Optogenetic activation of heterotrimeric G-proteins by LOV2GIVe, a rationally engineered modular protein
Figures
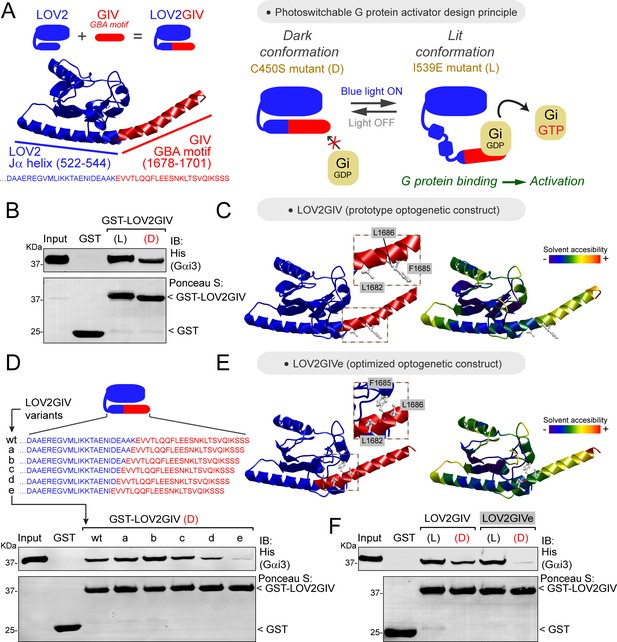
Design and optimization of LOV2GIV, an optogenetic activator of heterotrimeric G-proteins.
(A) Left, diagram depicting the design of the LOV2GIV fusion protein. The GBA motif of GIV (red) is fused to the C-terminus of the LOV2 domain (blue). Right, diagram depicting the design principle of the photoswitchable G-protein activator LOV2GIV. In the dark conformation (D, which is mimicked by the LOV2 C450S mutant), the C-terminus of LOV2 forms an α-helix (Jα) and the GBA motif is not readily accessible for G-proteins. In the lit conformation (L, which is mimicked by the LOV2 mutant I539E), the C-terminus of LOV2 is more disordered and the GBA motif becomes accessible for G-proteins, which in turn are activated upon binding to the GBA motif. (B) LOV2GIV (L) binds Gαi3 better than LOV2GIV (D). Approximately 20 µg of the indicated purified GST-fused constructs were immobilized on glutathione-agarose beads and incubated with 3 µg (~300 nM) of purified His-Gαi3. Resin-bound proteins were eluted, separated by SDS-PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) with the indicated antibodies. Input = 10% of the total amount of His-Gαi3 added in each binding reaction. (C) Ribbon representation of a LOV2GIV structure homology model generated using the I-TASSER server. On the left, the model is colored blue for LOV2 and red for GIV, whereas on the right it is colored according to solvent accessibility. Selected GIV residues known to be important for G-protein binding (L1682, F1685, L1686) (de Opakua et al., 2017; Kalogriopoulos et al., 2019) are displayed in stick representation and enlarged in the boxes. (D) LOV2GIV (D) variant ‘e’ displays greatly diminished Gαi3 binding compared to LOV2GIV (D) wt. Protein binding experiments were carried out as described in panel B, except that the indicated LOV2GIV variants were used. (E) Ribbon representation of a structure homology model of the LOV2GIVe variant was generated and displayed as described for LOV2GIV in panel C. (F) The dynamic range of Gαi3 binding to lit versus dark conformations is improved for LOV2GIVe compared to LOV2GIV. Protein binding experiments were carried out as described in panel B, except that the indicated LOV2GIV variants were used. For all protein binding experiments in this figure, one representative result of at least three independent experiments is shown.
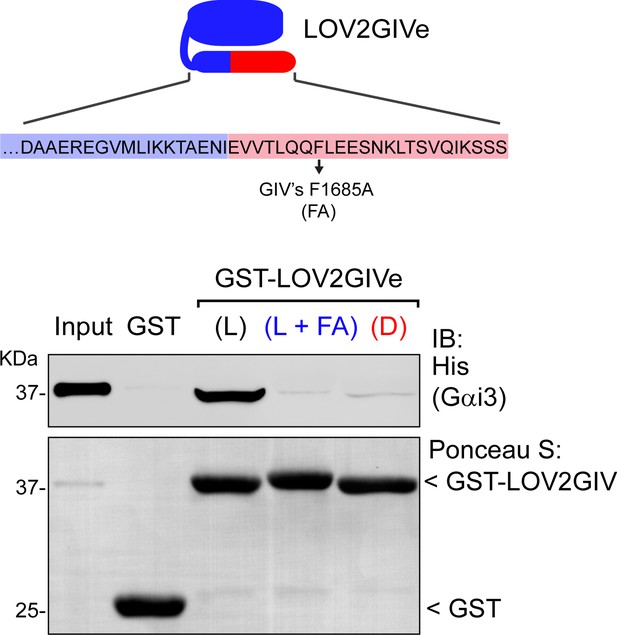
FA mutation in LOV2GIVe (L) impairs G-protein binding.
Top, diagram depicting the F1685A mutation of GIV’s GBA motinf in the context of LOV2GIVe. Bottom, approximately 20 µg of the indicated purified GST-fused constructs were immobilized on glutathione–agarose beads and incubated with 3 µg (~300 nM) of purified His-Gαi3. Resin-bound proteins were eluted, separated by SDS-PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) with the indicated antibodies. Input = 10% of the total amount of His-Gαi3 added in each binding reaction. One representative result of two independent experiments is shown.
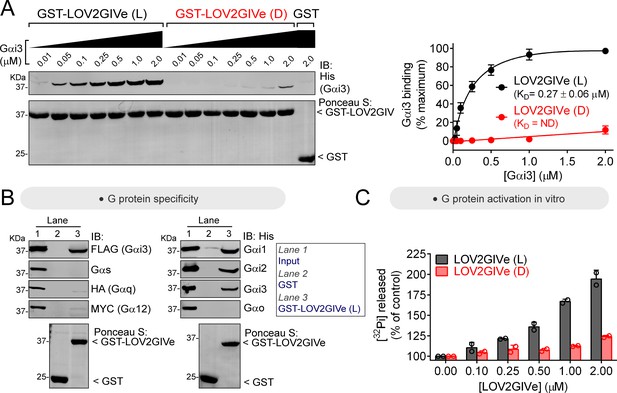
LOV2GIVe binds and activates Gαi3 in vitro in its lit conformation.
(A) LOV2GIVe binds with high affinity to Gαi3. Approximately 10 µg of the indicated purified GST-fused constructs were immobilized on glutathione-agarose beads and incubated with the indicated concentrations of purified His-Gαi3. Resin-bound proteins were eluted, separated by SDS-PAGE, and analyzed by Ponceau S-staining and immunoblotting (IB) with the indicated antibodies. One representative result is shown on the left, and the graph on the right corresponds to the quantification of three independent experiments presented as mean ± S.E.M. for each data point and a solid line for the fit to a single site binding curve used to determine the KD values. (B) LOV2GIVe binds specifically to Gαi compared to other Gα subunits. Approximately 20 µg of the indicated purified GST-fused constructs were immobilized on glutathione-agarose beads and incubated with the lysates of HEK293T cells expressing the indicated G-proteins (FLAG-Gαi3, Gαs, Gαq-HA and Gα12-MYC on the left panels) or purified His-tagged proteins (3 µg, ~300 nM of His-Gαi1, His-Gαi2, His-Gαi3 and His-Gαo on the right panels). One representative experiment of at least three is shown. (C) LOV2GIVe (L), but not LOV2GIVe (D), increases Gαi3 activity in vitro. Steady-state GTPase activity of purified His-Gαi3 was determined in the presence of increasing amounts (0–2 µM) of purified GST-LOV2GIVe (L) (black) or GST-LOV2GIVe (D) (red) by measuring the production of [32P]Pi at 15 min. Results are the mean ± S.D. of n = 2.
-
Figure 2—source data 1
Numerical data used for panel A.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig2-data1-v2.xlsx
-
Figure 2—source data 2
Numerical data used for panel C.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig2-data2-v2.xlsx
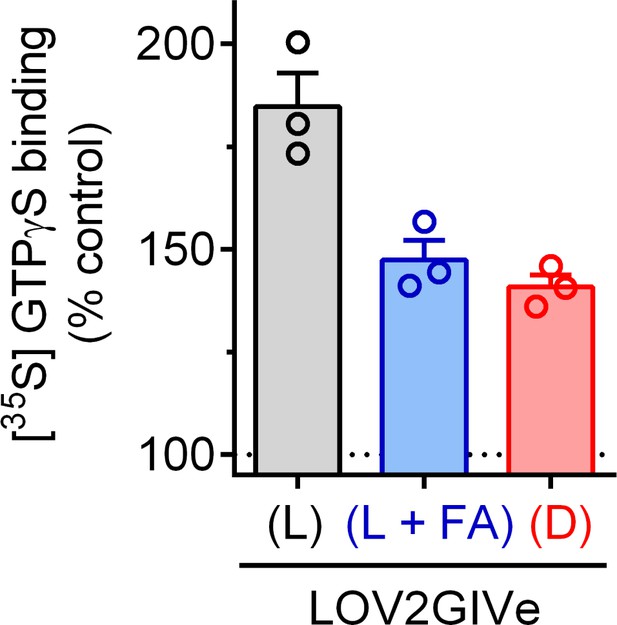
LOV2GIVe (L) promotes GTP binding to Gαi3 in vitro.
Binding of radiolabeled GTPγS to purified His-Gαi3 was determined in the absence or presence 2 µM of purified GST-LOV2GIVe (L) (black), GST-LOV2GIVe (L + FA) (blue) or GST-LOV2GIVe (D) (red). Results are expressed as a % of GTPγS binding to Gαi3 in the absence of LOV2GIVe (mean ± S.E.M., n = 3).
-
Figure 2—figure supplement 1—source data 1
Numerical data used to generate the graph.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig2-figsupp1-data1-v2.xlsx
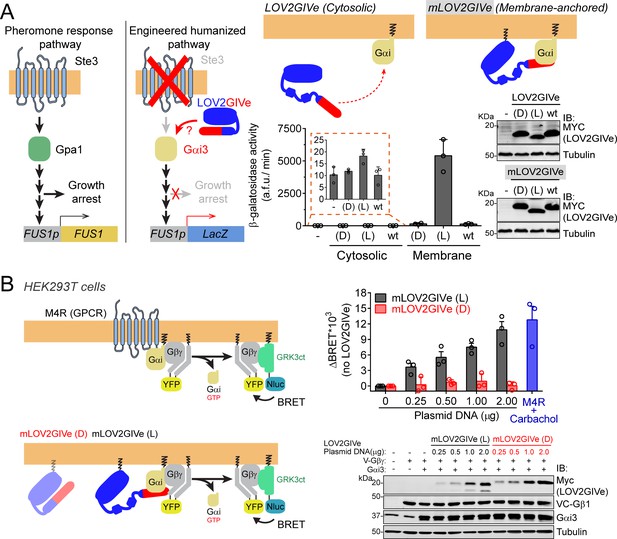
LOV2GIVe activates G-protein signaling in cells in its lit conformation.
(A) Left, diagram comparing key steps and components of the mating pheromone response pathway of Saccharomyces cerevisiae to those of an engineered, humanized strain used in the experiments shown in this figure. In the engineered strain, no pheromone responsive GPCR (Ste3) is expressed, the yeast G-protein Gpa1 is replaced by human Gαi3, and downstream signaling does not lead to growth arrest but promotes the activation of a reporter gene (LacZ) under the control of the pheromone-sensitive, G-protein-dependent promoter of FUS1 (FUS1p). Right, membrane-anchored LOV2GIVe (mLOV2GIVe), but not its untargeted parental version, leads to strong G-protein activation only in the lit conformation. Yeast strains expressing the indicated LOV2GIVe constructs ((D), (L) or wt) or an empty vector (-) were lysed to determine β-galactosidase activity using a fluorogenic substrate (mean ± S.E.M., n = 3) and to prepare samples for immunoblotting (IB)(one experiment representative of 3 is shown). (B) Left, diagrams showing the principle for the G-protein activity biosensor used in this panel. Upon action of a GPCR (top) or mLOV2GIVe (bottom), G-protein activation leads to the release of YFP-tagged Gβγ from Gαi, which then can associate with Nluc-tagged GRK3ct and results in the subsequent increase in BRET. Right, mLOV2GIVe (L) but not mLOV2GIVe (D), leads to increased G-protein activation similar to a GPCR as determined by BRET. BRET was measured in HEK293T cells transfected with the indicated amounts of mLOV2GIVe plasmid constructs along with the components of the BRET biosensor and the GPCR M4R. M4R was stimulated with 100 µM carbachol. Results in the graph on the top are expressed as difference in BRET (ΔBRET) relative to unstimulated cells not expressing mLOV2GIVe (mean ± S.E.M., n = 3). One representative immunoblot from cell lysates made after the BRET measurements is shown on the bottom.
-
Figure 3—source data 1
Numerical data used for panel A.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Numerical data used for panel B.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig3-data2-v2.xlsx
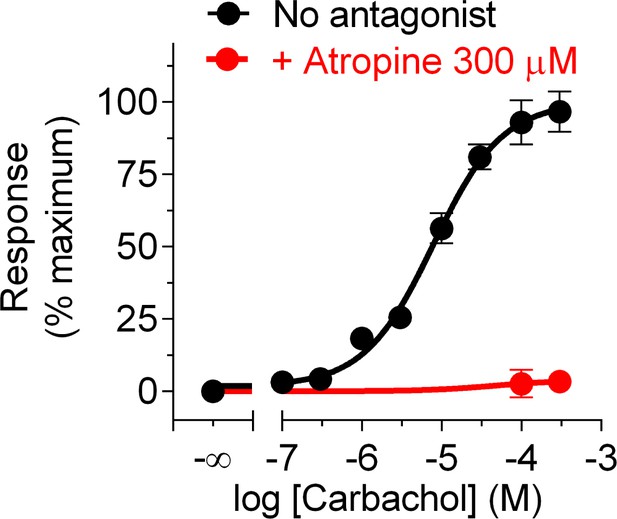
Carbachol dose-dependent G-protein activation and its blockade by atropine.
BRET was measured in HEK293T cells transfected with the components of the BRET biosensor described in Figure 3B after stimulation with the indicated concentrations of carbachol for 4 min in the presence or absence of atropine. Difference in BRET relative to cells not stimulated with carbachol was normalized to the maximal response and expressed as mean ± S.E.M. of n = 3.
-
Figure 3—figure supplement 1—source data 1
Numerical data used to generate the graph.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig3-figsupp1-data1-v2.xlsx
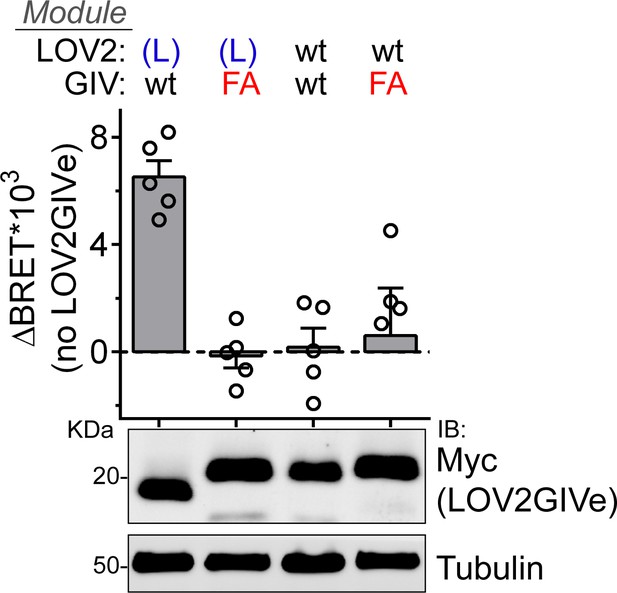
FA mutation abolishes G-protein activation by mLOV2GIVe (L) in cells.
BRET was measured in HEK293T cells transfected with 2 µg of the indicated mLOV2GIVe plasmid constructs along with the components of the BRET biosensor described in Figure 3. Results in the graph on the top are expressed as difference in BRET (ΔBRET) relative to cells not expressing mLOV2GIVe (mean ± S.E.M., n = 5). One representative immunoblot from cell lysates made after the BRET measurements is shown on the bottom.
-
Figure 3—figure supplement 2—source data 1
Numerical data used to generate the graph.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig3-figsupp2-data1-v2.xlsx
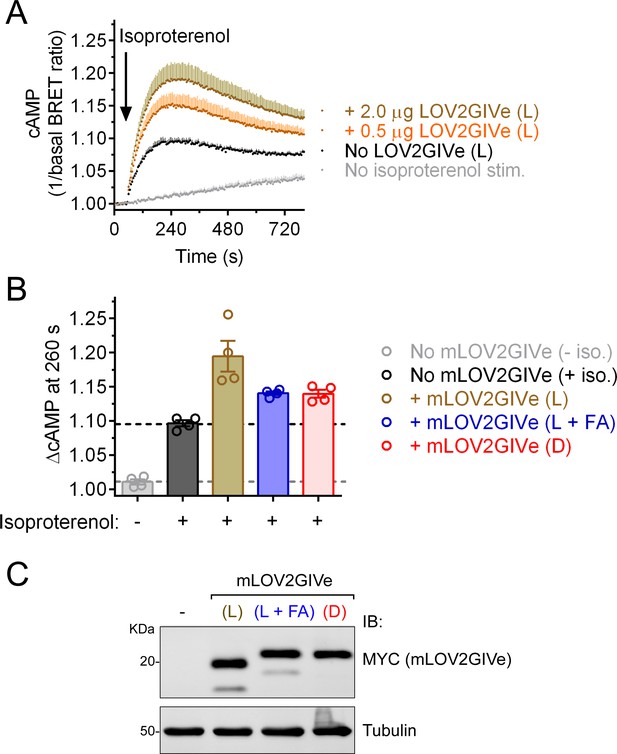
LOV2GIVe (L) expression enhances isoproterenol-induced cAMP levels in cells via Gi regulation.
(A) Changes in cAMP levels upon isoproterenol stimulation (100 nM) were measured in HEK293T cells transfected with the NLuc-EPAC-VV BRET biosensor and the indicated amounts of plasmids encoding mLOV2GIV (L). Results are expressed as mean ± S.E.M of n = 4. Error bars (shaded in a color tone lighter than the corresponding data points) are displayed only in the positive direction for clarity. (B) The peak change of cAMP (200 s after isoproterenol injection time) was quantified in HEK293T cells transfected with 2 µg of plasmid DNA encoding the indicated mLOV2GIVe constructs. Results are expressed as mean ± S.E.M of n = 4. (C) Representative immunoblot from cell lysates made after the BRET measurements done for panel (B).
-
Figure 3—figure supplement 3—source data 1
Numerical data used for panel A.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig3-figsupp3-data1-v2.xlsx
-
Figure 3—figure supplement 3—source data 2
Numerical data used for panel B.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig3-figsupp3-data2-v2.xlsx
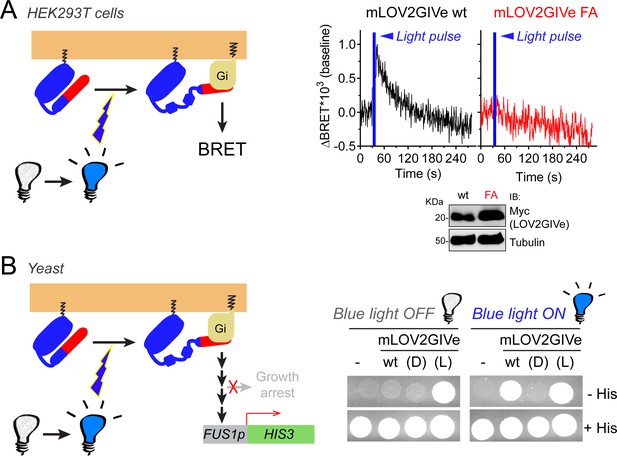
mLOV2GIVe activates G-protein signaling in cells upon illumination.
(A) mLOV2GIVe activates G-proteins in mammalian cells upon illumination. Kinetic BRET measurements were carried out in HEK293T cells transfected with 2 µg of plasmids for the expression of mLOV2GIVe wt or FA along with the components of the BRET biosensor. Results are expressed as baseline corrected BRET changes (ΔBRET) and representative traces of one experiment out of four are presented. (B) mLOV2GIVe activates G-protein signaling in cells upon blue light illumination. Yeast strains expressing the indicated mLOV2GIVe constructs (wt, (D) or (L)) or an empty vector (-) were spotted on plates with or without histidine as indicated and imaged after 4 days of incubation in the dark (blue light OFF) or in the presence of blue light (blue light ON). One experiment representative of 3 is shown.
-
Figure 4—source data 1
Numerical data used in panel A.
- https://cdn.elifesciences.org/articles/60155/elife-60155-fig4-data1-v2.xlsx

Binding of N-terminally truncated Gαi1 (Gαi1ΔN) to LOV2GIVe is detectable by Ponceau S staining.
Approximately 20 µg of the indicated purified GST or GST-LOV2GVe (L) were immobilized on glutathione-agarose beads and incubated with 3 µg (~300 nM) of purified Gαi3ΔN. Resin-bound proteins were eluted, separated by SDSPAGE, and analyzed by Ponceau S-staining and immunoblotting with the Gαi1/2 antibodies. One representative result of two independent experiments is shown.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Escherichia coli) | BL21(DE3) | Invitrogen | Cat# C600003 | |
| Strain, strain background (Escherichia coli) | DH5alpha | New England Biolabs | Cat# C2987I | |
| Genetic reagent (Saccharomyces cerevisiae) | CY7967 [MATα GPA1(1–41)-Gαi3 far1Δ fus1p-HIS3 can1 ste14:trp1:LYS2 ste3Δ lys2 ura3 leu2 trp1 his3] | Cismowski et al., 1999 | Provided by James Broach (Penn State University) | |
| Cell line (Homo sapiens) | HEK293T cells | ATCC | CRL3216 | |
| Antibody | α-tubulin (mouse monoclonal) | Sigma | T6074 | Immunoblotting Dilution (1: 2,500) |
| Antibody | FLAG tag (mouse monoclonal) | Sigma | F1804 | Immunoblotting Dilution (1: 2,000) |
| Antibody | His-tag (mouse monoclonal) | Sigma | H1029 | Immunoblotting Dilution (1: 2,500) |
| Antibody | Hemagglutinin (HA) tag (clone 12CA5) (mouse monoclonal) | Roche | Cat# 11583816001 | Immunoblotting Dilution (1: 1,000) |
| Antibody | MYC-tag (9B11) (mouse monoclonal) | Cell Signaling | Cat# 2276 | Immunoblotting Dilution (1: 1,000) |
| Antibody | Gαs (C-18) (rabbit polyclonal) | Santa Cruz Biotechnology | Cat# sc-383 | Immunoblotting Dilution (1: 500) |
| Antibody | Goat anti-rabbit Alexa Fluor 680 (goat polyclonal) | Life Technologies | Cat# A21077 | Immunoblotting Dilution (1:10,000) |
| Antibody | Goat anti-mouse IRDye 800 (goat polyclonal) | LiCor | Cat# 926–32210 | Immunoblotting Dilution (1:10,000) |
| Recombinant DNA reagent | pLIC-GST (plasmid) | Cabrita et al., 2006 | Provided by John Sondek (University of North Carolina- Chapel Hill) | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIV (plasmid) | This paper | ||
| Recombinant DNA reagent | pLIC-GST-LOV2GIV (L) (plasmid) | This paper | Contains the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIV (D) (plasmid) | This paper | Contains the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIVa (D) (plasmid) | This paper | Contains the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIVb (D) (plasmid) | This paper | Contains the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIVc (D) (plasmid) | This paper | Contains the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIVd (D) (plasmid) | This paper | Contains the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIVe (D) (plasmid) | This paper | Contains the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIVe (L) (plasmid) | This paper | Contains the lit-mimicking mutation I539E | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIVe (D) (plasmid) | This paper | Contains the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-GST-LOV2GIVe (L + FA) (plasmid) | This paper | Contains the lit-mimicking mutation I539E and the GEF-deficient mutation F1685A | |
| Recombinant DNA reagent | pET28b-Gαi3 (plasmid) | Garcia-Marcos et al., 2009 | For the bacterial expression of rat His-Gαi3 | |
| Recombinant DNA reagent | pET28b-Gαo (plasmid) | Garcia-Marcos et al., 2010 | For the bacterial expression of rat His-Gαo (isoform A) | |
| Recombinant DNA reagent | pLIC-His (plasmid) | Stols et al., 2002 | ||
| Recombinant DNA reagent | pLIC-His- Gαi1(int.6xHis) (plasmid) | This paper | For the bacterial expression of rat Gαi1 with an internal His-tag | |
| Recombinant DNA reagent | pET28b-Gαi2 (plasmid) | This paper | For the bacterial expression of rat His-Gαi2 | |
| Recombinant DNA reagent | p3XFLAG-CMV10-Gαi3 (plasmid) | Garcia-Marcos et al., 2009 | For the mammalian expression of rat FLAG-Gαi3 | |
| Recombinant DNA reagent | pcDNA3.1(+)-Gαs (plasmid) | Beas et al., 2012 | For the mammalian expression of human Gαs | |
| Recombinant DNA reagent | pcDNA3-Gαq-HA (plasmid) | Wedegaertner et al., 1993 | For the mammalian expression of mouse Gαq with an internal HA tag. Provided by P. Wedegaertner (Thomas Jefferson University) | |
| Recombinant DNA reagent | pcDNA3.1-Gα12-MYC (plasmid) | Ritchie et al., 2013 | For the mammalian expression of mouse Gα12 with an internal MYC-tag. Provided by T. Meigs (University of North Carolina, Asheville) | |
| Recombinant DNA reagent | pLIC-YES2 (plasmid) | Coleman et al., 2016 | ||
| Recombinant DNA reagent | pLIC-YES2-N9Gpa1 (plasmid) | Coleman et al., 2016 | ||
| Recombinant DNA reagent | pLIC-YES2-LOV2GIVe wt (plasmid) | This paper | For the yeast expression of cytosolic LOV2GIVe | |
| Recombinant DNA reagent | pLIC-YES2-LOV2GIVe (L) (plasmid) | This paper | For the yeast expression of cytosolic LOV2GIVe bearing the lit-mimicking mutation I539E | |
| Recombinant DNA reagent | pLIC-YES2-LOV2GIVe (D) (plasmid) | This paper | For the yeast expression of cytosolic LOV2GIVe bearing the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-YES2- N9Gpa1-LOV2GIVe wt (plasmid) | This paper | For the yeast expression of membrane-anchored (m)LOV2GIVe | |
| Recombinant DNA reagent | pLIC-YES2- N9Gpa1-LOV2GIVe (L) (plasmid) | This paper | For the yeast expression of membrane-anchored (m)LOV2GIVe bearing the lit-mimicking mutation I539E | |
| recombinant DNA reagent | pLIC-YES2- N9Gpa1-LOV2GIVe (D) (plasmid) | This paper | For the yeast expression of membrane-anchored (m)LOV2GIVe bearing the dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-myc (plasmid) | Cabrita et al., 2006 | Provided by John Sondek (University of North Carolina- Chapel Hill) | |
| Recombinant DNA reagent | pLIC-lyn11-myc (plasmid) | This paper | ||
| Recombinant DNA reagent | pLIC-lyn11-myc-LOV2GIVe (plasmid) | This paper | For the mammalian expression of mLOV2GIVe | |
| Recombinant DNA reagent | pLIC-lyn11-myc-LOV2GIVe (L) (plasmid) | This paper | For the mammalian expression of mLOV2GIVe bearing the lit-mimicking mutation I539E | |
| Recombinant DNA reagent | pLIC-lyn11-myc-LOV2GIVe (D) (plasmid) | This paper | For the mammalian expression of mLOV2GIVe dark-mimicking mutation C450S | |
| Recombinant DNA reagent | pLIC-lyn11-myc-LOV2GIVe (FA) (plasmid) | This paper | For the mammalian expression of mLOV2GIVe GEF-deficient mutation F1685A | |
| Recombinant DNA reagent | pLIC-lyn11-myc-LOV2GIVe (L) (FA) (plasmid) | This paper | For the mammalian expression of mLOV2GIVe lit-mimicking mutation I539E and the GEF-deficient mutation F1685A | |
| Recombinant DNA reagent | pcDNA3.1-Venus(155-239)-Gβ1 (plasmid) | Hollins et al., 2009 | For the mammalian expression of Gβ1 tagged with a fragment of Venus (VC-Gβ1). Provided by N. Lambert (Augusta University, GA) | |
| Recombinant DNA reagent | pcDNA3.1-Venus(1-155)-Gγ2 (plasmid) | Hollins et al., 2009 | For the mammalian expression of Gγ2 tagged with a fragment of Venus (VN-Gγ2). Provided by N. Lambert (Augusta University, GA) | |
| Recombinant DNA reagent | pcDNA3.1-Gβ1 (plasmid) | Hollins et al., 2009 | For the mammalian expression of untagged Gβ1. Provided by N. Lambert (Augusta University, GA) | |
| Recombinant DNA reagent | pcDNA3.1-Gγ2 (plasmid) | Hollins et al., 2009 | For the mammalian expression of untagged Gγ2. Provided by N. Lambert (Augusta University, GA) | |
| Recombinant DNA reagent | pcDNA3-Gαi3 (plasmid) | Garcia-Marcos et al., 2010 | For the mammalian expression of rat Gαi3 | |
| Recombinant DNA reagent | pcDNA3.1-masGRK3ct-Nluc (plasmid) | Masuho et al., 2015 | Provided by K. Martemyanov (Scripps Research Institute, FL) | |
| Recombinant DNA reagent | pcDNA3.1-Nluc-EPAC-VV (plasmid) | Masuho et al., 2015 | Provided by K. Martemyanov (Scripps Research Institute, FL) | |
| Recombinant DNA reagent | pcDNA3.1-3xHA-M4R (plasmid) | cDNA Resource Center at Bloomsburg University | Cat# MAR040TN00 | |
| Commercial assay or kit | QuikChange II Site-Directed Mutagenesis Kit | Agilent | Cat# #200523 | |
| Chemical compound, drug | NanoGlo Luciferase Assay System | Promega | Cat# N1120 | |
| Chemical compound, drug | Carbachol | Acros Organics | Cat# AC-10824 | |
| Chemical compound, drug | Atropine | Alfa Aesar | Cat# A10236 | |
| Chemical compound, drug | DL-isoproterenol hydrochloride | Alfa Aesar | Cat# J61788 | |
| Chemical compound, drug | [γ-32P]GTP | Perkin Elmer | NEG004Z250UC | |
| Chemical compound, drug | [32S]GTPγS | Perkin Elmer | NEG030H250UC | |
| Chemical compound, drug | Fluorescein di-β-D-galactopyranoside (FDG) | Marker Gene Technologies | Cat# M0250 | |
| Software, algorithm | I-TASSER | Yang et al., 2015 | https://zhanglab.ccmb.med.umich.edu/I-TASSER/ | Homology modeling server |
| Software, algorithm | ICM version 3.8–3 | Molsoft LLC., San Diego, CA | Protein structure visualization software |
Additional files
-
Supplementary file 1
Sequences of LOV2GIV constructs.
- https://cdn.elifesciences.org/articles/60155/elife-60155-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60155/elife-60155-transrepform-v2.pdf





