Ribosome collisions trigger cis-acting feedback inhibition of translation initiation
Figures
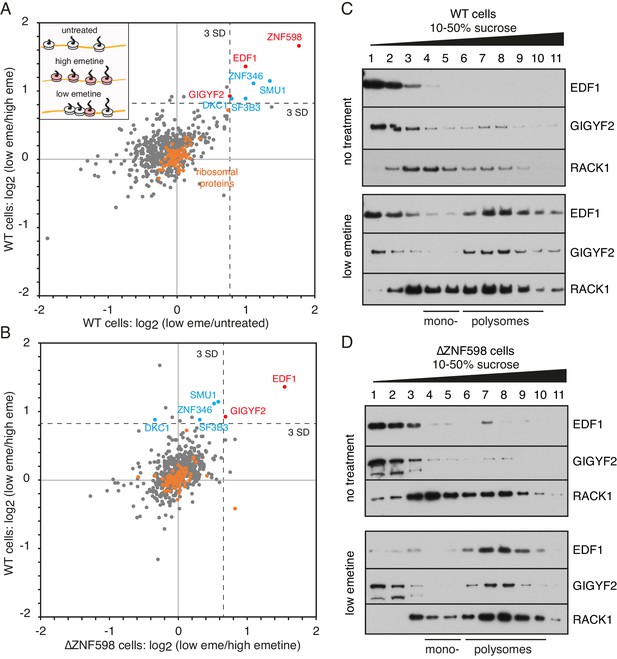
Proteomic identification of collision-specific ribosome interacting factors.
(A) Plot of proteins identified by quantitative mass spectrometry in the polysome fraction of cytosol separated by sucrose gradient centrifugation. The cells were either left untreated, treated with low-dose emetine (1.8 µM) to induce collisions (low eme), or treated with high-dose emetine (360 µM) to freeze ribosomes in place (high eme). Pairwise comparisons are plotted such that factors specifically associating with collided ribosomes (i.e., only in low-dose emetine samples) should fall in the upper-right quadrant. Ribosomal proteins are indicated in orange. The position of 3 standard deviations (SD) from the mean along each axis is indicated with a dashed line. Proteins that deviate 3 SD or more along both axes are labelled (in blue and red). (B) Cells knocked out for ZNF598 (∆ZNF598) were treated with low or high dose emetine as in panel A and the polysome fractions were analysed by mass spectrometry. These data (x-axis) are plotted relative to the same comparison in wild type cells (y-axis). The position of 3 SD along each axis is indicated with a dashed line, and the data points for the same proteins from panel A are labelled. Note that only EDF1 and GIGYF2 fall into the upper right quadrant in both plots. As expected, ZNF598 was not detected in ∆ZNF598 cells. (C, D) Sucrose gradient fractionation and immunoblots of cytosolic lysates prepared from untreated cells versus low-dose emetine treated cells. WT cells were analysed in panel C and ∆ZNF598 cells in panel D. RACK1 is a 40S-ribosomal protein. The positions of fractions containing mono- and polysomes are indicated. Source data are provided for the mass spectrometry analysis.
-
Figure 1—source data 1
Proteins identified by mass spectrometry of polysomes.
- https://cdn.elifesciences.org/articles/60038/elife-60038-fig1-data1-v2.xlsx
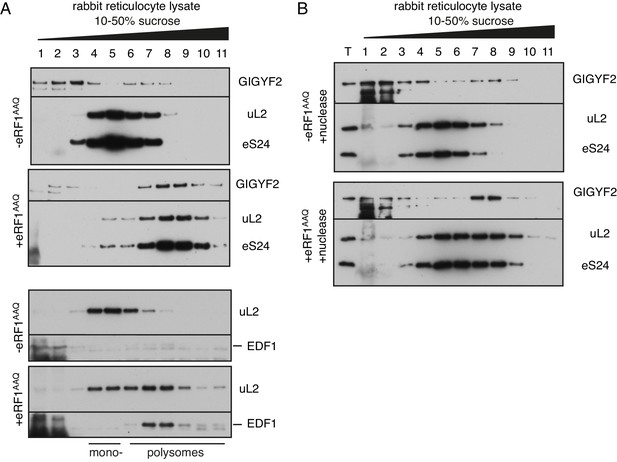
Analysis of GIGYF2 and EDF1 interaction with collided ribosomes in rabbit reticulocyte lysate.
(A) Endogenous native globin polysomes were elongated in rabbit reticulocyte lysate in the absence or presence of excess exogenously added mutant termination factor eRF1AAQ. This protein acts as a dominant-negative to stall ribosomes at the stop codon, thereby inducing ribosome collisions behind it. Samples were separated on 10–50% sucrose gradient and analysed by western blotting to assess migration of GIGYF2 and EDF1 relative to ribosomes (indicated by uL2 and eS24 ribosomal proteins). Note the noticeable enrichment of both proteins in fractions 7–9 in the samples containing eRF1AAQ. As previously characterised, these fractions contain ribosomes collided at the stop codon. Even without eRF1AAQ, endogenous RRL polysomes have some collisions due to their high density on globin mRNAs. The background smear observed in fractions 1 and 2 is due to a large amount of endogenous haemoglobin. (B) Reactions like in panel A were additionally treated with micrococcal nuclease after translation to digest mRNA, then separated on a sucrose gradient. T indicates total input sample. Note that GIGYF2 still co-migrates with collided ribosomes after nuclease digestion. This indicates that its binding is likely to be direct as opposed to indirect tethering by mRNA, and that it is interacting with collisions (which are resistant to separation by nuclease).
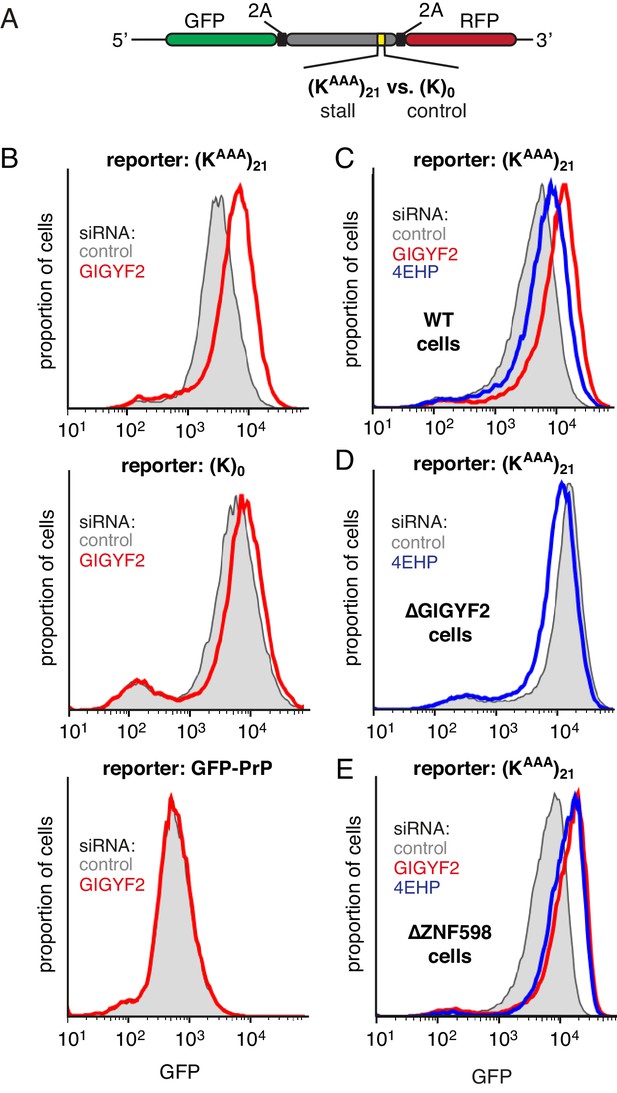
GIGYF2 is a collision-dependent cis-acting translational repressor.
(A) Schematic representation of the reporter construct containing GFP followed by a viral 2A sequence and either the (KAAA)21 stall sequence or a (K)0 control insert that does not stall. (B) Flp-In T-REx 293 cells containing the indicated reporter construct stably integrated at a doxycycline-inducible site were treated for 5 days with either non-targeting control siRNAs (gray shaded) or siRNAs targeting GIGYF2 (red traces). The reporter construct was then induced with doxycycline for 20 hr prior to analysis by flow cytometry. GFP-PrP was used as an irrelevant fluorescent protein control to exclude non-specific effects on translation. (C–E) Flp-In T-REx 293 cells with stably intergrated (KAAA)21 reporter were treated with siRNAs targeting GIGYF2 (red traces), 4EHP (blue traces) or control (gray shaded) as in panel A, induced with doxycycline for 20 hr, and analysed by flow cytometry. Panels D and E used cells that were knocked out for GIGYF2 or ZNF598, respectively. GFP expression in all graphs is plotted on a log scale as a histogram and represents around 20,000 cells.
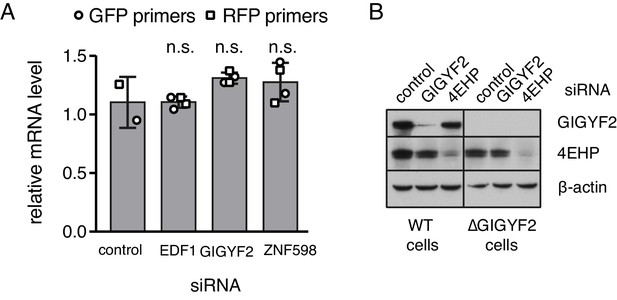
Additional characterisation of knockdown and knockout cells.
(A) Flp-In T-REx 293 with stably integrated (KAAA)21 stalling reporter were treated with siRNAs targeting the indicated genes for 72 hr before induction of the reporter with doxycycline for another 6 hr. Total RNA was isolated from each sample and analysed by quantititative RT-PCR. Two biological replicates, each containing three technical replicates, were analysed using two sets of primers targeting the same transcript. Relative mRNA levels were normalised to the standard curve and against reference genes and plotted for each condition (mean ± SD of four measurements). Levels of mRNA between the control and each of the other samples were compared using the non-parametric Mann-Whitney test and each was found to be non-significant (n.s., indicating a p-value greater than 0.05). (B) Total lysates from WT or ∆GIGYF2 cells treated for 5 days with siRNA targeting GIGYF2, 4EHP and control siRNA were analysed by western blotting for the expression of indicated proteins.
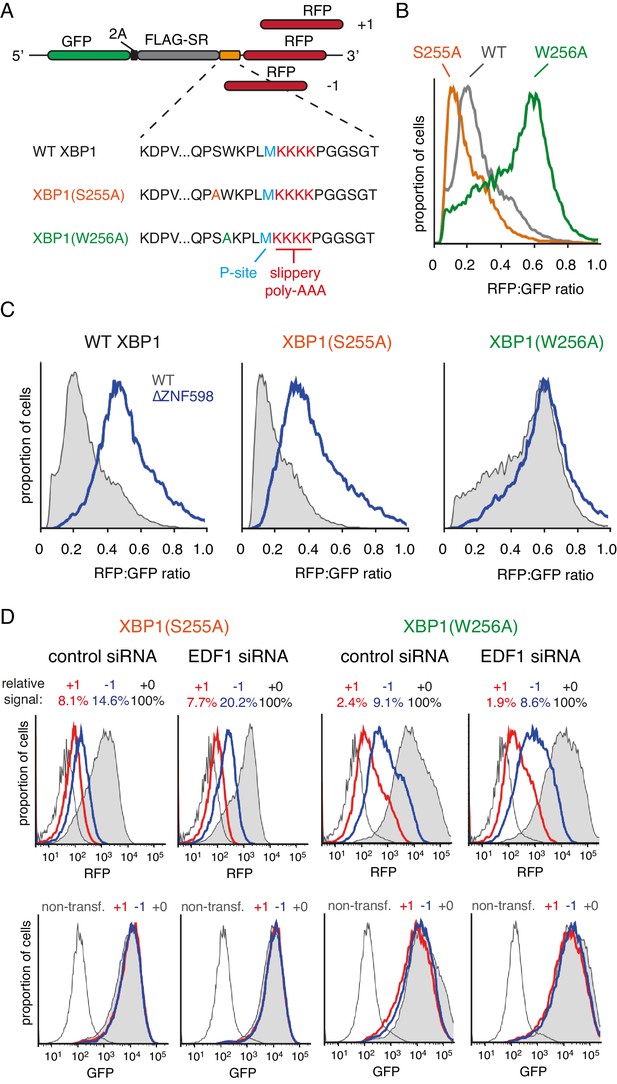
Effect of EDF1 on frameshifting at stall sites.
(A) Schematic representation of the reporter constructs used to analyse frameshifting. RFP was directly appended to each of the three tested XBP1 reporters (see amino acid sequences below) in three different reading frames. Note that S255A mutation exacerbates, whereas W256A reduces ribosomal stalling, relative to the wild type XBP1 sequence. (B) In-frame versions of the constructs from panel A were co-transfected with BFP-encoding plasmid into Flp-In T-REx 293 cells and analysed by flow cytometry after 20 hr of transgene expression. At least 20,000 transfected (BFP-positive) cells were analysed here and in all the subsequent experiments involving frameshifting reporters. The RFP:GFP ratio, an indicator of readthrough at the XBP1 sequence, is plotted as a histogram for each reporter. (C) The same in-frame constructs as in panel B were analysed in WT and ZNF598 KO cells. Note that the RFP:GFP ratio is elevated in ∆ZNF598 cells for WT XBP1 and XBP1(S255A), but not for XBP1(W256A). (D) XBP1(S255A) (left) or XBP1(W256A) (right) was transfected into cells pre-treated for 72 hr with non-targeting siRNA (control) or EDF1-targeting siRNA and analysed by flow cytometry after 20 hr of reporter expression. Expression of GFP (bottom panels) and RFP (top panels) was plotted as histograms. Gray shaded traces represent cells positively transfected with in-frame (+0) constructs, blue traces represent −1 constructs and red traces represent +1 constructs. Background fluorescence observed in non-transfected cells is indicated by the unshaded thin gray traces. The amount of RFP signal observed for out-of-frame constructs (+1,–1), represented as a percentage of the in-frame signal, is indicated above each graph. Note that GFP expression is not influenced by the RFP reading frame.
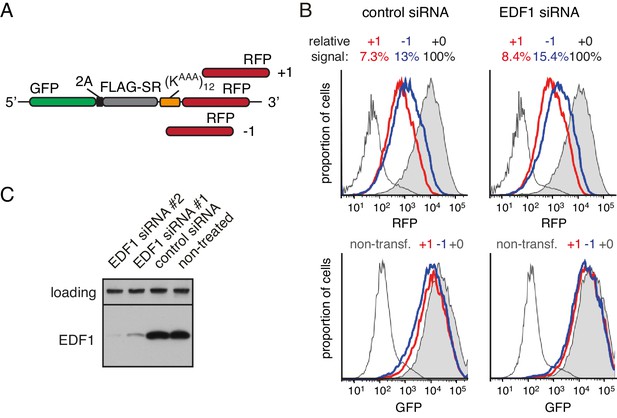
Effect of EDF1 on frameshifting at stall sites.
(A) Schematic representation of the reporter constructs used to analyse frameshifting. RFP was directly appended to (KAAA)12 stalling sequence in three different reading frames. (B) Constructs from panel A were transfected into cells pre-treated for 72 hr with non-targeting siRNA (control) or EDF1-targeting siRNA and analysed by flow cytometry after 20 hr of reporter expression. Expression of GFP (bottom panels) and RFP (top panels) was plotted as histograms. Gray shaded traces represent cells positively transfected with in-frame (+0) constructs, blue traces represent −1 constructs and red traces represent +1 constructs. Background fluorescence observed in non-transfected cells is indicated by the unshaded thin gray traces. The amount of RFP signal observed for out-of-frame constructs (+1,–1), represented as a percentage of the in-frame signal, is indicated above each graph. Note that GFP expression is not influenced by the RFP reading frame. (C) Total cell lysates from cells depleted for EDF1 using two different siRNA sequences were analysed by western blotting alongside the lysates from control siRNA-treated and non-treated cells. The siRNA treatment here and in other experiments which used siRNA-mediated depletion of EDF1 was for 72 hr.
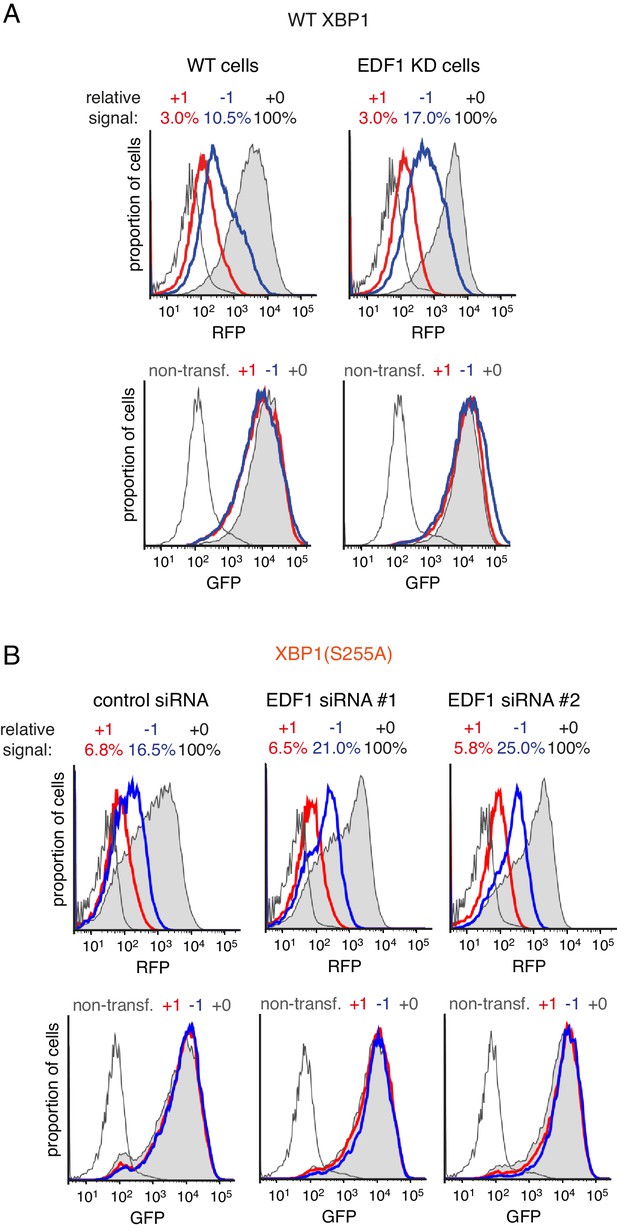
Effect of EDF1 on frameshifting at stall sites.
(A) Analysis of the frameshifting reporters containing WT XBP1 exactly as in Figure 3D. (B) Analysis of the frameshifting reporters containing XBP1(S255A) as in Figure 3D using two different siRNAs targeting EDF1. In both panel A and panel B, the reporter constructs were transfected into cells pre-treated for 72 hr with non-targeting siRNA (control) or EDF1-targeting siRNA and analysed by flow cytometry after 20 hr of reporter expression. Expression of GFP (bottom panels) and RFP (top panels) was plotted as histograms. Gray shaded traces represent cells positively transfected with in-frame (+0) constructs, blue traces represent −1 constructs and red traces represent +1 constructs. Background fluorescence observed in non-transfected cells is indicated by the unshaded thin gray traces. The amount of RFP signal observed for out-of-fram constructs (+1,–1), represented as a percentage of the in-frame signal, is indicated above each graph. Note that GFP expression is not influenced by the RFP reading frame.
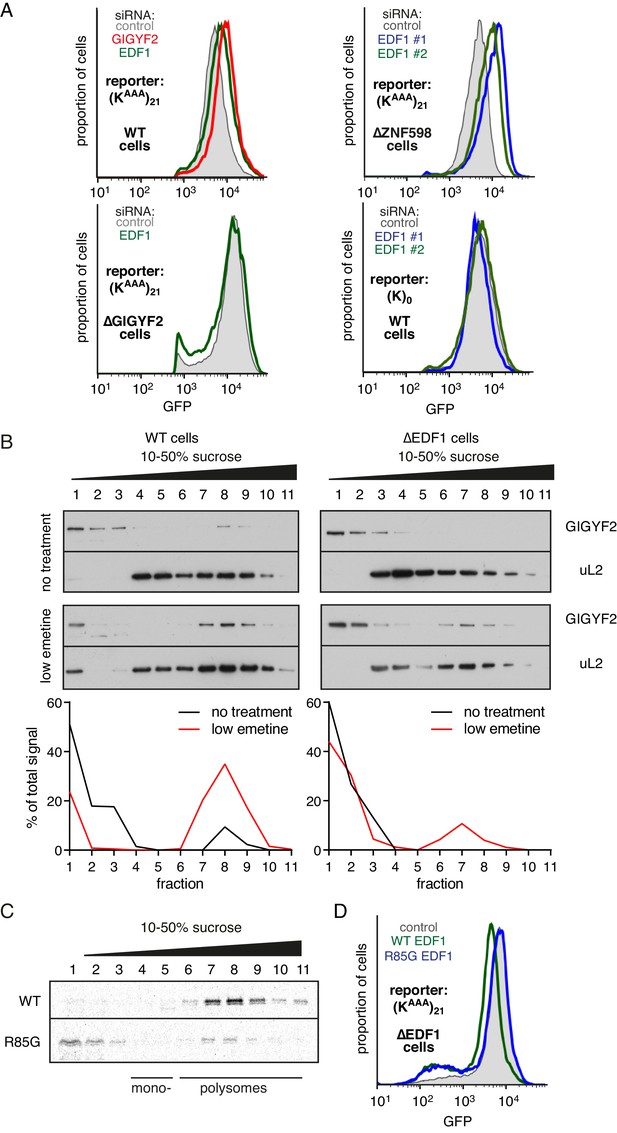
EDF1 facilitates GIGYF2 function at collided ribosomes.
(A) The indicated cell lines bearing stably integrated translation reporters were treated with the indicated siRNAs for 72 hr. After additional 20 hr of doxycycline-induced reporter expression, cells were collected and analysed by flow cytometry. Histograms representing GFP expression were plotted on a logarithmic scale. (B) WT cells (left panel) and ∆EDF1 cells (right panel) were treated with low dose emetine (1.8 µM) for 1 hr and their cytosolic extracts were analysed by sucrose gradient centrifugation followed by western blotting. The profiles of GIGYF2 across the gradient, quantified by densitometry, are depicted below the blots. (C) Radiolabelled and immunopurified WT EDF1 or R85G mutant EDF1 produced by in vitro translation was added to separate non-labelled in vitro translation reactions in which ribosomes on native globin mRNA were stalled and collided at the stop codon with eRF1AAQ. The samples were then separated by sucrose gradient centrifugation and EDF1 detected by autoradiography. Note that almost 100% of the WT EDF1 comigrates with collided ribosomes, whereas only ~40% of the R85G mutant is found in polysomal fractions with remainder at the top of the gradient. (D) ∆EDF1 cells with a stably integrated stalling reporter were transfected with a control plasmid (expressing BFP; shaded gray), WT EDF1 (green traces) and R85G EDF1(blue traces) and expression of transgenes was allowed for 24 hr. After additional 20 hr of doxycycline-induced expression of the stalling reporter, GFP expression was analysed by flow cytometry. Note that only WT, but not the R85G mutant reduces the expression of GFP to the level comparable to WT cells.
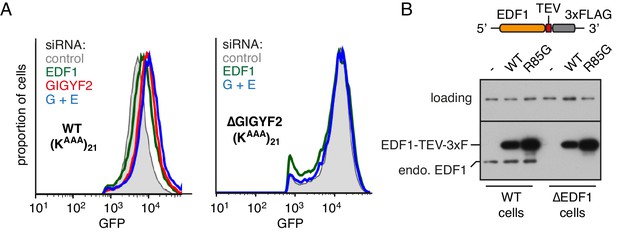
Additional characterisation of EDF1 function.
(A) WT cells (left graph) and ∆GIGYF2 cells (right graph) with stably integrated (KAAA)21 reporter were treated with the indicated siRNAs for 72 hr. ‘G+E’ indicates both GIGYF2 and EDF1 siRNAs were used. After additional 20 hr of doxycycline-induced expression of the reporter, cells were collected, analysed by flow cytometry and GFP expression was plotted as histograms. (B) WT and ∆EDF1 cells were transfected with plasmids encoding either WT EDF1 or R85G mutant EDF1 (see schematic). Expression of the transgenes relative to endogenous EDF1 was analysed by western blotting using antibody against EDF1.
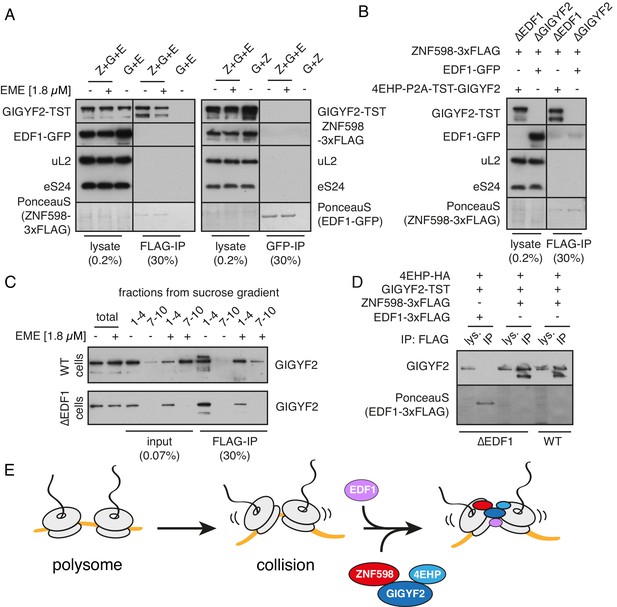
Analysis of interactions between EDF1, ZNF598, and GIGYF2.
(A) WT HEK293 cells were transfected with constructs encoding indicated proteins (Z- ZNF598-3xFLAG; G - 4EHP-P2A-TST-GIGYF2; E - EDF1-GFP) and allowed to express for 20 hr. Cytosol was depleted of ribosomes by sedimentation and subjected to immunoprecipitation (IP) of 3xFLAG-ZNF598 (left panel) or EDF1-GFP (right panel). When indicated, transfected cells were additionally pre-treated with 1.8 μM emetine for 30 min prior to lysis. Total lysate as well as eluates after IP were analysed by western blotting. The stained blot verifies that the immunoprecipitated proteins were recovered. The IP lanes for each antigen (and the stained blot) are from the same gel and exposure as the lysate lanes, with intervening lanes removed digitally. The relative amounts of lysate and IP samples are indicated on the figure. (B) Cells lacking EDF1 (∆EDF1) or GIGYF2 (∆GIGYF2) were transfected with indicated plasmids and analysed as in panel A. (C) WT (top panel) or ∆EDF1 (bottom panel) cells were transfected with ZNF598-3xFLAG and 4EHP-P2A-TST-GIGYF2 encoding plasmids. After 20 hr of expression and, when indicated, pre-treatment with 1.8 μM emetine, cell lysates were fractionated on a sucrose gradient. Ribosome free fractions 1–4 and heavy polysome fractions 7–10 were pooled and each was analysed by FLAG-IP and western blotting. (D) WT or ∆EDF1 cells were transfected with indicated plasmids and IP was performed as in panel B. Note that EDF1 tagged with 3xFLAG does not recover GIGYF2 (lane 2), unlike ZNF598 containing the same tag (lanes 4 and 6). (E) ZNF598-GIGYF2-4EHP form a complex independent of the ribosome. This complex does not include or require EDF1. However, both EDF1 and the ZNF598-GIGYF2-4EHP engage collided ribosomes. Without EDF1, the ZNF598-GIGYF2-4EHP complex is less stably bound to collided ribosomes.
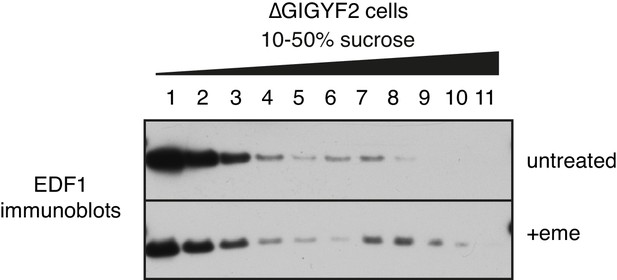
EDF1 recruitment to collided ribosomes in cells lacking GIGYF2.
GIGYF2 KO cells were treated with nothing (top) or 1.8 μM emetine prior to lysis and sucrose gradient fractionation. Individual fractions were analysed by western blotting to detect EDF1.
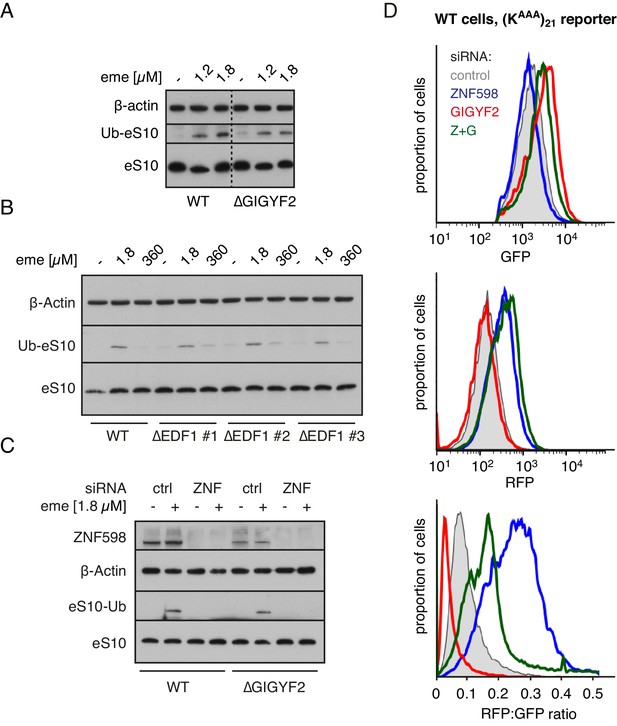
ZNF598 and GIGYF2 mediate alternate responses to ribosome collisions.
(A) Total cell lysates from WT and ∆GIGYF2 cells treated with nothing or with collision-inducing low-dose emetine (1.2 µM or 1.8 µM) were analysed by western blotting to monitor ubiquitination of eS10, the primary target of ZNF598. Note that due to its low abundance, the strip of the blot containing ubiquitinated eS10 (Ub-eS10) was detected with higher concentration of primary antibody, but is otherwise from the same membrane and gel used for the other antigens. (B) WT and ∆EDF1 cells were treated with nothing, collision-inducing low dose of emetine (1.8 µM) or stall inducing high dose of emetine (360 µM). The lysates were analysed by western blotting to assess ubiquitination of eS10 as in panel A. (C) WT and ∆GIGYF2 cells were treated with indicated siRNAs for 72 hr. Where indicated, cells were treated with 1.8 µM emetine for 15 min just prior to lysis. Shown are total cell lysates analysed by western blotting to asses ubiquitination of eS10. Note that in both cell types, eS10 ubiquitination is completely abolished when ZNF598 is knocked down. (D) Flp-In T-REx 293 with stably integrated (KAAA)21 reporter were treated with the indicated siRNAs for 3 days. After an additional 20 hr of doxycycline-induced expression of the fluorescent reporter, the cells were analysed by flow cytometry. Expression of GFP (top panel) and RFP (middle panel) was represented as a histogram for each condition plotted on a log scale. The RFP:GFP ratio (bottom panel) was calculated for each cell line using FlowJo software and also represented as a histogram for each condition.
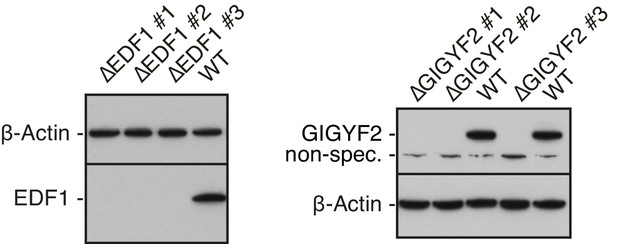
Validation of EDF1 and GIGYF2 knockout cell lines.
Total cell lysates from different clonal cell lines obtained after CRISPR-Cas9-mediated gene disruption of EDF1 (∆EDF#1-#3) or GIGYF2 (∆GIGYF2 #1-#3) were analysed alongside WT cell lysate by western blotting to confirm efficient depletion of the protein products of targeted genes. β-actin served as a loading control. A non-specific band detected by anti-GIGYF2 antibody is indicated.
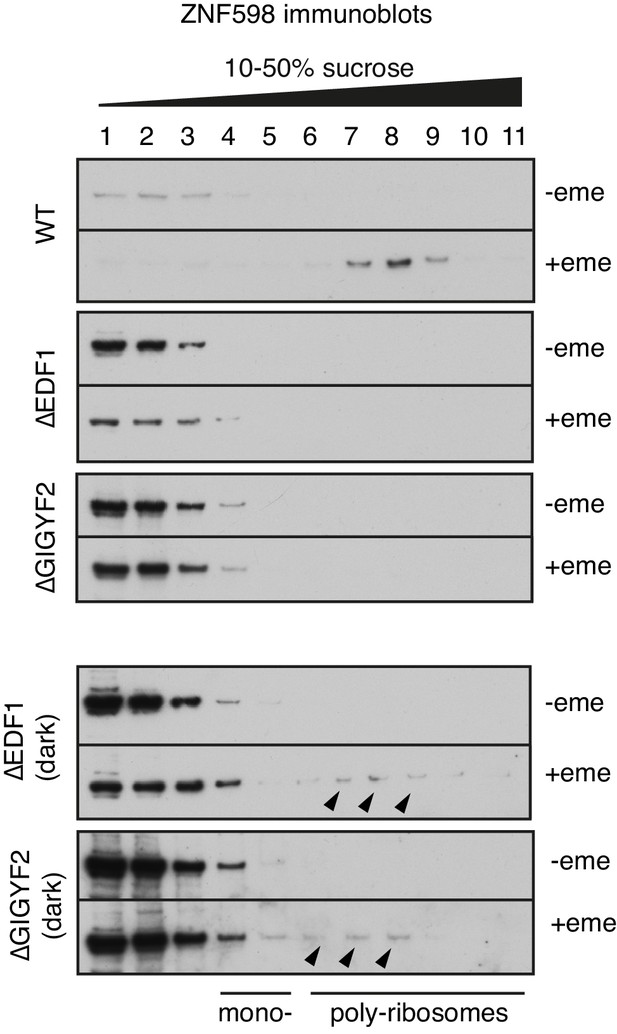
ZNF598 recruitment to collided ribosomes in different cell lines.
Indicated cell lines were treated with nothing or 1.8 μM emetine for 30 min prior to lysis. The cytosol was separated by sucrose gradient sedimentation and individual fractions were analysed by western blotting using anti-ZNF598 antibody. The two bottom panels are dark exposures of the ∆EDF1 and ∆GIGYF2 gradients. Black arrowheads on the dark exposures indicate faint bands corresponding to ZNF598 migrating in fractions containing collided poly-ribosomes even in the absence of either EDF1 or GIGYF2. This explains why ZNF598-mediated ubiquitination of eS10 is still able to occur in these cell lines.
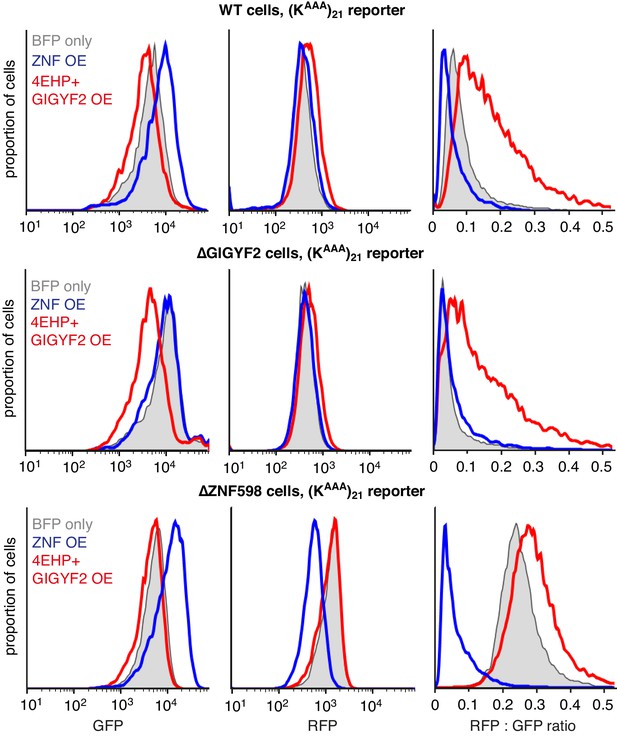
Effect of ZNF598 and GIGYF2 overexpression.
WT (upper panel), ∆GIGYF2 (middle panel) or ∆ZNF598 (bottom panel) cells with stably integrated (KAAA)21 reporter were co-transfected with either ZNF598 or 4EHP-P2A-GIGYF2 together with BFP encoding plasmids. After 8 hr of recombinant protein expression, reporter expression was induced for another 15 hr and cells were analysed by flow cytometry. Cells with high expression of BFP (i.e., transfected cells) were gated and analysed for the expression of GFP (left panels), RFP (middle panels) and the RFP:GFP ratio (right panels).
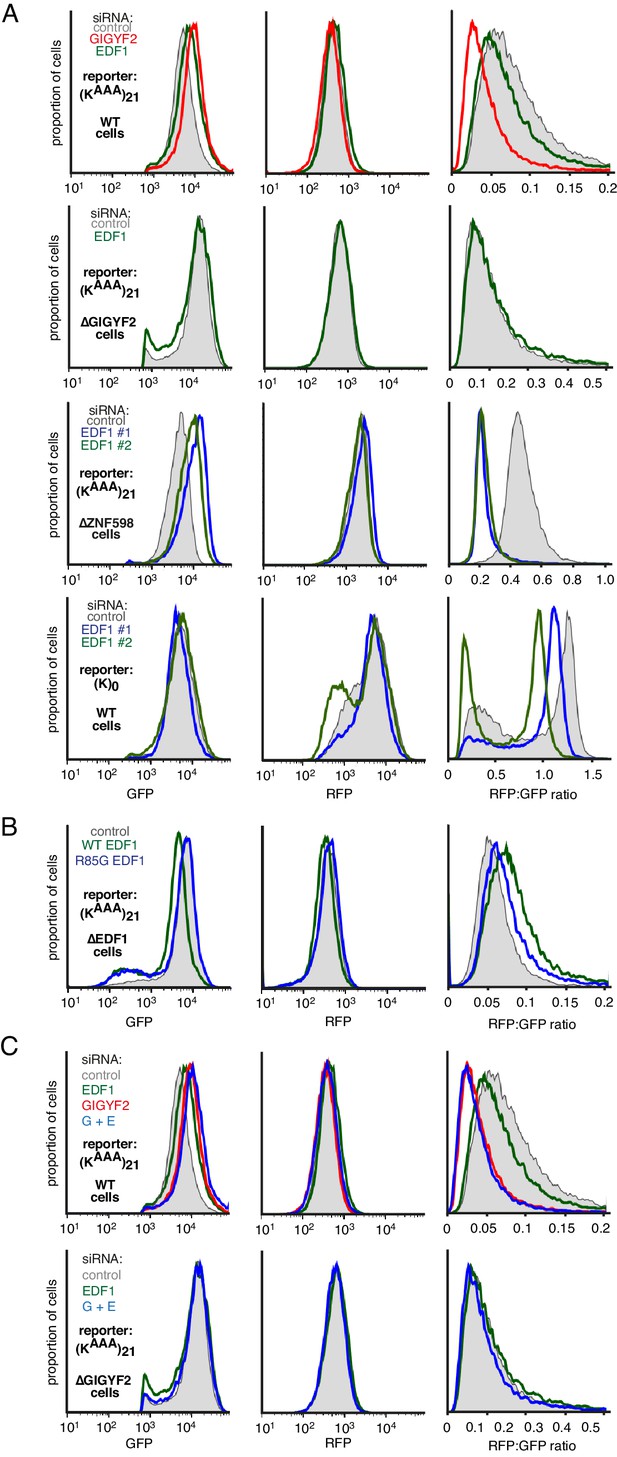
Additional analysis of dual-colour stalling reporters.
(A) GFP, RFP, and RFP:GFP histograms for the experiment in Figure 4A. Note that the GFP histogram is identical to that shown in Figure 4A. (B) GFP, RFP, and RFP:GFP histograms for the experiment in Figure 4D. Note that the GFP histogram is identical to that shown in Figure 4D. (C) GFP, RFP, and RFP:GFP histograms for the experiment in Figure 4—figure supplement 1A. Note that the GFP histogram is identical to that shown in Figure 4—figure supplement 1A.
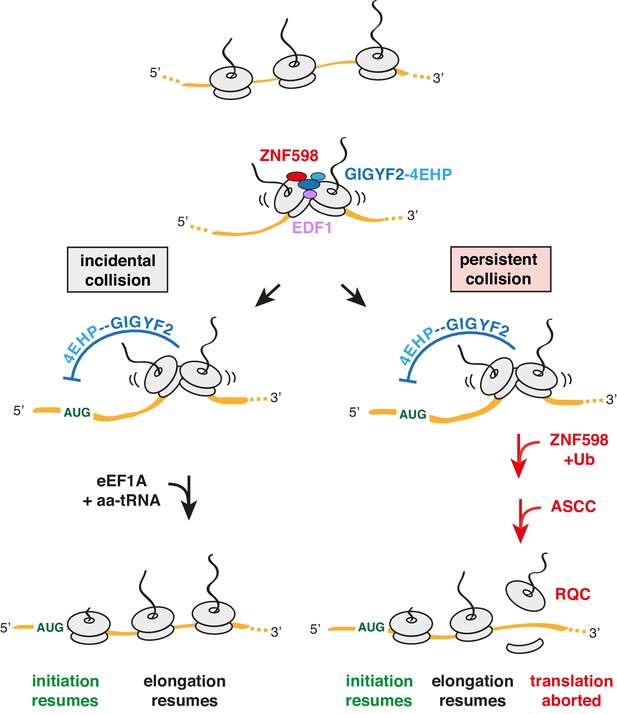
Model for tiered response to ribosome collisions.
When two ribosomes collide, EDF1 is most likely to engage the collision first due to its high abundance. Because downstream responses require recruitment of additional factors, no action is taken if the collision resolves quickly by elongation of the lead ribosome. This allows for random bumping of ribosomes on a crowded message. However, if resolution is not immediate, EDF1 stabilises the ZNF598-GIGYF2-4EHP complex. 4EHP inhibits initiation via sequestration of the 5’ cap to avoid further increase in ribosome density. This inhibition is relieved in one of two ways. If the collision is incidental (left limb), as might occur at sites of physiologic ribosome pausing, elongation resumes through the action of eEF1A in complex with aminoacyl-tRNA (aa-tRNA). If the collision is persistent (right limb), as would occur at sites of pathologic stalling, ZNF598 ubiquitinates the ribosome, which signals the ASC-1 complex (ASCC) to disassemble the lead ribosome. The 60S complex of the disassembled lead ribosome engages the ribosome quality control (RQC) complex. Because the stalled ribosome and ensuing collision has been resolved, elongation of trailing ribosomes resumes, causing dissociation of EDF1 and the ZNF598-GIGYF2-4EHP complex. Loss of 4EHP permits initiation to resume. Ub is ubiquitin.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Cell line (H. sapiens) | HEK293T | ATCC | CRL-3216; RRID:CVCL_0063 | |
| Cell line (H. sapiens) | Flp-In T-REx 293 | Thermo Fisher | R78007; RRID:CVCL_U427 | |
| Cell line (H. sapiens) | Flp-In T-REx 293 GFP-P2A-(KAAA)21-P2A-RFP | Juszkiewicz and Hegde, 2017 | ||
| Cell line (H. sapiens) | Flp-In T-REx 293 TRex GFP-P2A-(K)0-P2A-RFP | Juszkiewicz and Hegde, 2017 | ||
| Cell line (H. sapiens) | Flp-In T-REx 293 GFP-P2A-(KAAA)21-P2A-RFP ∆ZNF598 | Juszkiewicz and Hegde, 2017 | ||
| Cell line (H. sapiens) | Flp-In T-REx 293 GFP-P2A-(KAAA)21-P2A-RFP ∆GIGYF2 | This paper | CRISPR/Cas9 targeting GIGYF2, clonal selection | |
| Cell line (H. sapiens) | Flp-In T-REx 293 GFP-P2A-(KAAA)21-P2A-RFP ∆EDF1 | This paper | CRISPR/Cas9 targeting EDF1, clonal selection | |
| Antibody | anti-ZNF598 (rabbit polyclonal) | Abcam | Cat #ab80458; RRID:AB_2221273 | WB (1:500) |
| Antibody | anti-uL2 (rabbit polyclonal) | Abcam | Cat #ab169538; RRID:AB_2714187 | WB (1:10000) |
| Antibody | anti-eS24 (rabbit polyclonal) | Abcam | Cat #ab196652; RRID:AB_2714188 | WB (1:2500) |
| Antibody | HRP-conjugated anti-FLAG (mouse monoclonal) | Sigma | cat #A8592; RRID:AB_439702 | WB (1:5000) |
| Antibody | anti-eS10 (rabbit monoclonal) | Abcam | cat #ab151550; RRID:AB_2714147 | WB (1:5000 – unmodified eS10; 1:250 – Ub-eS10) |
| Antibody | HRP-conjugated anti-beta-Actin (mouse monoclonal) | Sigma | cat #A3854; RRID:AB_262011 | WB (1:10000) |
| Antibody | anti-GIGYF2 (rabbit polyclonal) | Bethyl | cat #A303-732A; RRID:AB_11205815 | WB (1:2000) |
| Antibody | anti-EDF1 (rabbit polyclonal) | Bethyl | cat #A304-039A; RRID:AB_2621288 | WB (1:1000) |
| Antibody | anti-RACK1 (rabbit polyclonal) | Bethyl | cat #A302-545A; RRID:AB_1999012 | WB (1:2000) |
| Antibody | anti-4EHP (rabbit polyclonal) | GeneTex | cat #GTX103977; RRID:AB_2036842 | WB (1:250) |
| Antibody | anti-GFP (rabbit polyclonal) | Chakrabarti and Hegde, 2009 | WB (1:5000) | |
| Antibody | HRP-conjugated anti-mouse (goat polyclonal) | Jackson ImmunoResearch | cat #115-035-003; RRID:AB_10015289 | WB (1:5000) |
| Antibody | HRP-conjugated anti-rabbit (goat polyclonal) | Jackson ImmunoResearch | cat# 111-035-003; RRID:AB_2313567 | WB (1:5000) |
| Recombinant DNA reagent | pX459-EDF1- sgRNA1 | This paper | sgRNA targeting EDF1 | |
| Recombinant DNA reagent | pX459-EDF1- sgRNA2 | This paper | sgRNA targeting EDF1 | |
| Recombinant DNA reagent | pX459-GIGYF2- sgRNA1 | This paper | sgRNA targeting GIGYF2 | |
| Recombinant DNA reagent | pX459-GIGYF2- sgRNA2 | This paper | sgRNA targeting GIGYF2 | |
| Recombinant DNA reagent | pcDNA3.1-ZNF598-TEV-3xFLAG | Juszkiewicz and Hegde, 2017 | RRID:Addgene_105690 | Human ZNF598 with TEV-3xFLAG tag at the C-terminus for mammalian expression |
| Recombinant DNA reagent | pcDNA3.1-EDF1-TEV-3xFLAG | This paper | Human EDF1 tagged with TEV-3xFLAG at the C-terminus for mammalian expression | |
| Recombinant DNA reagent | pcDNA3.1-EDF1-GFP | This paper | Human EDF1 tagged with GFP at the C-terminus for mammalian expression | |
| Recombinant DNA reagent | pCMV-4EHP-HA | This paper | Human 4EHP tagged with HA at the C-terminus for mammalian expression | |
| Recombinant DNA reagent | pcDNA3.1-4EHP-P2A-Twin-Strep-GIGYF2 | This paper | Human 4EHP and Twin-Strep tagged GIGYF2 for mammalian expression | |
| Recombinant DNA reagent | pcDNA3.1-EDF1(R85G)-TEV-3xFLAG | This paper | Human EDF1 R85G mutant tagged with TEV-3xFLAG at the C-terminus for mammalian expression | |
| Recombinant DNA reagent | pSP64-EDF1-TST | This paper | Human EDF1 tagged with C-terminal TST for in vitro transcription and translation | |
| Recombinant DNA reagent | pSP64-EDF1(R85G)-TST | This paper | Human EDF1 R85G mutant with C-terminal TST for in vitro transcription and translation | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-K(AAA)12+0-RFP | Juszkiewicz and Hegde, 2017 | Frameshifting reporters based on K12 poly sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-K(AAA)12+1-RFP | Juszkiewicz and Hegde, 2017 | Frameshifting reporters based on K12 poly sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-K(AAA)12–1-RFP | Juszkiewicz and Hegde, 2017 | Frameshifting reporters based on K12 poly sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-WT_XBP1_4AAA+0-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-S255A_XBP1_4AAA+0-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-W256A_XBP1_4AAA+0-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-WT_XBP1_4AAA+1-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-S255A_XBP1_4AAA+1-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-W256A_XBP1_4AAA+1-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-WT_XBP1_4AAA-1-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | pCMV-GFP-P2A-3XFLAG-VHPb-S255A_XBP1_4AAA-1-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | GFP-P2A-3XFLAG-VHPb-W256A_XBP1_4AAA-1-RFP | This paper | Frameshifting reporterts based on XBP1 stalling sequence | |
| Recombinant DNA reagent | pRSETA 6xHIS-TEV-eRF1(AAQ) | Brown et al., 2015 | Human mutant eRF1(AAQ) for expression in E. coli | |
| Sequence-based reagent | CRISPR: EDF1 sgRNA1 | IDT | 5' - ATCTTAGCGGCACAGAGACG - 3' | |
| Sequence-based reagent | CRISPR: EDF1 sgRNA1 | IDT | 5' - GAGCAAGGGGCTTACGCAGA - 3' | |
| Sequence-based reagent | CRISPR: GIGYF2 sgRNA1 | IDT | 5' - GGGAACACATGGAACGACGT - 3' | |
| Sequence-based reagent | CRISPR: GIGYF2 sgRNA2 | IDT | 5' - GGCGACTAGCTGGATCAAGG - 3' | |
| Sequence-based reagent | siRNA: control | Thermo Fisher | 4390843 | Silencer Select |
| Sequence-based reagent | siRNA: EDF1 #1 | Thermo Fisher | #16610 | Silencer Select |
| Sequence-based reagent | siRNA: EDF1 #2 | Thermo Fisher | #s225027 | Silencer Select |
| Sequence-based reagent | siRNA: GIGYF2 #1 | Thermo Fisher | #s25033 | Silencer Select |
| Sequence-based reagent | siRNA: GIGYF2 #2 | Thermo Fisher | #s25034 | Silencer Select |
| Sequence-based reagent | siRNA: ZNF598 | Thermo Fisher | #s40509 | Silencer Select |
| Sequence-based reagent | siRNA: 4EHP | Thermo Fisher | #s18149 | Silencer Select |
| Sequence-based reagent | siRNA: 4EHP | Thermo Fisher | #s18150 | Silencer Select |
| Sequence-based reagent | GAPDH Forward primer for RT-PCR | IDT | 5’-AGCTCATTTCCTGGTATGACA-3’ | |
| Sequence-based reagent | GAPDH Reverse primer for RT-PCR | IDT | 5’-AGGGGAGATTCAGTGTGGTG-3’ | |
| Sequence-based reagent | RPLP1 Forward primer for RT-PCR | IDT | 5’-CTCACTTCATCCGGCGACTAG-3’ | |
| Sequence-based reagent | RPLP1 Reverse primer for RT-PCR | IDT | 5’-GCAGAATGAGGGCCGAGTAG-3’ | |
| Sequence-based reagent | GFP Forward primer for RT-PCR | IDT | 5’-GGCAAGCTGACCCTGAAGTT-3’ | |
| Sequence-based reagent | GFP Reverse primer for RT-PCR | IDT | 5’-CTTGTAGTTGCCGTCGTCCT-3’ | |
| Sequence-based reagent | RFP Forward primer for RT-PCR | IDT | 5’-AGCAAGGGCGAGGAGGATAA-3’ | |
| Sequence-based reagent | RFP Reverse primer for RT-PCR | IDT | 5’- TAGGCCTTGGAGCCGTACAT-3’ | |
| Peptide, recombinant protein | S7 Micrococcal Nuclease | Roche | Cat #11873580001 | purified protein |
| Chemical compound, drug | Hygromycin B | Millipore | Cat #400051-100KU | Selection antibiotic |
| Chemical compound, drug | Blasticidin S | Santa Cruz Biotechnology | Cat #sc204655 | Selection antibiotic |
| Chemical compound, drug | Emetine | Calbiochem | Cat #324693 | Used to induce ribosomal stalling |
| Software, algorithm | FlowJo | FlowJo | RRID:SCR_008520 | Analysis of FACS data |
| Software, algorithm | GraphPad Prism | GraphPad Prism | RRID:SCR_008520 | Statistical analysis, graphs |
| Other | Complete EDTA-free protease inhibitor cocktail | Roche | Cat #118735800001 |





