Puromycin reactivity does not accurately localize translation at the subcellular level
Figures
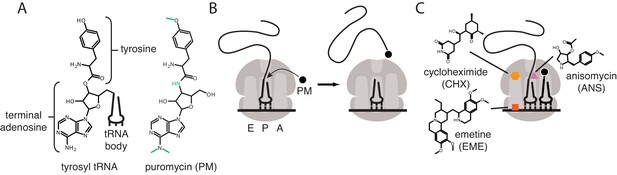
Mechanisms of action of puromycin and other translational inhibitors.
(A) Comparison of structure of 3′ terminus of tyrosyl tRNA with that of puromycin. Key differences are highlighted in green. tRNA body not drawn to scale. (B) Scheme for reaction of puromycin with peptidyl P-site tRNA on the ribosome, leading to dissociation of puromycylated peptide. (C) Structures and schematicized ribosome binding sites of translational inhibitors cycloheximide, anisomycin and emetine. Binding sites are based on Garreau de Loubresse et al., 2014; Wong et al., 2014.
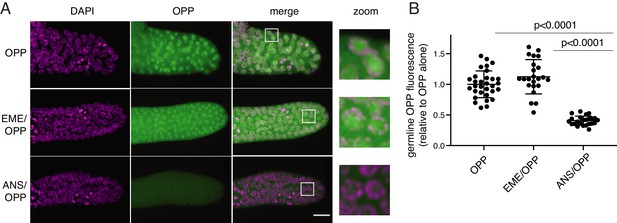
O-propargyl-puromycin (OPP) labels nuclei in the distal germline of C. elegans in the presence or absence of emetine.
(A) Representative photomicrographs of germlines labeled for 5 min with 20 µM OPP, and pre-treated for 15 min with control buffer (top row), 45 µM emetine (second row), or 37 µM anisomycin (bottom row). DAPI labels chromosomes. Post-fixation, click labeling of OPP with Alexa Fluor 488 picolyl azide revealed OPP throughout the cytoplasm and concentrated in nuclei. Scale bar = 10 µm. (B) Quantification of OPP-Alexa 488 signal in distal germlines. Each dot represents the average fluorescence of the mitotic zone of one worm germline. Values are normalized to the average obtained for germlines pre-treated with control buffer (OPP alone). P values were obtained through an unpaired t-test. Experiment performed in duplicate.
-
Figure 2—source data 1
Source data for Figure 2B.
- https://cdn.elifesciences.org/articles/60303/elife-60303-fig2-data1-v2.xlsx
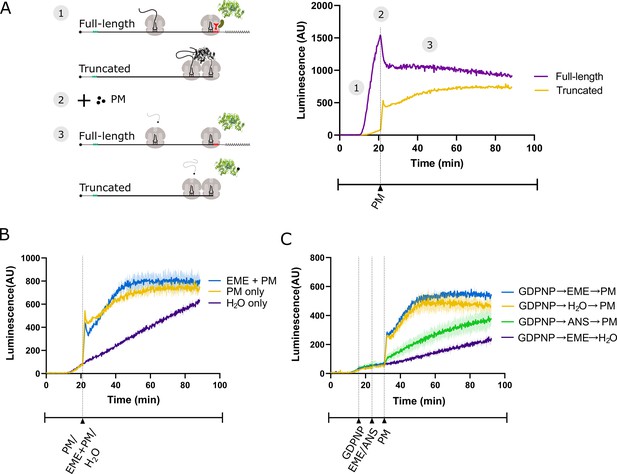
Emetine does not prevent release of puromycylated luciferase from rabbit reticulocyte ribosomes.
(A) Schematic of the real-time translation monitoring assay in rabbit reticulocyte lysate. (1) (Purple trace) Ribosomes translate the full-length luciferase mRNA and release luciferase which becomes enzymatically active and results in an increase in luminescence. (Yellow trace) Ribosomes stall at the 3’ end of a truncated luciferase mRNA and little to no luminescence is observed as the ribosome-bound luciferase peptides are in an enzymatically inactive conformation. (2) Puromycin (PM) is added to the system, stopping further translation and causing all nascent peptides to release from the ribosomes. (3) (Yellow) The luciferase rapidly folds into an enzymatically active conformation and a substantial increase in luminescence is observed. (B) Either puromycin (blue), H2O (purple) or a mixture of emetine (EME) and puromycin (yellow) was added to a reaction containing truncated luciferase mRNA at t = 21 min. Experiment was performed in duplicate; mean traces shown as solid lines and range of replicates shaded. (C) GDPNP was added to a reaction containing truncated luciferase mRNA at t = 16 min for 5 min to inhibit translation across samples. Then, either emetine (blue, purple), anisomycin (ANS) (green) or H2O (yellow) was added to the reaction followed by puromycin (blue, yellow, green) or H2O (purple) 5 min later. Experiment was performed in duplicate; mean traces shown as solid lines and range of replicates shaded. Note that the experiments in (A and B), and Figure 3—figure supplement 1B were done in the same batch, and the yellow traces (PM treated) in these panels are the same.
-
Figure 3—source data 1
Source data for Figure 3A, B and C.
- https://cdn.elifesciences.org/articles/60303/elife-60303-fig3-data1-v2.xlsx
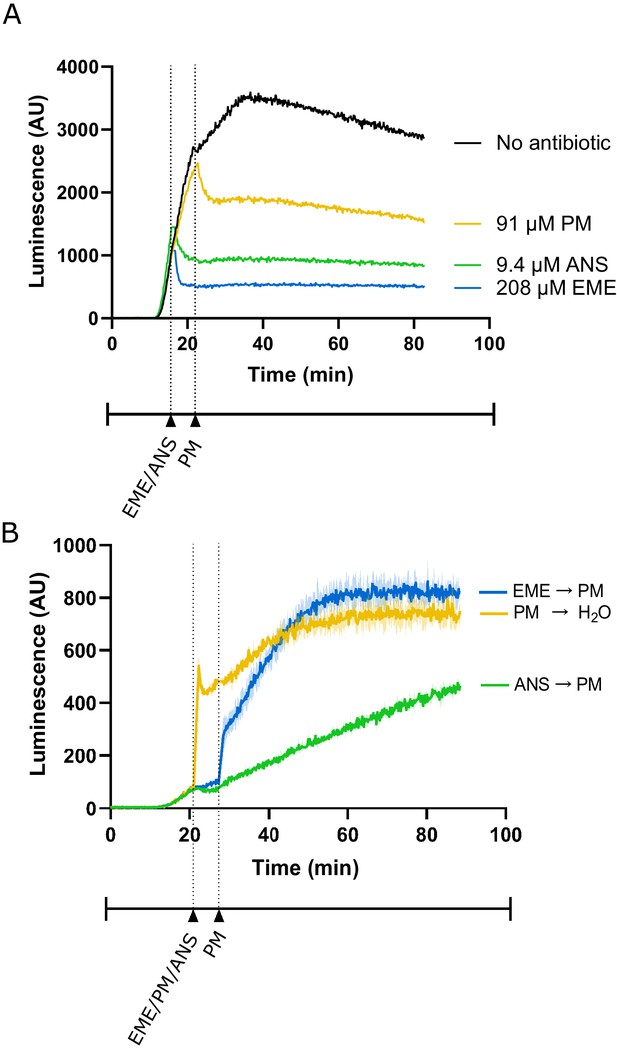
Additional control experiments for lysate-based luciferase assays.
(A) Luminescence measured from translation of full-length luciferase mRNA in rabbit reticulocyte lysate. Either emetine (blue), anisomycin (green) or puromycin (yellow) was added to the reaction at indicated concentrations. Due to experimental restrictions, emetine and anisomycin were added at t = 16 min and puromycin was added at t = 22 min. Experiment performed once. (B) Luminescence measured from translation of truncated luciferase mRNA in rabbit reticulocyte lysate. Either emetine (blue), puromycin (yellow) or anisomycin (green) was added to the reaction at t = 21 min for 5 min to inhibit translation across the samples. Then, either puromycin (blue, green) or H2O (yellow) was added to the reaction. Experiment was performed in duplicate; mean traces shown as solid lines and range of replicates shaded.
-
Figure 3—figure supplement 1—source data 1
Source data for Figure 3—figure supplement 1A and B.
- https://cdn.elifesciences.org/articles/60303/elife-60303-fig3-figsupp1-data1-v2.xlsx
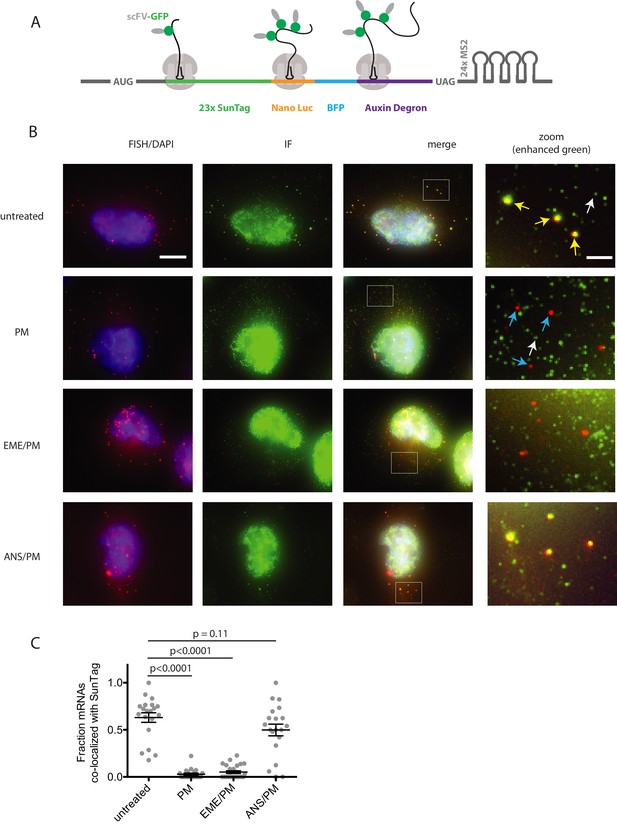
Puromycin treatment causes loss of nascent peptide-mRNA co-localization, independent of elongation inhibitors.
(A) SunTag reporter schematic. In addition to the tandem SunTag repeats and the auxin-inducible degron, this reporter encodes nano luciferase and BFP, which are not used in the present experiments. The 3’ UTR also encodes tandem repeats of the MS2 stem loop, which can be used to label the mRNA red. However, since we detect mRNA by FISH, we do not use the MS2 stem loops in the present experiments. (B) Example cells imaged by FISH-IF. Cells were either untreated (top row), treated with 91 μM puromycin for 5 min (second row), pre-treated with 208 μM emetine for 15 min followed by 91 μM puromycin for 5 min (third row), or pre-treated with 37 μM anisomycin for 5 min followed by 91 μM puromycin for 5 min (last row). Yellow arrows: examples of translating mRNAs; White arrows: example of single fully synthesized SunTag polypeptide (released from the ribosome); Blue arrows: examples of untranslating mRNAs. Scale bar in top left image: 10 microns. Scale bar in top right image: two microns. (C) Fraction of mRNAs co-localized with SunTag signal. Each dot represents one cell. Cells are only included in the analysis if they have more than five and fewer than 36 mRNAs. 20–27 cells and 313–513 mRNAs per condition were analyzed. Black lines indicate mean with standard error of the mean. P values were calculated by two-sample t-test. Experiment performed once.
-
Figure 4—source data 1
Source data for Figure 4C.
- https://cdn.elifesciences.org/articles/60303/elife-60303-fig4-data1-v2.xlsx
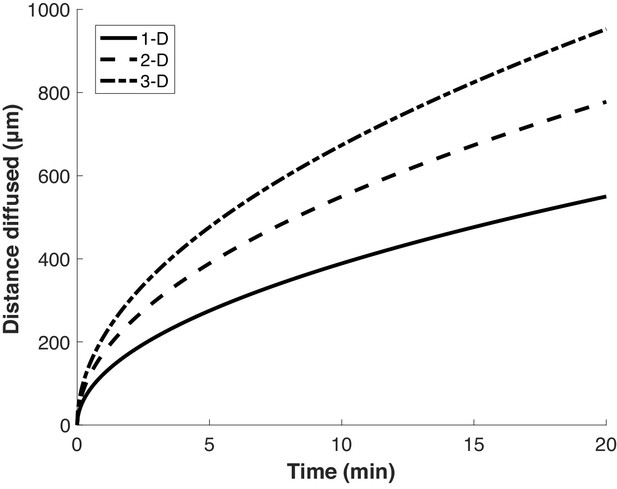
Proteins diffuse over long distances in the cell during common puromycin labeling times.
Calculation of expected displacement by diffusion as a function of time, using the equation < x2 > = 2nDt where n is the dimensionality, D is the diffusion coefficient (126 µm2/s Di Rienzo et al., 2014) and t is time. The calculation is shown for 1, 2 and 3 dimensions.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Recombinant DNA reagent | pGEM-luc (plasmid) | Promega | GenBank X65316.2 | Firefly luciferase cassette vector |
| Recombinant DNA reagent | pSL312 (plasmid) | This paper | Full-length firefly luciferase template; can be obtained from Green Lab | |
| Recombinant DNA reagent | P3.35_pGEM_luc_trunc_kozak_RC (plasmid) | This paper | Truncated firefly luciferase template; can be obtained from Green Lab | |
| Peptide, recombinant protein | StuI (restriction enzyme) | NEB | R0187S | Linearization of pSL312 for SP6 transcription |
| Peptide, recombinant protein | HpaI (restriction enzyme) | NEB | R0105S | Linearization of P3.35 for SP6 transcription |
| Sequence-based reagent | Full-length luciferase mRNA | This paper | SP6 transcribed from pSL312 | |
| Sequence-based reagent | Truncated luciferase mRNA | This paper | SP6 transcribed from P3.35 | |
| Commercial assay or kit | mMESSAGE mMACHINE SP6 transcription kit | Invitrogen | AM1340 | |
| Commercial assay or kit | Nuclease-treatedrabbit reticulocyte lysate translation reactions | Promega | L4960 | |
| Chemical compound, drug | Luciferin | PerkinElmer | 122799 | |
| Peptide, recombinant protein | Superase-In RNase Inhibitor | Invitrogen | AM2696 | |
| Chemical compound, drug | 5'-guanylyl imidodiphosphate (GDPNP) | Jena Bioscience | NU-401–50 | |
| Chemical compound, drug | Emetine | Cayman Chemical | 21048 | |
| Chemical compound, drug | Puromycin | Sigma Aldrich | P7255 | |
| Chemical compound, drug | Anisomycin | Sigma | A9789 | |
| Genetic reagent (C. elegans) | N2 | Caenorhabditis Genetics Center (CGC) | ||
| Commercial assay or kit | Click-iT Plus OPP Alexa Fluor 488 Protein Synthesis Assay kit | Invitrogen | C10456 | |
| Cell line (human) | U-2OS cells containing Flp-In locus | Andrew Holland lab (Johns Hopkins University) | ||
| Chemical compound, drug | amino-11–12 ddUTP | Lumiprobe | A5040 | |
| Peptide, recombinant protein | deoxynucleotidyl transferase | Thermo Fisher | EP0162 | |
| Chemical compound, drug | doxycycline hyclate | Millipore Sigma | D9891 | |
| Chemical compound, drug | Cy3-NHS ester | Lumiprobe | 41020 | |
| Chemical compound, drug | 3-indole acetic acid | Sigma Aldrich | I2886 | |
| Chemical compound, drug | paraformaldehyde | Electron Microscopy Sciences | 50-980-492 | |
| Peptide, recombinant protein | rat tail collagen I | Gibco | A1048301 | |
| Peptide, recombinant protein | BSA | VWR | VWRV0332-25G | |
| Chemical compound, drug | SSC buffer | Corning | 46–020 CM | |
| Chemical compound, drug | formamide | Sigma Aldrich | F9037-100ML | |
| Sequence-based reagent | E. coli tRNA | Sigma Aldrich | 10109541001 | |
| Chemical compound, drug | dextran sulfate | Sigma Aldrich | D8906-100G | |
| Chemical compound, drug | ribonucleoside vanadyl complex | NEB | S1402S | |
| Antibody | Chicken polyclonal anti-GFP antibody | Aves Labs | RRID:AB_2307313 | 1:1000 dilution |
| Antibody | Goat anti-chicken polyclonal IgY secondary antibody | Thermo Fisher | RRID:AB_2534096 | 1:1000 dilution |
| Chemical compound, drug | ProLong Diamond antifade reagent | Invitrogen | P36962 | |
| Recombinant DNA reagent | pubc-OSTIR1-IRES-scFv-sfGFP-NLS (plasmid) | Reference 32 | ||
| Software, algorithm | FISH-quant | Reference 45 | ||
| Software, algorithm | Custom MATLAB scripts for processing FISH-quant output | This paper | Scripts for quantifying number of translating ribosomes per mRNA from FISH-IF data; available asSource code 1 in supplementary files | |
| Sequence-based reagent | Oligonucleotides used to generate FISH probes | Reference 32 | See Supplementary file 1 |
Additional files
-
Source code 1
Script for analysis of FISH-IF data.
- https://cdn.elifesciences.org/articles/60303/elife-60303-code1-v2.m.zip
-
Supplementary file 1
Oligonucleotide sequences used to generate smFISH probes.
- https://cdn.elifesciences.org/articles/60303/elife-60303-supp1-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60303/elife-60303-transrepform-v2.docx





