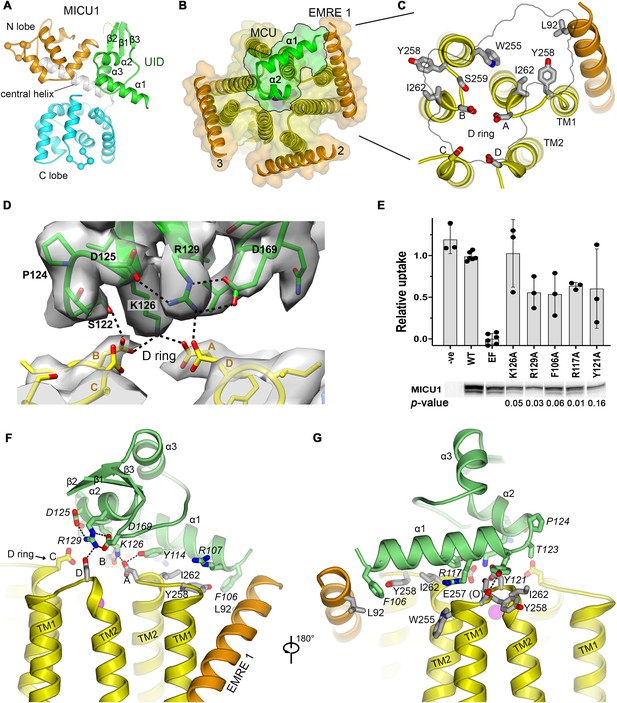
The UID and its receptor site.
(A) Domain architecture of MICU1. Cartoon representation colored according to subdomains. Spheres indicate the Cα positions of amino acids that would bind Ca2+ in EF-hands EF1 and EF4. (B) The UID-receptor interface. A semitransparent surface-cartoon rendition of MCU and EMRE is shown, viewed from the IMS. An outline marks the boundaries of the interface of the UID with the channel. α1 and α2 of the UID are depicted as green ribbons. (C) Close up view of the receptor site, with residues that interact with the UID drawn as sticks. (D) Cryo-EM density and interactions at the IMS mouth of the pore. The interface region between MICU1 and the D ring is shown (MICU1, green; MCU, yellow), with density depicted as a semitransparent surface. Dashed lines indicate hydrogen bonds. (E) Mutagenesis of the UID and the effect on mitochondrial Ca2+ uptake. The indicated mutants of human MICU1, made in the background of disrupted EF-hand domains (EF), were transiently expressed in MICU1 knockout cells and uptake was quantified relative to wild type (WT) MICU1 and disrupted EF-hand (EF) controls. Western blots demonstrate expression. ‘-ve’ indicates untransfected cells. (mean ±s.d., independent experiments: n = 6 for WT and EF controls, and n = 3 for the remainder. Student’s t-Test p-values for mutants relative to the EF construct, calculated using the two-tailed distribution with unequal variance method, are listed). (F and G) Views showing UID-receptor site interactions (sticks). MICU1 residues are in italics.
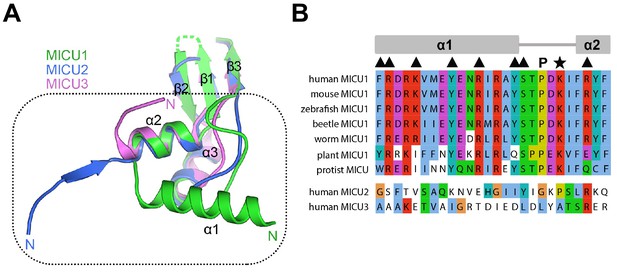
The UID is unique to MICU1.
(A) Comparison of the UID of MICU1 (green) to corresponding regions of MICU2 and MICU3 from of X-ray structures (PDB: 6EAZ and 6AGJ). A box highlights differences with α1 of MICU1. (B) Sequence alignment of various UIDs and with corresponding regions of MICU2 and MICU3. Residues that interact with the channel are denoted with triangles; Lys 126 as a star.
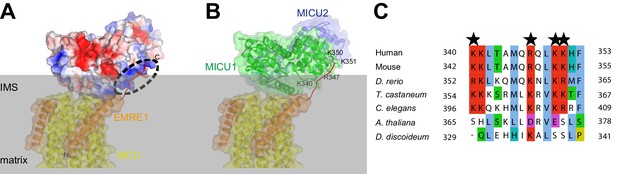
Electrostatic surface of MICU1-MICU2 and possible interaction site for the C-terminal tail of EMRE.
(A) A view of holocomplex parallel to the membrane. MCU and EMRE are shown as cartoon representations and colored in gold and orange, respectively. The MICU1-MICU2 heterodimer is shown as a molecular surface and colored according to electrostatic potential (light gray regions are neutral; red, −4 kT e−1; blue, +4 kT e−1). A hypothetical model of the acidic C-terminal tail of EMRE1 is drawn as a red line and docked on a positive-charged surface on MICU1 (dashed oval). For clarity, the matrix domains of MCU are omitted. Mitochondrial inner membrane is depicted as transparent gray bar. (B) The same perspective as (A) with MICU1 and MICU2 depicted as cartoons in transparent surfaces. Basic amino acids that constitute the positive-charged surface highlighted in (A) are drawn as sticks and labeled. (C) Sequence alignment showing conservation of the basic region among of MICU1 proteins. The residues indicated in (B) are marked by black stars. This region is conserved among metazoan species (from C. elegans to human) but not with A. thaliana and D. discoideum, which lack EMRE homologs.
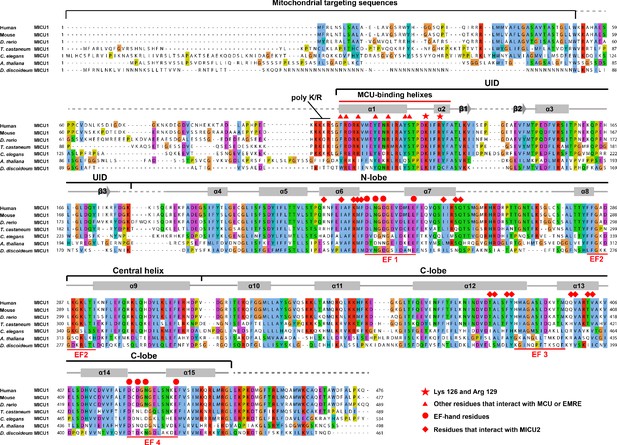
Sequence alignment of MICU1 proteins.
The amino acid sequences of human, M. musculus, D. rerio, T. castaneum, C. elegans, A. thaliana, and D. discoideum MICU1 proteins are aligned and coloured according to the ClustalW convention (UniProt accession numbers: Q9BPX6, Q8VCX5, A4IG32, D6WJK2, Q95PZ2, Q9SZ45 and Q54JS1, respectively). Secondary structures are indicated with cylinders representing α-helices, arrows representing beta sheets, solid gray lines representing structured loops, and dashed gray lines representing disordered regions.
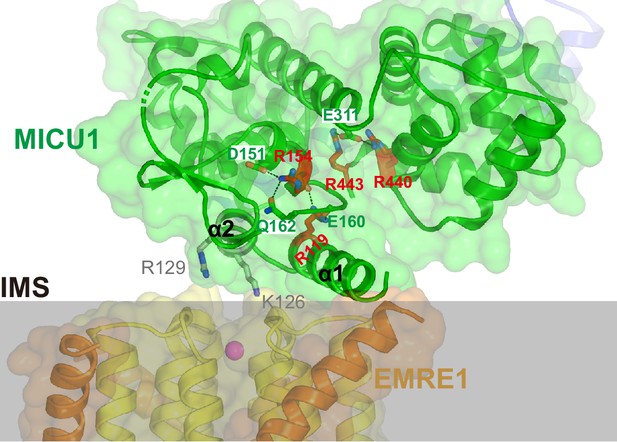
Locations of MICU1 residues that were previously proposed to interact with the channel.
A portion of the holocomplex is shown from the side. MCU, EMRE, MICU1 and MICU2 are shown as cartoons in transparent surfaces and colored in gold, orange, green and blue, respectively. Ca2+ in the ‘E’ ring of the selectivity filter is depicted as a purple sphere. MICU1-Lys 126 and MICU1-Arg 129 are shown as gray sticks. A portion of the mitochondrial inner membrane is depicted as transparent gray bar. Basic residues of MICU1 that have been proposed on the basis of previous studies to interact with MCU are drawn as red sticks (Paillard et al., 2018, Phillips et al., 2019). Amino acids that coordinate these are drawn as green sticks, with hydrogen bonds indicated by dashed lines. Although the proposed residues don’t interact with MCU or EMRE, one can imagine how their mutation could disrupt the structure of the UID and its binding to the channel. MICU1-Arg 119 and MICU1-Arg 154, which were proposed to interact with MCU-Asp 261 on the basis of one study (Phillips et al., 2019), contribute to stabilization of the UID but do not coordinate MCU or EMRE directly. MICU1-Arg 440 and MICU1-Arg 443, which were proposed to interact with MCU on the basis of another study (Paillard et al., 2018). These residues are located within the C-lobe of MICU1, approximately 25 Å from the pore, where they form hydrogen bonds with MICU1-Glu 311 from the N-lobe; their mutation would be expected to destabilize the overall structure of MICU1.
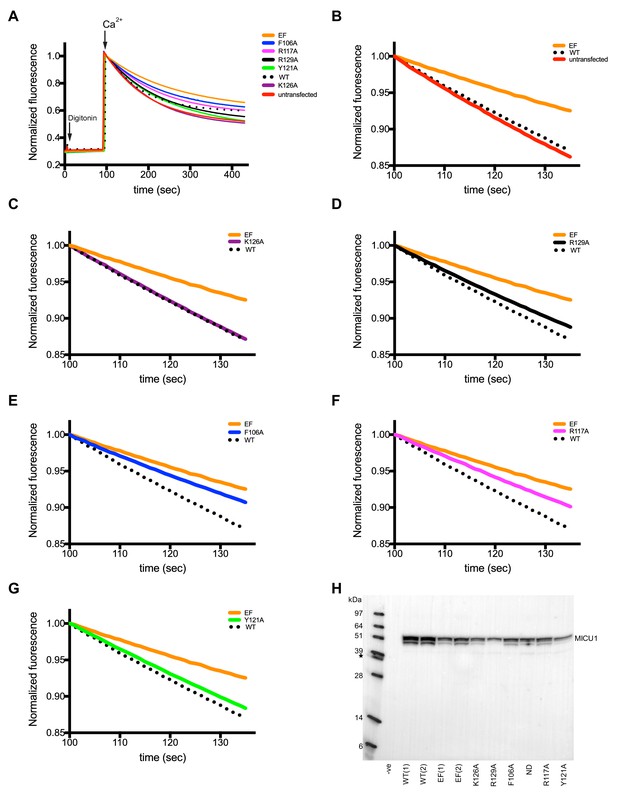
Functional analysis of the UID interactions in human mitochondria.
(A) Representative mitochondrial Ca2+ uptake experiments for digitonin-permeabilized MICU1 knockout cells expressing various human MICU1 constructs as indicated. ‘WT’ indicates wild type MICU1, ‘EF’ indicates MICU1 with disrupted EF-hands. The other indicated mutations were made on the background of the EF-hand mutant. Fluorescence data are normalized to the value at 100 s, where fluorescence readings are within ±5% for all curves. (B–G) Close up views of the 100 to 135 s time window showing the region used to determine slope. (H) Western blot analysis using a Rho-1D4 antibody, showing MICU1 protein expression levels. ‘-ve’ represents untransfected cells; ‘ND’ is a construct for which uptake was not studied; an asterisk represents a western control (MCU labeled with a 1D4 tag) that was included with molecular weight standards in the first lane.
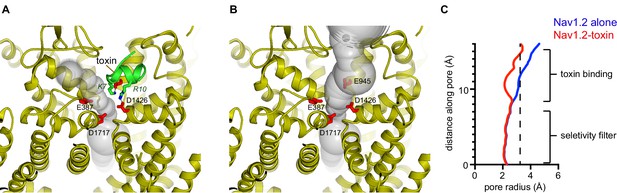
Toxin-Nav1.2 channel interaction.
(A–B) Depictions of Nav1.2 alone (B) and with bound μ-conotoxin KIIIA (A) (PDB:6J8E), showing the transmembrane and extracellular regions, viewed from the extracellular side above the selectivity filter. The Nav1.2 channel is colored gold and μ-conotoxin KIIIA is green; both are shown as cartoon representations. The pore (a semitransparent gray surface) is depicted as the minimal radial distance from its center to the nearest van der Waals protein contact. Lys 7 and Arg 10 residues of μ-conotoxin KIIIA (green sticks) coordinate two of four similarly positioned acidic amino acids at the mouth of the selectivity filter (red sticks). (C) Pore dimensions of Nav1.2 with and without μ-conotoxin KIIIA (red and blue, respectively). A dashed line indicates the radius of a hydrated Na+ ion. The distance along the pore (y-axis) is relative to a point at the lower end of the selectivity filter.
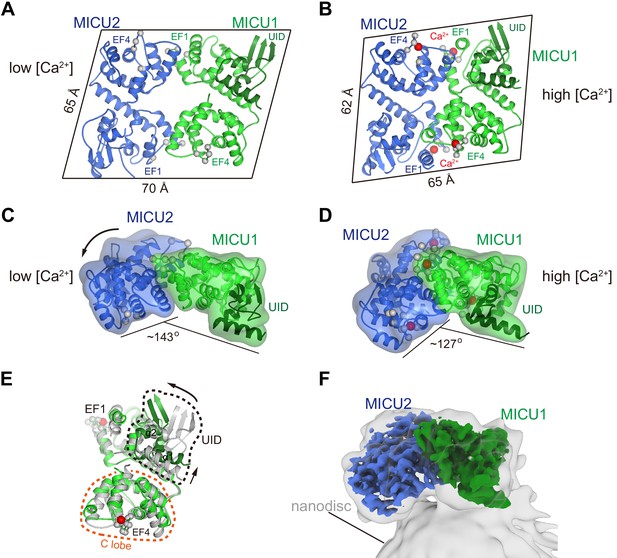
Ca2+-dependent changes in MICU1-MICU2.
(A) Cartoon representation of the apo MICU1-MICU2 heterodimer. Gray spheres denote EF-hand residues. (B) Overall structure of Ca2+-bound MICU1-MICU2. Red spheres indicate bound Ca2+ ions. (C–D) Bend between MICU1 and MICU2 induced by Ca2+ binding (C, apo; D, Ca2+-bound). Overall structures are shown from the side with semitransparent molecular surfaces. (E) Superposition of apo (gray) and Ca2+-bound (green) MICU1 highlighting a rotation of the UID upon Ca2+ binding. (F) Cryo-EM density depicting the Ca2+-bound MICU1-MICU2 complex associated with a lipid nanodisc.
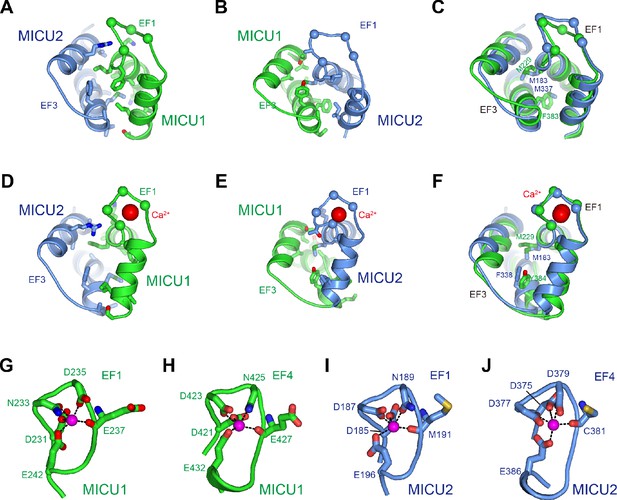
MICU1-MICU2 interfaces and Ca2+-binding.
(A and B) Interfaces between MICU1 and MICU2 in the Ca2+-free heterodimer. Depictions are for the interface between MICU1-EF1 and MICU2-EF3 (A) and for the interface between MICU1-EF3 and MICU2-EF1 (B). Residues within the interfaces are shown as sticks. Spheres represent Cα locations of amino acids within EF-hands that would coordinate Ca2+. (C) Superposition of the interfaces from (A–B), with amino acids at their centers labeled. (D–F) MICU1-MICU2 interfaces in the Ca2+ bound complex, depicted as for (A–C). Ca2+ ions are shown as red spheres. (G – J) Coordination of Ca2+ ions in the Ca2+-bound MICU1-MICU2 structure. The four EF-hand domains are shown as cartoons with amino acids that coordinate Ca2+ as sticks. Ca2+ ions are shown as purple spheres, with Ca2+-oxygen interactions depicted as dashed lines.
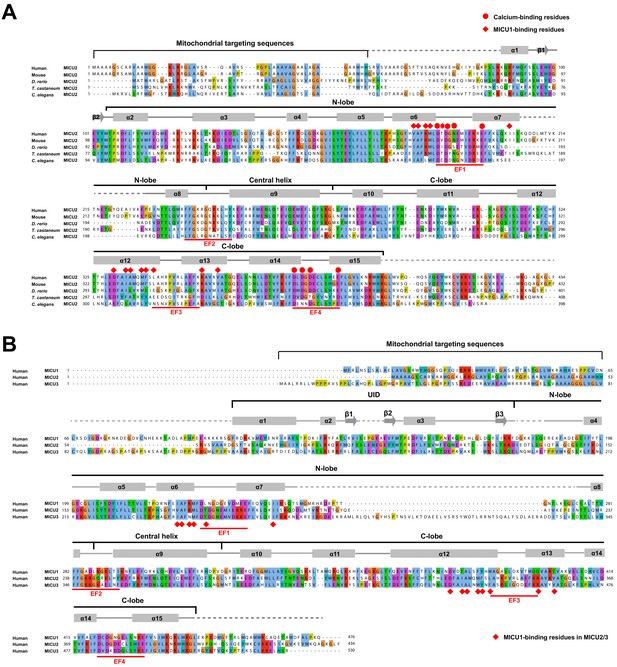
Structure based sequence alignments of MICU proteins.
(A) Sequence alignment of MICU2 orthologs. The amino acid sequences of human, mouse (M. musculus), zebrafish (D. rerio), T. castaneum, and C. elegans MICU2 are aligned and coloured according to the ClustalW convention (UniProt accession numbers: Q8IYU8, Q8CD10, Q1LCY6, D6WQH5 and D7SFN6, respectively). Secondary structures are indicated with cylinders representing α-helices, arrows representing beta sheets, solid gray lines representing structured loops and dashed gray representing disordered loops. All the structural elements (N-lobe, Central helix and C-lobe) and 4 EF-hand motifs are highlighted. Residues involved in MICU1-binding and calcium-binding are highlighted with red dots and red diamonds, respectively. (B) Sequence alignment of human MICU1, MICU2 and MICU3. The secondary structure elements of human MICU1 are above the alignment. MICU2 residues that are involved in binding to MICU1 are conserved in MICU3 (red diamonds).
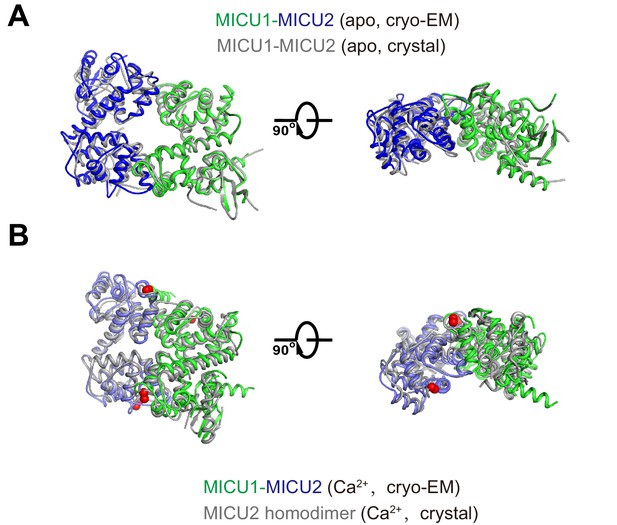
Comparisons of MICU1, MICU2 and MICU3 in apo and Ca2+-bound conformations.
(A) Superposition of the MICU1-MICU2 heterodimer from the holocomplex (apo; green and blue) with an X-ray structure of an isolated apo MICU1-MICU2 heterodimer (gray, PDB: 6LE5). The RMSD is 0.8 Å for Cα atoms. Two orthogonal views are shown. (B) Superposition of the Ca2+-bound conformation of the MICU1-MICU2 heterodimer from our cryo-EM analysis (green and blue) with a Ca2+-bound X-ray structure of a MICU2-MICU2 homodimer (gray, PDB: 6IIH, Cα RMSD = 2.3 Å). Ca2+ ions are drawn as purple spheres.
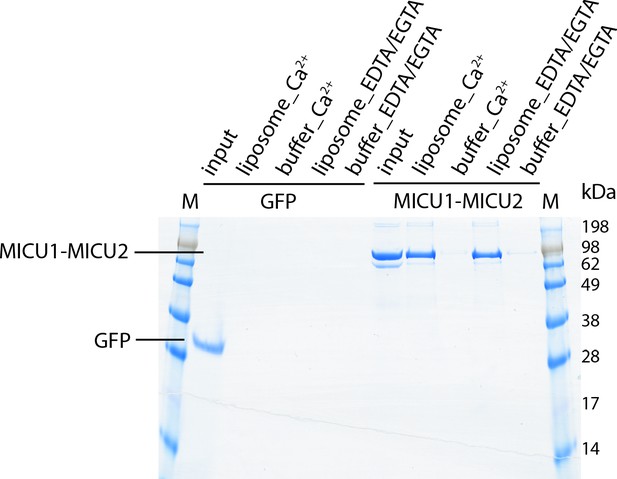
The MICU1-MICU2 regulatory complex binds to liposomes in both Ca2+ and Ca2+-free conditions.
Following ultracentrifugation of purified MICU1-MICU2 in the presence or absence of liposomes, and with and without Ca2+, pellets were resuspended and analyzed by Coomassie-stained SDS-PAGE. MICU1-MICU2 pellets with liposomes but remains in the supernatant when liposomes are not present (buffer). GFP is used as a control that does not associate with liposomes. Refer to Methods for details.
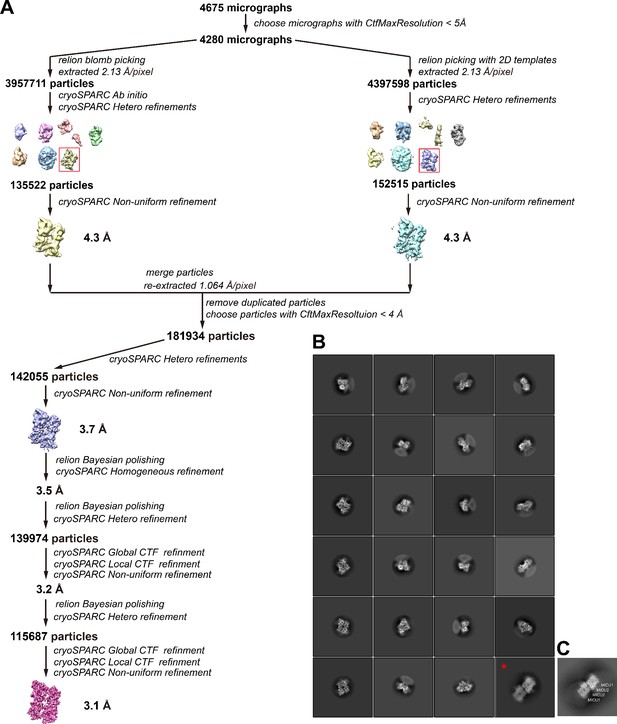
Flowchart of the cryo-EM data processing of the Ca2+-bound MICU1-MICU2 complex.
(A) Flowchart of data processing and 3D reconstruction. (B) Representative 2D class averages. An asterisk denotes a class containing two MICU1-MICU2 assemblies (representing ~3% of particles). (C) A 2D projection of a dimer of MICU1-MICU2 heterodimers from the cryo-EM map of the human holocomplex under high [Ca2+] conditions (EMD-21643). The channel (MCU/EMRE) densities were removed for the projection. MICU2:MICU2 interfaces mediate a dimerization of MICU1-MICU2 heterodimers in that structure (Fan et al., 2020). The projection resembles the 2D class from (B) with two MICU1-MICU2 heterodimers (asterisk), which suggests, but does not prove, that a MICU2:MICU2 interface mediates the dimerization of MICU1-MICU2 heterodimers in a fraction of our cryo-EM data.
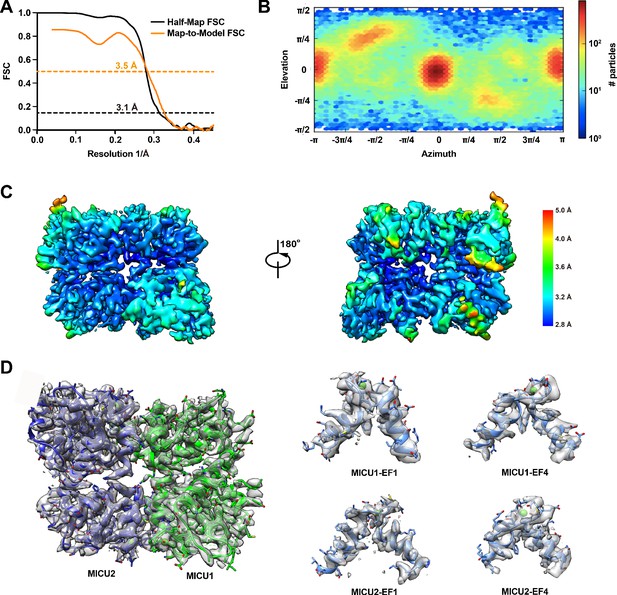
Cryo-EM analysis of the Ca2+-bound MICU1-MICU2 complex.
(A) FSC curves of the final 3D reconstruction. The half-map (black) and map-to-model (orange) FSC curves are shown (dotted lines indicate 0.143 and 0.5 levels and corresponding resolutions). (B) Euler angle distribution plot of the final 3D reconstruction. (C) Local resolution estimation (calculated with Resmap). The unsharpened map is colored as indicated. Two views are shown. (D). EM densities for indicated regions of the atomic model. Calcium ions are depicted as green spheres.
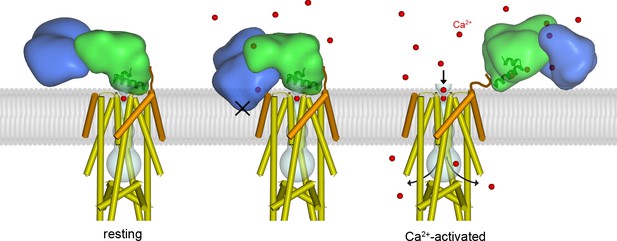
Proposed mechanism of Ca2+-dependent control.
Left, structure of the holocomplex under resting [Ca2+]IMS conditions. MCU and EMRE are depicted with cylindrical helices; MICU1 and MICU2 are represented as semitransparent surfaces, with α1 and α2 helices of the UID as ribbons. The lipid membrane (gray) is based upon an atomistic model (Figure 5—figure supplement 1) . The UID blocks the pore (gray tube). Following elevation of [Ca2+]IMS, Ca2+ binding causes the MICU1-MICU2 heterodimer to bend (the Ca2+-bound MICU1-MICU2 structure is depicted with the channel in the center and right panels). The center panel indicates that bending would dislodge the UID from its receptor site in order to avoid thermodynamically unfavorable interactions of MICU2 with the membrane (‘X’). The dislodged Ca2+-bound MICU1-MICU2 heterodimer would no longer block the pore, thereby allowing Ca2+ permeation through the channel, and it would be free to interact with the membrane (right). Upon the return of resting [Ca2+], the heterodimer would resume its blocking conformation (left). One MICU1-MICU2 heterodimer is depicted and only one can bind to the receptor on the channel at a time but multiple MICU1-MICU2 heterodimers may be associated with the channel. A brown line depicts a hypothetical interaction of the acidic C-terminus of EMRE with MICU1.
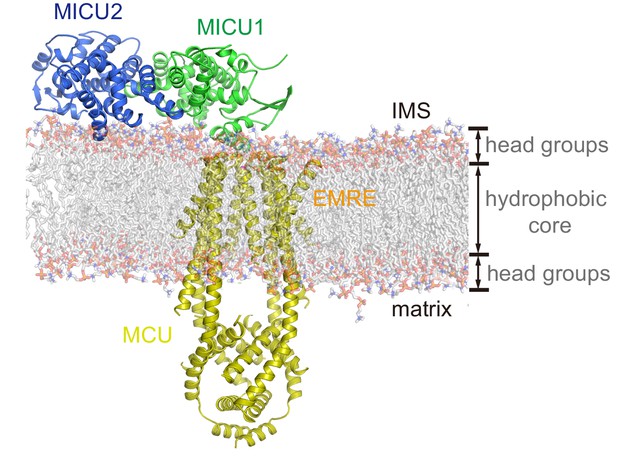
Atomistic model of the resting holocomplex in a lipid bilayer.
The holocomplex was modeled into a DPPC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine) lipid bilayer, using CHARMM-GUI (http://www.charmm-gui.org/). MCU, EMRE, MICU1 and MICU2 are shown as cartoons. DPPC molecules are shown as transparent sticks.
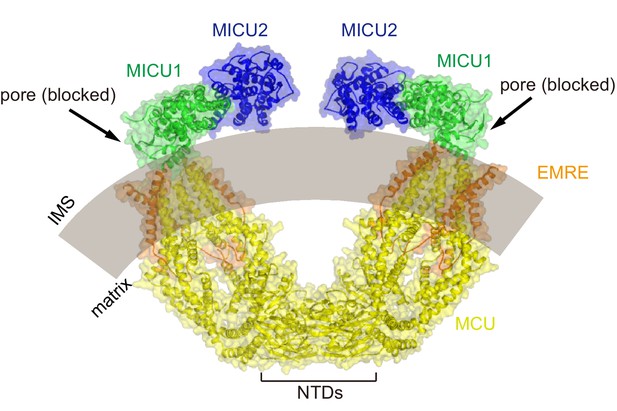
Model of a dimeric human MCU-EMRE-MICU1-MICU2 complex under resting [Ca2+] conditions.
The complex was modeled based on holocomplex and the dimeric human MCU-EMRE complex (PDB:6O58). MCU, EMRE, MICU1 and MICU2 are shown as cartoons with transparent molecular surfaces and colored gold, orange, green and blue, respectively. A curved mitochondrial inner membrane is depicted as a transparent gray arc. Dimerization of channels is due to the NTDs of human MCU. As shown, MICU1-MICU2 regulatory complexes could bind to both channels of the dimer without sterically interfering.
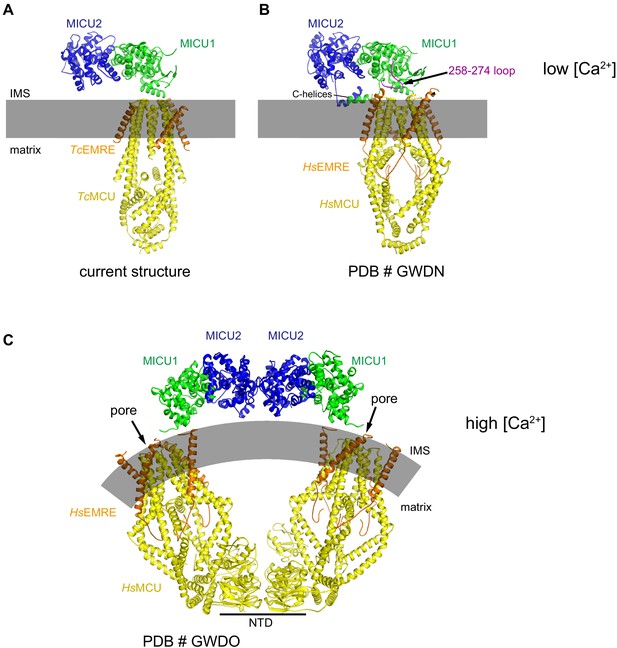
Comparisons of the TcMCU-EMRE-MICU1-MICU2 holocomplex structure with structures of human MCU-EMRE-MICU1-MICU2 holocomplexes.
(A–B) Low [Ca2+] structures. (A) TcMCU-EMRE-MICU1-MICU2 holocomplex structure reported here. (B) Holocomplex containing human MCU, EMRE, MICU1, and MICU2 subunits (PDB # GWDN). An ordered loop of MICU1 (amino acids 258–274) is indicated, as are the C-helices of MICU1 and MICU2, which are also ordered in that structure. (C) Structure of a holocomplex comprising human MCU, EMRE, MICU1, and MICU2 that was determined at high (2 mM) [Ca2+] (PDB # GWDO). A dimer of channels present in the cryo-EM sample is formed by interactions between the NTDs. For each channel, a Ca2+-bound MICU1-MICU2 regulatory complex is located adjacent to the pore and no longer occludes it.
Tables
Data collection, refinement, and validation statistics.
| Holocomplex at low [Ca2+] PDB: 6XQN EMDB: EMD-22290 | Ca2+-bound MICU1-MICU2 PDB: 6XQO EMDB: EMD-22291 | |
|---|---|---|
| Data collection and processing | ||
| Microscope | FEI Titan Krios (at MSKCC) | FEI Titan Krios (at MSKCC) |
| Camera | Gatan K3 | Gatan K3 |
| Magnification | 22,500× | 22,500× |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 71 | 71 |
| Defocus range (μm) | −1.0 ~ −3.0 | −0.8 ~ −2.3 |
| Pixel size (Å) | 1.064 (0.532)* | 1.064 (0.532)* |
| Software | RELION 3.0, cryoSPARC v2 | RELION 3.0, cryoSPARC v2 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 17,440,131 | 4,397,598 |
| Final particle images (no.) | 350,160 | 115,687 |
| Overall map resolution (Å) FSC threshold 0.143 | 3.3 | 3.1 |
| Local map resolution range (Å) | 3.0–5.0 | 2.8–5.0 |
| Refinement | ||
| Software | Phenix 1.13 real-space-refine | Phenix 1.13 real-space-refine |
| Initial model used (PDB code) | N/A | N/A |
| Model resolution (Å) FSC threshold 0.5 | 3.6 | 3.5 |
| Map sharpening B factor (Å2) | −77 | −38 |
| Model composition | ||
| Non-hydrogen atoms | 9433 | 4225 |
| Protein residues | 1290 | 566 |
| Ligands | 1 (Calcium ion) | 4 (Calcium ions) |
| Water | 0 | 0 |
| B factors (Å2) | ||
| Protein | 124.29 | 88.3 |
| Ligand | 70.1 | 94.9 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.006 | 0.004 |
| Bond angles (°) | 1.027 | 0.569 |
| Validation | ||
| MolProbity score | 1.85 | 2.25 |
| Clashscore | 7.13 | 10.75 |
| Ramachandran plot | ||
| Favored (%) | 97.45 | 95.65 |
| Allowed (%) | 2.55 | 4.35 |
| Disallowed (%) | 0.0 | 0.0 |
| *Super-resolution pixel size. |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Tribolium castaneum) | TcMCU | IDT Inc | ||
| Gene (Tribolium castaneum) | TcEMRE | IDT Inc | ||
| Gene (Homo sapiens) | HsMICU1 | IDT Inc Genewiz, Inc. | ||
| Gene (Homo sapiens) | HsMCIU2 | IDT Inc | ||
| Cell line (H. sapiens) | Expi293F | Sigma | A14527 | |
| Cell line (H. sapiens) | HEK-293T-MICU1-KO | Kamer and Mootha, 2014 | DOI: 10.1002/embr.201337946 | |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine | Avanti Polar Lipids | 850757 | |
| Chemical compound, drug | 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (sodium salt) | Avanti Polar Lipids | 840457 | |
| Chemical compound, drug | 1',3'-bis[1,2-dioleoyl-sn-glycero-3-phospho]-glycerol (sodium salt) | Avanti Polar Lipids | 710335 | |
| Chemical compound, drug | Polyethylenimine, Linear, MW 25000, Transfection Grade (PEI 25K) | Polysciences, Inc | 23966–1 | |
| Chemical compound, drug | Sodium Butyrate | Sigma | 8451440100 | |
| Chemical compound, drug | n-Dodecyl-β-D-maltopyranoside | Anatrace | O310S | |
| Chemical compound, drug | (2α,3β,5α,15β,25R)−2,15-dihydroxyspirostan-3-yl O-β -D-glucopyranosyl-(1→3)-O-β-D-galactopyranosyl-(1→2)-O-[β-D-xylopyranosyl-(1→4)-β-D-galactopyranoside | Cayman Chemical Company | 14952 | |
| Chemical compound, drug | Membrane Scaffold Protein 1D1 | Sigma | M6574 | |
| Chemical compound, drug | 5-Methyl-2-oxo-4-imidazo lidinehexanoic acid | Sigma | D1411 | |
| Chemical compound, drug | Lipofectamine 3000 Transfection Reagent | Thermo Fisher | L3000008 | |
| Chemical compound, drug | Calcium Green−5N, Hexapotassium Salt | Thermo Fisher | C3737 | |
| Software, algorithm | MotionCor2 | Zheng et al., 2017 | RRID:SCR_016499 | |
| Software, algorithm | CtfFind 4.1.10 | Rohou and Grigorieff, 2015 | RRID:SCR_016731 | |
| Software, algorithm | RELION 3.0 | Zivanov et al., 2018 | http://www2.mrc-lmb. cam.ac.uk/relion RRID:SCR_016274 | |
| Software, algorithm | SerialEM | Glover, 2004 | RRID:SCR_017293 | |
| Software, algorithm | cryoSPARC v2 | Structura Biotechnology | https://cryosparc.com/ RRID:SCR_016501 | |
| Software, algorithm | PHENIX | Adams et al., 2010 | https://www.phenix-online.org/ RRID:SCR_014224 | |
| Software, algorithm | COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ RRID:SCR_014222 | |
| Software, algorithm | PyMOL | Schrödinger, 2020 | https://pymol.org/2/ RRID:SCR_000305 | |
| Software, algorithm | UCSF Chimera | Pettersen et al., 2004 | https://www.cgl.ucsf.edu/chimera RRID:SCR_004097 | |
| Software, algorithm | GraphPad Prism 7 | GraphPad Software | https://cryosparc.com/ RRID:SCR_016501 | |
| Software, algorithm | Hole | Smart et al., 1996 | http://www.holeprogram.org | |
| Software, algorithm | UCSF ChimeraX | Goddardnet al. 2018 | https://www.cgl.ucsf.edu/chimerax/ | |
| Others | QUANTIFOIL R1.2/1.3 holey carbon grids | Quantifoil | ||
| Others | FEI Vitrobot Mark IV | FEI Thermo Fisher |





