Emx2 regulates hair cell rearrangement but not positional identity within neuromasts
Figures
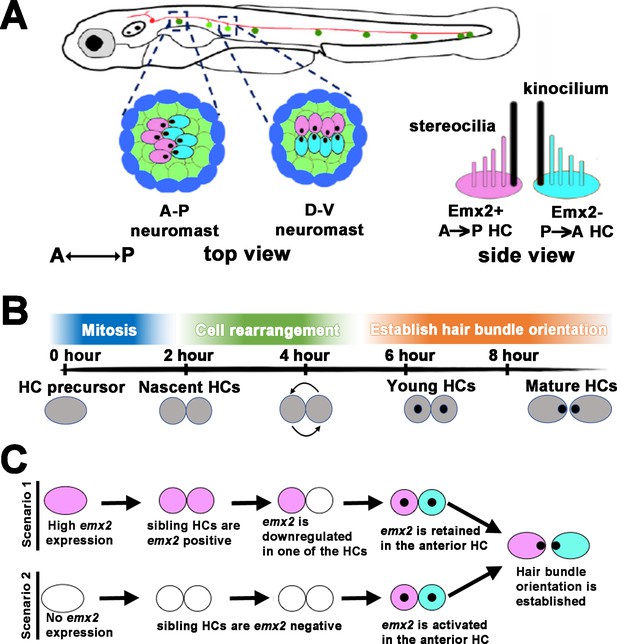
Schematic of bidirectional hair cell (HC) establishment in the zebrafish neuromast.
(A) The lateral line system in zebrafish showing the top view of an anterior-posterior (A-P) and dorsal-ventral (D-V) neuromast, based on their hair bundle orientation. Side view of two hair bundles, each comprises a kinocilium (black) and a stereociliary staircase (pink or blue), arranged in opposite orientation atop of an Emx2-positive (pink) and Emx2-negative (blue) HC. (B) An approximate developmental time-line of HC formation in zebrafish neuromast. A HC precursor (gray) divides to form two daughter HCs, which roll to exchange positions with each other 50% of the time before differentiating into mature HCs with opposite hair bundle orientation. The entire process takes approximately 8 hr from detectable Gfp signal driven by the myosin6b promoter to HCs with polarized hair bundles based on our live-imaging analyses. (C) Emx2, which is important for establishing the bidirectional HC pattern, can be expressed early in the HC precursors, which could affect the HC rearrangement (scenario 1) or later during hair bundle establishment after HCs are formed (scenario 2). A combination of both scenarios is also possible (not shown). Black dots represent the position of the kinocilium.
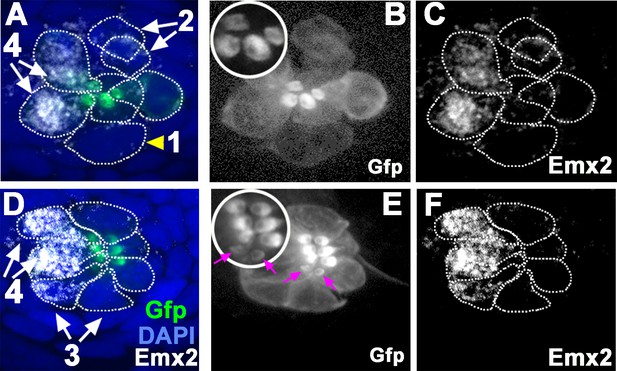
Emx2 immunostaining in wild-type (WT) (myo6b:actb1-Gfp) neuromasts.
(A and D) Merged image of DAPI, (B and E) β-actin-GFP, and (C and F) anti-Emx2 staining of neuromasts. Emx2 immunoreactivity is not detected in dividing hair cell (HC) precursor (#1, arrowhead) or daughter HCs during Rock and Roll (#2). However, it is detectable in immature daughter HCs after Rock and Roll, in which the oriented hair bundle is not yet evident (#3, E, magenta arrows), as well as in mature HCs (#4). Both immature (#3) and mature HCs (#4, A→P) that are located in the anterior side of the neuromast are Emx2-positive. Insets in B and E are higher magnifications showing the hair bundle orientation pattern at the apex of HCs.
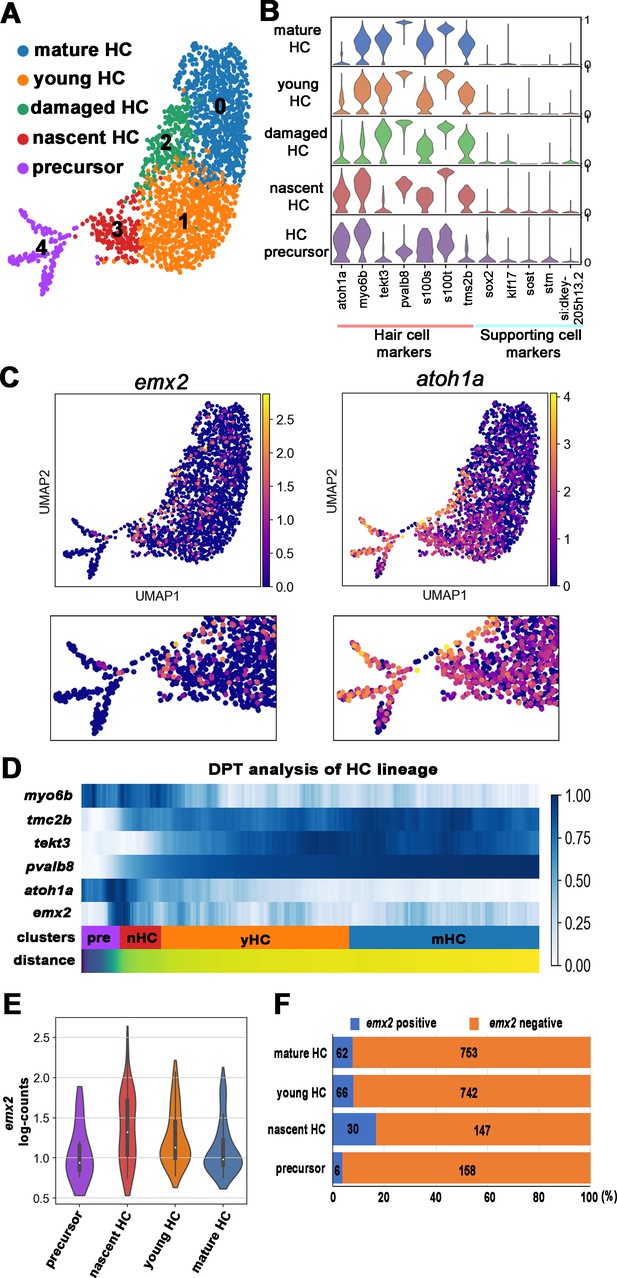
scRNA-seq analysis of the hair cell (HC) lineage in neuromasts.
(A) Uniform manifold approximation and projection (UMAP) plot colored by Louvain clustering annotated with cell types in the HC lineage. (B) Violin plots of selected HC (red bar) and supporting cell (blue bar) marker genes in each cluster. (C) Distribution of cells expressing emx2 and atoh1a on UMAP. Clusters of precursors (#4) and nascent HCs (#3) are enlarged in bottom panels. (D) Heat map representation of selected gene expression dynamics in pseudotime. (E) A violin plot showing significantly higher read counts of emx2 in the nascent HC cluster than other clusters (p=0.0069, two tail Student's t-test, source data 1). (F) A bar graph showing a significantly higher number of emx2-positive cells in the nascent HC cluster than others (X2 = 22.44, df = 3, p<0.0001, source data 2). A post-hoc 2 × 2 chi-squared test was also performed for multiple comparisons: precursor HC cluster vs nascent HC cluster, X2 (df = 1)=15.92, p<0.0001; nascent HC cluster vs young HC cluster, X2 (df = 1)=12.73, p=0.0004; nascent HC cluster vs mature HC, X2 (df = 1)=15.08, p<0.0001. The following figure supplements are available for Figure 3—figure supplement 1. Identification and annotation of cell types from Louvain clustering.
-
Figure 3—source data 1
Comparison of the emx2 log-counts in nascent hair cell (HC) cluster versus other clusters.
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Comparison of emx2-positive and -negative cells among four hair cell (HC) clusters.
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig3-data2-v2.xlsx
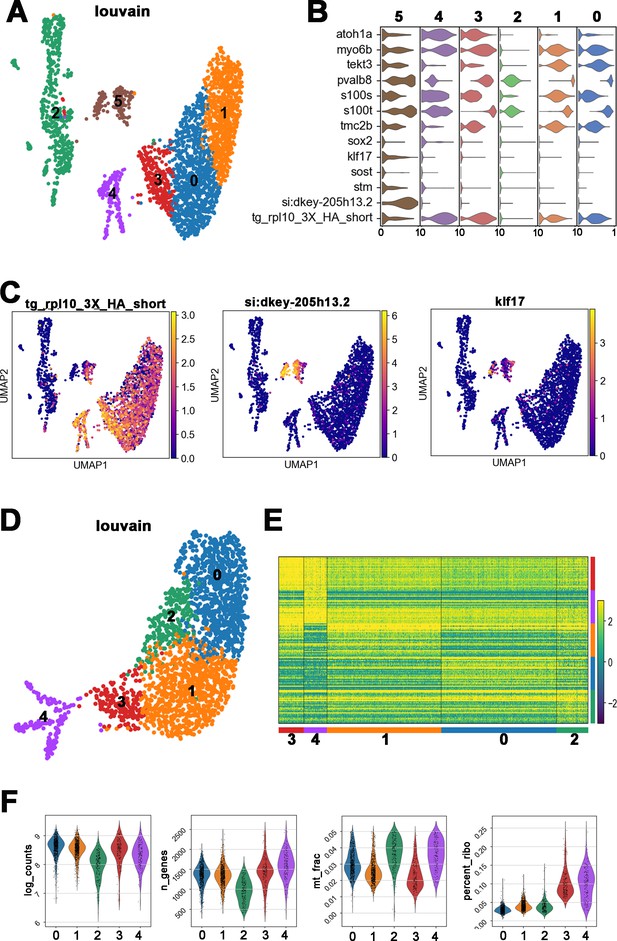
Identification and annotation of cell types from Louvain clustering.
(A) Uniform manifold approximation and projection (UMAP) plot colored by Louvain clustering showing six clusters that included rpl10-HA (RiboTag) positive and negative cells. (B) Violin plots of selected marker genes of hair cell (HC) and SC. High levels of transgene tg_rpl10_3X_HA_short transcripts were concentrated in cluster #0, 1, 3, and 4 indicating they are cells within the HC lineage. Relative lower levels of rpl10-HA transcripts were found in cluster #2 of unknown identity, and #5, which expresses a high level of a SC marker, si:dkey-205h13.2. (C) Distribution of selected marker genes in UMAP. tg_rpl10_3X_HA_short is distributed in all the clusters except cluster #2. Supporting cell markers, si:dkey-205h13.2 and klf17 positive cells are concentrated in cluster #5. Thus, clusters #2 and #5 were excluded from the reclustering. (D) UMAP plot of reclustering of #0, 1, 3, and 4 shown in (A). (E) Heatmap showing the top 30 genes expressed in each cluster after the reclustering. (F) Violin plot showing distribution of n-counts, n-genes, mitochondrial fraction, and percentage of ribosomal genes of each cluster. The lower count depth, lower transcripts, and higher fraction of mitochondrial genes in cluster #2 indicate that they are damaged HCs. High percentages of ribosomal genes in clusters #3 and #4 are features of HC precursors and nascent HCs (Lush et al., 2019).
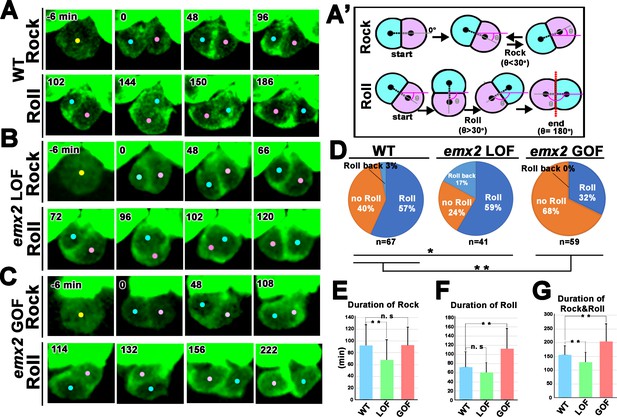
Quantification of frequency and duration of Rock and Roll among hair cells (HCs) in wild-type (WT) and emx2 mutants.
(A) In WT, a HC precursor (yellow dot) divides to form two daughter HCs (pink and blue dots), which first rock (Rock phase) and then frequently roll to exchange positions with each other (Roll phase). (A’) Definition of the Rock and Roll phase (see Materials and methods). (B and C) Compared to WT, emx2 loss of function (LOF) (B) and gain of function (GOF) (C) HC pairs took a shorter time in Rock and Roll and a longer time in the Roll phase, respectively. (D) Frequencies of Roll, No Roll, and Roll back of nascent HC pairs in WT (n = 67 from seven larvae), Emx2 LOF (n = 41 from six larvae), and GOF (n = 59 from seven larvae). Significance was assessed by using chi-squared test with 3 × 3 contingency table (X2 (df = 4)=30.00, p<0.0001, source data 1). A post-hoc 2 × 3 chi-squared test was performed for multiple comparisons. WT vs. LOF; (X2 (df = 2)=7.95, p=0.018), WT vs. GOF; (X2 (df = 2)=10.39, p=0.0055). Post-hoc chi-squared tests were performed for pairwise comparisons with FDR correction. (E–G) Duration of Rock (E), Roll (F), and Rock and Roll (G) of sibling HCs that underwent Rock and Roll in WT (n = 42), emx2 LOF (n = 35), and GOF (n = 19) larvae. Significance was assessed by using MANOVA (Rock, F(df1 = 2, df2 = 93)=5.349, p=0.0063, Wilks’ λ = 0.897; Roll, F(df1 = 2, df2 = 93)=15.638, p<0.0001, Wilks’ λ = 0.748; Rock and Roll, F(df1 = 2, df2 = 93)=20.10, p<0.0001, Wilks’ λ = 0.698), with post-hoc Tukey’s test for pairwise comparisons. *p<0.05, **p<0.01, n.s., not significant.
-
Figure 4—source data 1
Multiple comparisons of Roll frequencies among wild-type (WT), emx2 loss of function (LOF), and emx2 gain of function (GOF) hair cells (HCs).
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Quantification of Rock and Roll movements in wild-type (WT), emx2 loss of function (LOF), and emx2 gain of function (GOF) hair cells (HCs).
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig4-data2-v2.xlsx
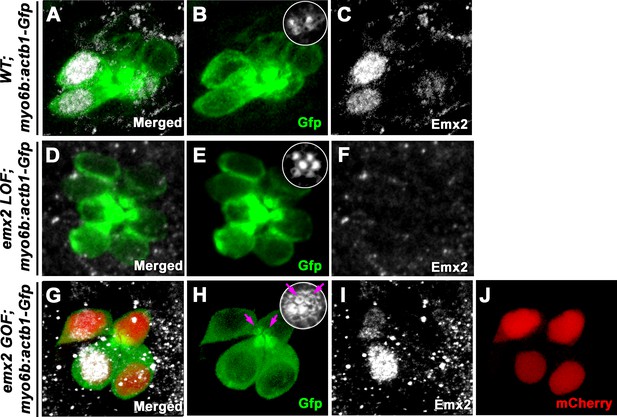
Localization of Emx2 expression in wild-type (WT), loss of function (LOF), and gain of function (GOF) neuromasts at 2.0 dpf.
(A–C) Only hair cells (HCs) (B, Gfp) in the anterior neuromast of WT are positive for anti-Emx2 staining (C). Inset in B shows the bidirectional hair bundle orientation pattern. (D–F) No HCs (E) are positive for anti-Emx2 staining (F) in emx2 LOF neuromast and all hair bundles are oriented in P→A direction (E). (G–J) An emx2 GOF neuromast. (G) Merged image of Gfp (H), anti-Emx2 (I), and mCherry (J) expression. Anti-Emx2 staining only detected endogenous Emx2 in the anterior HCs (I), whereas exogenous Emx2 expression as indicated by mCherry expression is present in all HCs (J). All hair bundles are oriented in A→P direction and two immature HCs show the kinocilium located in the center (magenta arrows).
Time-lapse video of the wild-type (WT) (myo6b:actb1-Gfp) hair cell (HC) pair shown in Figure 4A.
The HC precursor (yellow dot) divides to form two sibling HCs (pink and blue dots) that first rock and then roll to exchange positions with each other. Zero minute represents the beginning of Rock, immediately after cell division. Rock duration: 0–96 min, Roll duration: 102–186 min.
Time-lapse video of a hair cell (HC) pair in wild-type (WT) (myo6b:actb1-Gfp) neuromast.
The HC precursor (yellow dot) divides to form two sibling HCs (pink and blue dots) that do not roll to exchange positions with each other. Zero minute represents the beginning of Rock immediately after cell division.
Time-lapse video of the emx2 loss of function (LOF);myo6b:actb1-Gfp hair cell (HC) pair shown in Figure 4B.
The HC precursor (yellow dot) divides to form two sibling HCs (pink and blue dots) that rock briefly and then roll to exchange positions with each other. Zero minute represents the beginning of Rock, immediately after cell division. Rock duration: 0–66 min, Roll duration: 72–120 min.
Time-lapse video of the emx2 gain of function (GOF); myo6b:actb1-Gfp hair cell (HC) pair shown in Figure 4C.
The HC precursor (yellow dot) divides to form two sibling HCs (pink and blue dots) that first rock and then roll to exchange positions with each other, which takes longer than the wild-type (WT) in Figure 4—video 1. Zero minute represents the beginning of Rock, immediately after cell division. Rock duration: 0–108 min, Roll duration: 114–222 min.
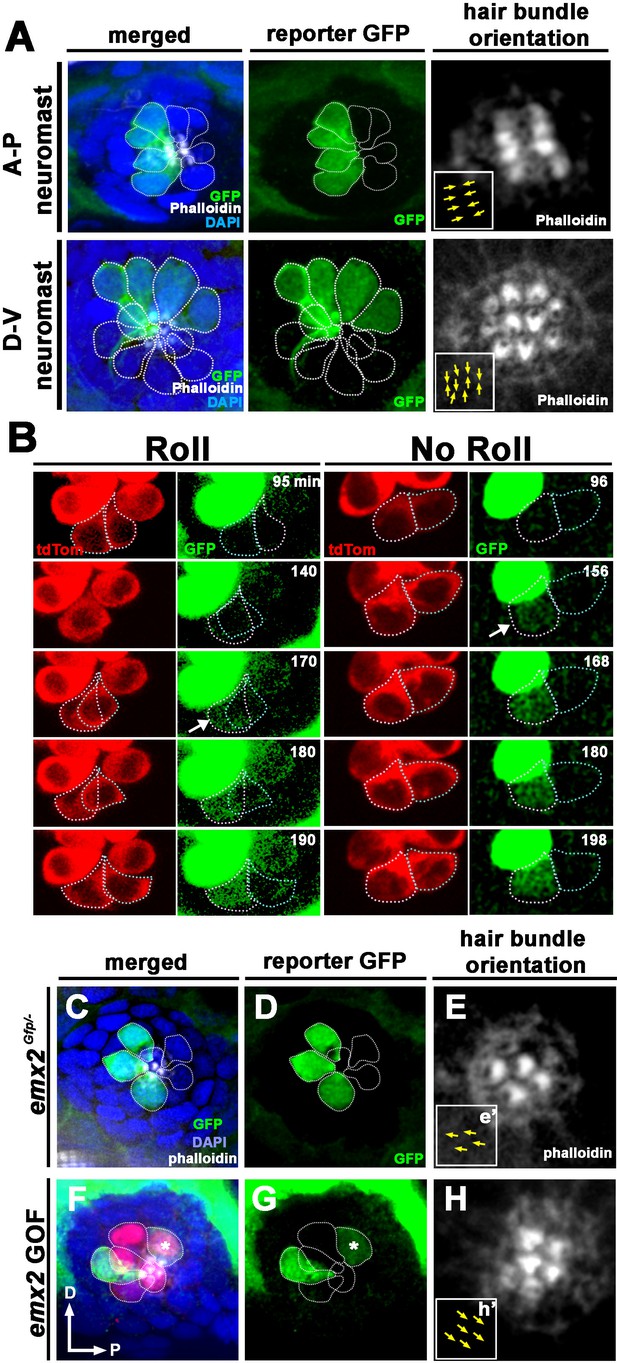
Spatiotemporal reporter activity of emx2 in wild-type (WT) and emx2 mutants.
(A) In an emx2:Gfp zebrafish, Gfp-positive hair cells (HCs) are located in the anterior region of an A-P neuromast (top) and in the dorsal region of a D-V (bottom) neuromast. Phalloidin staining shows the hair bundle orientation (yellow arrows). (B) Time-lapse imaging of nascent HC pairs undergoing Roll or No Roll movement in emx2:Gfp; myo6b:dtTomato neuromasts. Left panel (Roll): A tdTomato-positive, nascent HC pair undergoing roll movement shows detectable Gfp expression in the posterior HC as it rolls into the anterior position (white arrow, 170 min into the Rock and Roll process). Right panel (No Roll): A HC pair that did not roll and Gfp appears in the anterior HC by 156 min into the Rock phase (white arrow). (C–E) The distribution of Gfp-positive HCs in an emx2gfp/- (loss of function [LOF]) neuromast, showing a merged image (C) of DAPI, GFP (D) and phalloidin staining (E). Outline of HCs are dotted. Gfp-positive HCs are located in the anterior region but hair bundles are pointing in P→A direction (yellow arrows in e’). (F–H) The distribution of Gfp-positive HCs in an emx2 gain of function (GOF);emx2Gfp/+ neuromast, showing a merged image (F) of DAPI, GFP (G), mCherry, and phalloidin staining (H). Among the total six pairs of GOF HCs analyzed, only two are Gfp-positive (G), one anterior- and one posterior (asterisk)-located. All hair bundles are in A→P direction (yellow arrows in h’).
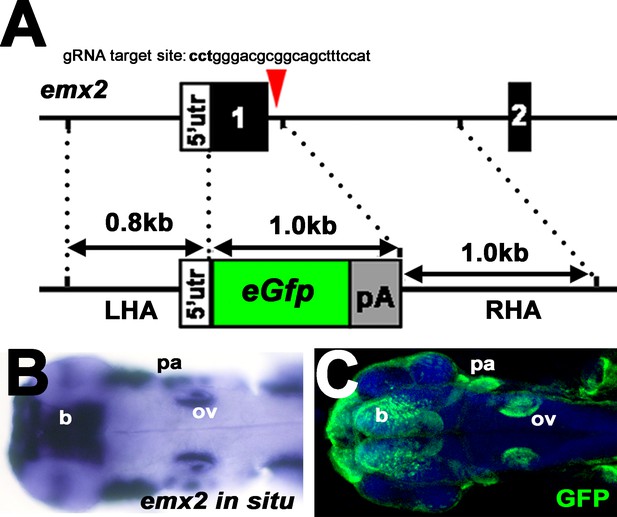
Generation of the emx2:Gfp transgenic line.
(A) Schematic of the emx2 genomic structure and its replacement with donor DNA using CRISPR. Red arrowhead indicates the target position for the guide RNA. The reporter eGfp and a polyA sequence in the targeting vector are flanked by left and right homologous arm (LHA and RHA) of the emx2 allele as indicated with dotted line. The targeting position for the guide RNA is not included in either the LHA or RHA. (B) Hybridization signals of emx2 transcripts in the brain (b), otic vesicles (ov), and pharyngeal arches (pa). (C) Gfp-positive regions in emx2:Gfp reporter larvae are similar to emx2 positive regions indicated by in situ hybridization.
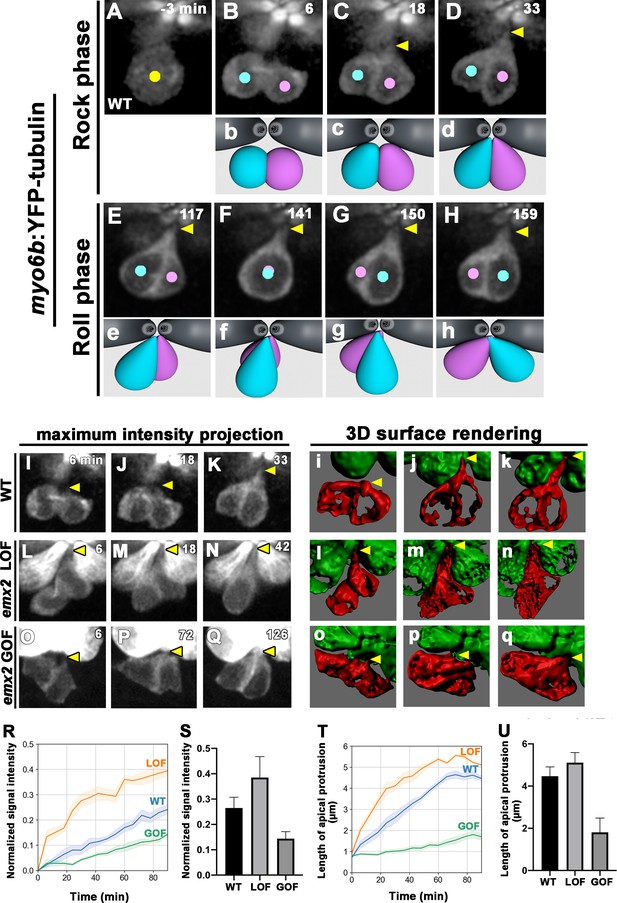
Emx2 affects apical protrusion formation in nascent hair cells (HCs).
(A–H) Time-lapse images of myo6b:YFP-tubulin (wild-type [WT]) transgenic line showing nascent HCs undergoing Rock and Roll. After precursor cell divides (A), sibling HCs form an apical protrusion within 30 min of the Rock phase (B–D, yellow arrowhead, n = 6). When they exchange positions at the Roll phase, they appear to be pivoted at the apex (E–H, yellow arrowheads). Animation of images is shown in respective panels (Rock phase: b–d, Roll phase: e–h). (I–Q) Maximum intensity projection images generated by selected slices along z-axis in WT (I–K, same sample of A–H, n = 3), emx2 loss of function [LOF] (L–N, n = 3) and emx2 gain of function (GOF) (O–Q, n = 3). (i–q) Three-dimensional surface rendering of selected time points of live-imaging of HC pairs (red) in WT (i–k, n = 3), emx2 LOF (l–n, n = 3), and emx2 GOF (o–q, n = 3) neuromasts. Mature HCs are labeled in green. (R) Increases of YFP-tubulin signal intensity at the apical region of nascent HCs of WT (n = 5), emx2 LOF (n = 5), and emx2 GOF (n = 6) during the Rock phase (90 min). The shaded area represents the standard error of the mean (SEM). (S) YFP-tubulin signal intensities of the apical protrusion at the end of Rock phase were significantly different among WT (n = 5), emx2 LOF (n = 5), and emx2 GOF HCs (n = 6, one-way ANOVA, F = 26.97, p<0.0001); post-hoc Dunnett’s multiple comparisons test for WT vs. LOF, p=0.0073, WT vs. GOF, p=0.0052. (T) Increases over time in the length of apical protrusion in WT (n = 5), emx2 LOF (n = 5), and emx2 GOF (n = 5) during the Rock phase (90 min). The shaded area represents the SEM. (U) The length of apical protrusion at the end of the Rock phase in WT (n = 5), emx2 LOF (n = 5), and emx2 GOF (n = 5) were compared. Significance was assessed by using one-way ANOVA, F = 52.15, p<0.0001. Post-hoc Dunnett’s multiple comparisons test for WT vs. LOF is not significant (p=0.148) but significant for WT vs. GOF (p<0.0001). The following figure supplement, source data, videos are available for Figure 6—figure supplement 1. Measurements of signal intensity and length of the apical protrusion using Image J Fiji.
-
Figure 6—source data 1
Quantification of changes in signal intensity of apical protrusion of hair cells (HCs) during the Rock Phase.
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig6-data1-v2.xlsx
-
Figure 6—source data 2
Quantification of signal intensity of apical protrusion at the end of Rock phase among wild-type (WT), emx2 loss of function (LOF), and emx2 gain of function (GOF) hair cells (HCs).
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig6-data2-v2.xlsx
-
Figure 6—source data 3
Quantification of changes in the length of apical protrusion during the Rock Phase.
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig6-data3-v2.xlsx
-
Figure 6—source data 4
Quantification of the length of apical protrusion at the end of Rock phase among wild-type (WT), emx2 loss of function (LOF), and emx2 gain of function (GOF) hair cells (HCs).
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig6-data4-v2.xlsx
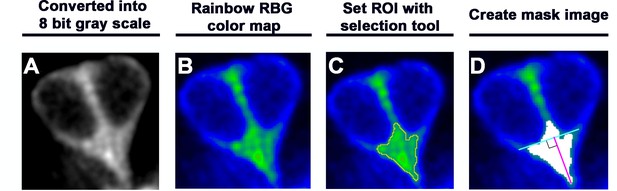
Measurements of signal intensity and length of the apical protrusion using ImageJ Fiji.
(A) A selected maximum intensity projection image was converted into an 8-bit gray scale image. (B) The gray scale image in (A) was converted to Rainbow RBG color mode to visualize the bright green color of the accumulated YFP-tubulin within the apical protrusion. (C) Based on the signal intensity, the region of interest (ROI, yellow outline) was automatically defined by using the wand selection tool of ImageJ Fiji and the intensity measured. (D) The ROI was converted into a mask image (white area). A blue line was drawn across the top of the two hair cell (HC) nuclei, and a pink line joining the tip of the apical protrusion to the blue line at right angle indicates the length of the apical protrusion.
Time-lapse video of the myo6b:yfp-tubulin hair cell (HC) pair in Figure 6A–H.
The nascent HC pair (pink and blue dots) extends a protrusion to the apex and the tip of the protrusion is pivoted at the apical surface of the neuromast. The yellow dot indicates the HC precursor.
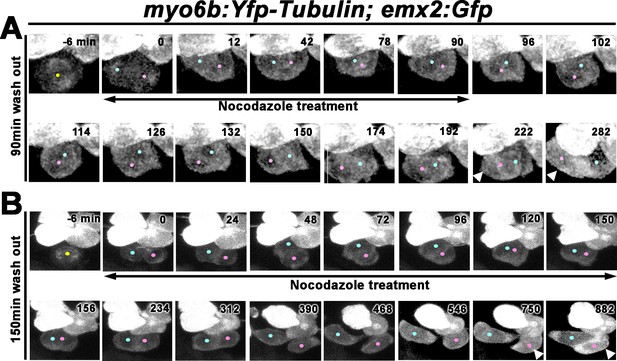
Nocodazole disrupts Rock and Roll and positional acquisition of hair cells (HCs).
(A) Nascent sibling HCs in myo6b:Yfp-tubulin; emx2:Gfp larvae were treated with nocodazole for 90 min immediately after the HC precursor was observed to divide under live-imaging (0 min). After nocodazole removal, the nascent HC pair started to roll and they exchanged positions (96–174 min). The HC located at the anterior (pink dot) showed stronger Gfp signal due to the presence of emx2:Gfp and Yfp-tubulin alleles (222–282 min, arrowheads) than the emx2:Gfp-negative HC at the posterior (blue dot), which only expresses Yfp-tubulin. (B) Nascent HCs were treated with nocodazole for 150 min after HC precursor divided (0–150 min). After nocodazole removal, HCs failed to exchange their positions (156–312 min), which resulted in the mislocation of the emx2:Gfp-positive HC to the posterior (arrowheads, 618–882 min, pink dots).
Time-lapse video of nocodazole treatment for 90 min of the myo6b:Yfp-Tubulin; emx2:Gfp hair cell (HC) pair shown in Figure 7A.
Nocodazole was added at 0 min immediately after the HC precursor (−6 min, yellow dot) divided and they were incubated with Nocodazole for 90 min (0–90 min). After removal of nocodazole, the two HCs initiated roll movements to exchange positions with each other (96–174 min). emx2 reporter positive HC (pink dot) was evident as the brighter GFP signal expressing both YFP-tubulin and GFP driven by the emx2 promoter versus the emx2 reporter negative HC (blue dot) expressing only YFP-tubulin (222–282 min).
Time-lapse video of nocodazole treatment for 150 min of the myo6b:Yfp-Tubulin; emx2:Gfp hair cell (HC) pair shown in Figure 7B.
Nocodazole was added at 0 min immediately after the HC precursor (−6 min, yellow dot) divided and they were incubated with nocodazole for 150 min (0–150 min). After nocodazole removal, the HC pair did not undergo the Roll phase to exchange their positions. As a result, the brighter GFP-positive, Emx2 HC (pink dot) was mispositioned in the posterior position, whereas the emx2 reporter-negative HC (blue dot) was located in the anterior position (750–882 min).
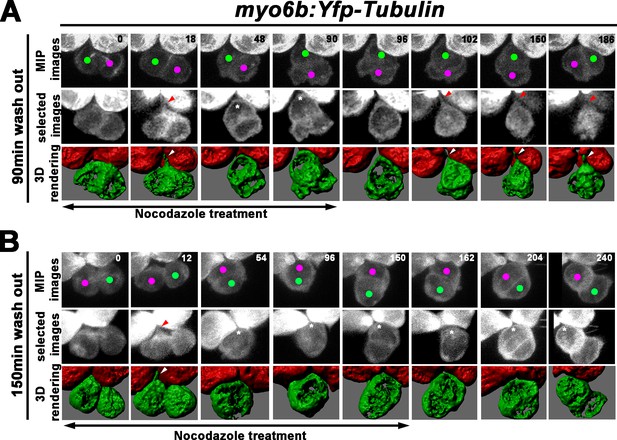
Formation of the apical protrusion is correlated with the Roll movement and hair cell (HC) positioning.
(A) Maximum intensity projection and 3-D rendering of selected images of nascent sibling HCs in myo6b:Yfp-tubulin larvae (magenta and green dots) that was treated with nocodazole for 90 min immediately after the precursor divided. The apical protrusion that normally forms within 18 min was still evident initially (18 min; red and white arrowheads) but disappeared quickly between 30 and 36 min (asterisks, 48 min). After nocodazole removal, the apical protrusion reappeared after 12 min (102 min, red and white arrowheads) and the two nascent HCs rolled to their respective positions after 84 min (186 min). (B) A nascent HC pair (red and green dots) that was treated with nocodazole for 150 min immediately after the precursor divided. Similar to 90 min treatments, the apical protrusion was evident between 12 and 18 min (12 min, red and white arrowheads) after HC precursor divided (0 min) but disappeared shortly within 30–36 min (54 min, asterisk). However, after nocodazole removal at 150 min, sibling HCs did not form an apical protrusion and exchange positions (162–240 min, asterisks).
Time-lapse video of nocodazole treatment for 90 min of a myo6b:Yfp-Tubulin hair cell (HC) pair shown in Figure 8A.
Nocodazole was added at 0 min immediately after the HC precursor (−6 min, yellow dot) divided and they were incubated with nocodazole for 90 min (0–90 min). After nocodazole was removed, the HC pair underwent the Roll phase to exchange positions with each other (96–186 min).
Time-lapse video of nocodazole treatment for 150 min of a myo6b:Yfp-Tubulin hair cell (HC) pair shown in Figure 8B.
Nocodazole was added at 0 min immediately after the HC precursor (−6 min, yellow dot) divided and they were incubated with Nocodazole for 90 min (0–150 min). After nocodazole removal, the HC pair did not undergo the Roll phase to exchange positions with each other (162–240 min).
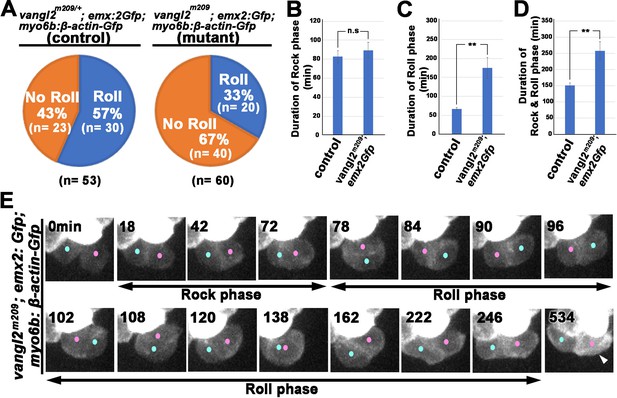
Rock and Roll is affected in the core planar cell polarity (cPCP) mutant, trilobite.
(A) Frequencies of Roll and No Roll of nascent hair cell (HC) pairs in control (vangl2m209/+; emx2gfp/+) and mutant (vangl2m209/m209; emx2gfp/+) larvae. Significance was assessed by using chi-squared test with a 2 × 2 contingency table (X2[(df = 1)]=6.1774, p<0.013, source data 1). (B–D) Duration of Rock (B), Roll (C), and Rock and Roll (D) of sibling HCs that underwent Rock and Roll in control (n = 28 from seven larvae) and vangl2m209/m209; emx2gfp/+ (n = 19 from seven larvae) larvae. Significance was assessed by using Student's t-test (*p<0.05, **p<0.001, source data 2). The error bars represent SEM. (E) Time-lapse images of a HC pair in trilobite (vangl2m209/m209); emx2gfp/+ mutant, which undergoes a prolong Roll phase from 78 to 246 min with several rolls (first roll: 78–96 min, second roll: 102–120 min, third partial roll: 138–162 min) and resulted in the emx2:Gfp-positive HC located to the posterior. The following figure supplement, source data, and video are available for Figure 9—figure supplement 1. Formation of apical protrusion in nascent HCs of trilobite.
-
Figure 9—source data 1
Frequencies of Roll and No Roll movements in vangl2 mutants.
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig9-data1-v2.xlsx
-
Figure 9—source data 2
Comparison of duration of Rock, Roll, and R and R between controls and vangl2 mutants.
- https://cdn.elifesciences.org/articles/60432/elife-60432-fig9-data2-v2.xlsx
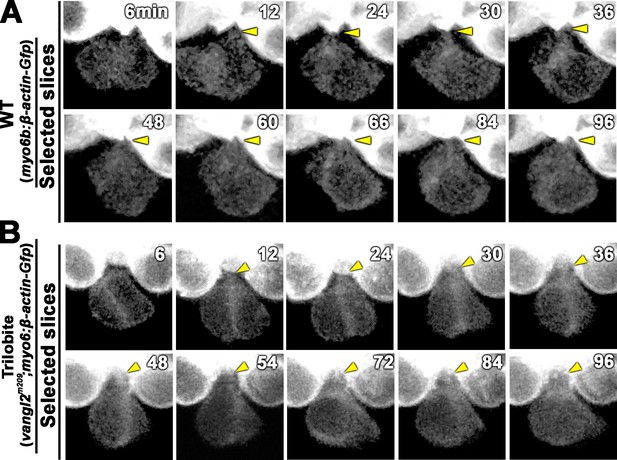
Formation of apical protrusion in nascent hair cells (HCs) of trilobite.
Time-lapse images of a nascent HC pair during Rock phase in (A) wild-type (WT) (myo6:β-actin-Gfp) and (B) trilobite larvae. The apical protrusion (yellow arrowhead) was evident by 12 min after the HC precursor divided in both WT and mutant.
Time-lapse video of a vangl2m209; emx2: Gfp; myo6b: β-actin-Gfp hair cell (HC) pair during Rock and Roll in Figure 9E.
The HC precursor (yellow dot) divides to form two sibling HCs (pink and blue dots), which underwent the Rock movement (0–72 min) and then rolled to exchange positions with each other (78–96 min). After the first exchange, the sibling HCs underwent another exchange (102–246 min). By the end of the Roll phase, the emx2 reporter-positive HC (pink dot) was mispositioned at the posterior position, as evident by its brighter GFP signal, compared to the emx2 reporter-negative HC (blue dot) located in the anterior position (534 min).
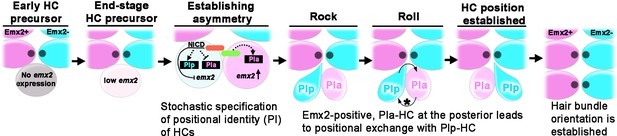
Model of positional acquisition of nascent hair cells (HCs) in the neuromast.
Unlike scenarios 1 and 2 illustrated in Figure 1C, our results show a model in which an end-stage HC precursor before division expresses low levels of emx2 (light pink). After the precursor divided, Notch signaling mediated through the Notch ligand (green color) and Notch1a receptor (red color) generates asymmetry between the two sibling HCs. This asymmetry results in stochastic specification of anterior (PIa) and posterior (PIp) positional identity (PI) to sibling HCs, which includes regulation of emx2 expression. Thus, an emx2-positive, Pla-HC formed at the posterior position undergoes positional exchange with its sibling Plp HC, whereaswhen an emx2-positive, Pla-HC formed in the anterior position, the two sibling HCs do not exchange positions. In addition to the delay in apical protrusion mediated by Emx2, the Rock and Roll is also regulated by the intercellular core planar cell polarity (cPCP) pathway (asterisk).
Tables
Quantification of emx2:Gfp positive cells in wild-type (WT) and emx2 mutant neuromasts.
| Genotype | Fish | Neuromast | Total hair cells (HCs) | Gfp+ cells | Anterior Gfp+ HCs | Posterior Gfp+ HCs | Mislocated Gfp+ HCs |
|---|---|---|---|---|---|---|---|
| WT | 7 | 20 | 160 | 78 (49%) | 78/78* (100%) | 0/78 (0%) | 0/78 (0%) |
| emx2Gfp/-(loss of function [LOF]) | 3 | 10 | 70 | 33 (47%) | 32/33 (97%) | 1/33 (3%) | 1/33 (3%) |
| emx2GOF(F1); emx2Gfp/+ | 10 | 18 | 123 | 30 (24%) | 22/30* (73%) | 8/30 (27%) | 8/30 (27%) |
| emx2GOF(F3); emx2Gfp/+ | 29 | 29 | 246 | 100 (41%) | 98/100* (98%) | 2/100 (2%) | 2/100 (2%) |
-
*All Gfp+ cells are also positive for Emx2 immunostaining.
Summary of emx2:Gfp-positive hair cell (HC) positions in neuromasts after Rock and Roll.
| | Positions of emx2:Gfp HCs | |||||||
|---|---|---|---|---|---|---|---|---|
| Roll | No Roll | |||||||
| Anterior | Posterior | Dorsal | Ventral | Anterior | Posterior | Dorsal | Ventral | |
| Control* (%) | 30/30 (100%) | 0/30 (0%) | 0/30 (0%) | 0/30 (0%) | 19/19 (100%) | 0/19 (0%) | 0/19 (0%) | 0/19 (0%) |
| vangl2m209 ;emx2:Gfp; myo6:βactin-Gfp** (%) | 5/17 (29.4%) | 6/17 (35.3%) | 4/17 (23.5%) | 2/17 (11.7%) | 13/39 (33.3%) | 13/39 (33.3%) | 7/39 (17.9%) | 6/39 (15.4%) |
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Danio rerio) | Tg(myo6b:actb1-GFP) | PMC3426295 | RRID:ZFIN_ZDB-GENO-120926-20 | Kindt group (NIDCD, NIH) |
| Genetic reagent (Danio rerio) | Tg(myo6b:emx2-p2a-nls-mCherry) | PMC5388538 | RRID:ZFIN_ZDB-GENO-170619-3 | Wu group (NIDCD, NIH) |
| Genetic reagent (Danio rerio) | Tg(myo6b:YFP-Tubulin) | N.A | N.A. | Kindt group (NIDCD, NIH) |
| Genetic reagent (Danio rerio) | Tg(myo6b: tdTomato) | N.A | RRID:ZFIN_ZDB-TGCONSTRCT-160316-7 | Kindt group (NIDCD, NIH) |
| Genetic reagent (Danio rerio) | emx2 ko | PMC5388538 | RRID:ZFIN_ZDB-ALT-170606-5 | Kindt group (NIDCD, NIH) |
| Genetic reagent (Danio rerio) | Emx2:GFP | this paper | N.A. | Wu group (NIDCD, NIH) |
| Genetic reagent (Danio rerio) | Tg(myo6b: RiboTag) | PMC5939014 | PRID:ZFIN_ZDB-ALT-180129–10 | Kindt group (NIDCD, NIH) |
| Genetic reagent (Danio rerio) | Tg(myo6b:actb1-GFP) | PMC3426295 | RRID:ZFIN_ZDB-GENO-120926-20 | Kindt group (NIDCD, NIH) |
| Sequence-based reagent | tracrRNA | IDT | Cat# 1072533 | 25 μM |
| Sequence-based reagent | emx2_guide RNA 5’- atggaaagctgccgcgtcccagg-3’ | IDT | this paper | 50 μM |
| Sequence-based reagent | tracrRNA | IDT | Cat# 1072533 | 25 μM |
| Recombinant DNA | pT3TS-nls-zCas9-nls | Addgene | Cat# 46757 | |
| Recombinant DNA | pKHR4 | PMC4806538 | ||
| Recombinant DNA | pKHR4-LHA-emx2-RHA | this paper | this paper | Wu group (NIDCD, NIH) |
| Antibodies | Rabbit anti-Emx2 | Trans Genic (fukuoka, japan) | KO609 | |
| Antibodies | Mouse anti-GFP | Thermo Fisher Scientific | A11120 | |
| Antibodies | Rabbit anti-GFP | Abcam | Ab6556 | |
| Antibodies | Alexa Fluor 647 phalloidin | Themo Fisher Scientific | A22287 | |
| Chemical compound, drug | Nocodazole | Sigma-Aldrich | M1404 | 10 μg/ml |
| Recombinant proteins | I-SceI | NEB | R0694S | |
| Software and algorithms | ImageJ | NIH | https://imagej.net/Fiji/Downloads | |
| Software and algorithms | ImageJ Fiji | NIH | https://imagej.net/Fiji/Downloads | |
| Software and algorithms | PoorMan3Dreg | http://sybil.ece.ucsb.edu/pages/software.html | ||
| Software and algorithms | MTrack J | Meijering et al., 2012 | https://imagescience.org/meijering/software/mtrackj/ | |
| Software and algorithms | Fluorender 2.16 | University of Utah, SCI | http://www.sci.utah.edu/software/fluorender.html |





