Qki regulates myelinogenesis through Srebp2-dependent cholesterol biosynthesis
Figures
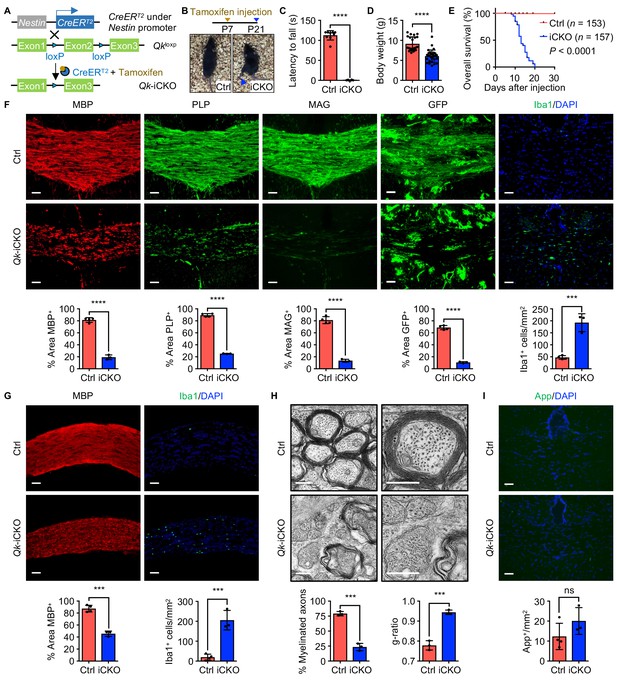
Deletion of Qk in mouse neural stem cells leads to hypomyelination in the central nervous system.
(A) Schema of the generation of Qk-Nestin-iCKO mice. (B) Representative images of severe hind limb paralysis in Qk-Nestin-iCKO mice 2 weeks after tamoxifen injection. Ctrl: control. (C) Latency of mice falling off the rotarod at a constant speed (5 rpm). n = 6 mice in the Qk-Nestin-iCKO group; n = 9 mice in the control group. (D) Body weights of Qk-Nestin-iCKO mice (n = 29) and control mice (n = 22) 12 days after tamoxifen injection. (E) Kaplan–Meier curves of and log-rank test results for overall survival in Qk-Nestin-iCKO mice (n = 157) and control mice (n = 153). (F) Representative images of and quantification of immunofluorescent staining of MBP, PLP, MAG, GFP, and Iba1 in the corpus callosum tissues in Qk-Nestin-iCKO mice (n = 3) and control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. (G) Representative images of and quantification of immunofluorescent staining of MBP and Iba1 in the optic nerves in Qk-Nestin-iCKO mice (n = 3) and control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. (H) Representative electron micrographs of the optic nerves in Qk-Nestin-iCKO mice and control mice with quantification of the percentage of myelinated axons and g-ratios 2 weeks after tamoxifen injection (n = 3 mice/group). Scale bars, 500 nm. (I) Representative images and quantification of immunofluorescent staining of amyloid precursor protein (App) in the corpus callosum tissues in Qk-Nestin-iCKO mice (n = 3) and control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. ***p<0.001; ****p<0.0001; ns: not significant.
-
Figure 1—source data 1
Exact p-values for statistical analysis.
- https://cdn.elifesciences.org/articles/60467/elife-60467-fig1-data1-v2.xlsx
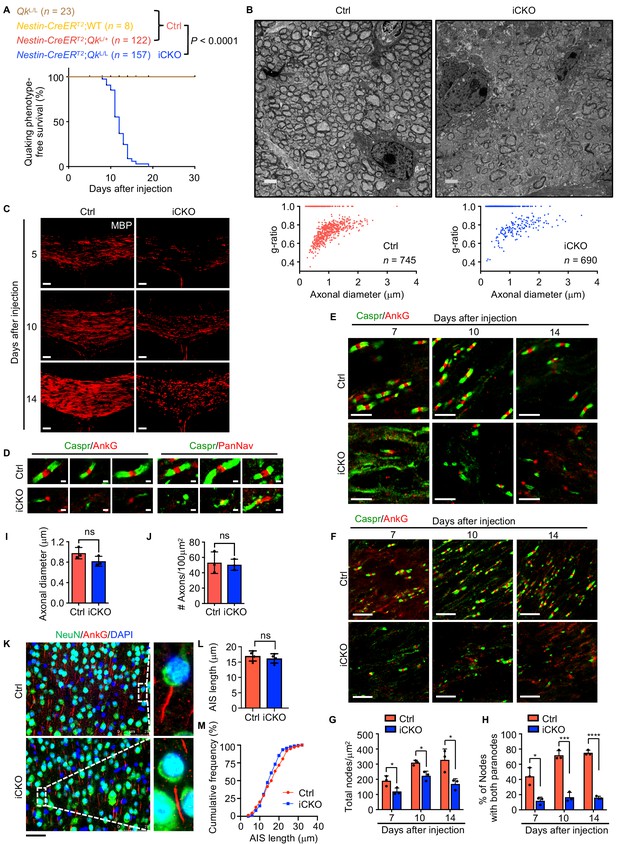
Deletion of Qk in mouse neural stem cells leads to hypomyelination in the central nervous system.
(A) Kaplan–Meier curves of and log-rank test results for quaking phenotype-free survival of QkL/L mice (n = 23), Nestin-CreERT2;WT mice (n = 8), Nestin-CreERT2;QkL/+ mice (n = 122), and Nestin-CreERT2;QkL/L mice (n = 157). (B) Representative electron micrographs of and quantification of the g-ratio in the optic nerves in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection (n = 3 mice/group). Scale bars, 2 μm. (C) Representative images of immunofluorescent staining of MBP in the corpus callosum tissues in Qk-Nestin-iCKO mice and control mice 5, 10, and 14 days after tamoxifen injection (n = 3 mice/group). Scale bar, 50 μm. (D) Representative images of immunofluorescent staining of Caspr, AnkG, and PanNav in the optic nerves of Qk-Nestin-iCKO mice and controls 2 weeks after tamoxifen injection (n = 3 mice/group). Scale bars, 1 μm. (E, F) Representative images of immunofluorescent staining of Caspr and AnkG in the optic nerves of Qk-Nestin-iCKO mice and control mice 7, 10, and 14 days after tamoxifen injection (n = 3 mice/group). Scale bar, (E) 5 μm, (F) 10 μm. (G, H) Quantification of total number of nodes (G) and % of nodes with both paranodes (H) in the optic nerves of Qk-Nestin-iCKO mice and control mice 7, 10, and 14 days after tamoxifen injection (n = 3 mice/group). Data are shown as mean ± s.d. and were analyzed using Student's t test. *p<0.05; ***p<0.001; ****p<0.0001. (I, J) Quantification of axonal diameter (I) and density of axon (J) in the mice in (B). Data are shown as mean ± s.d. and were analyzed using Student's t test. ns: not significant. (K–M) Representative images (K) of and quantification of axon initial segment length (L) and cumulative frequency (M) in the cortex tissues in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection (n = 3 mice/group). Scale bars, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. ns: not significant.
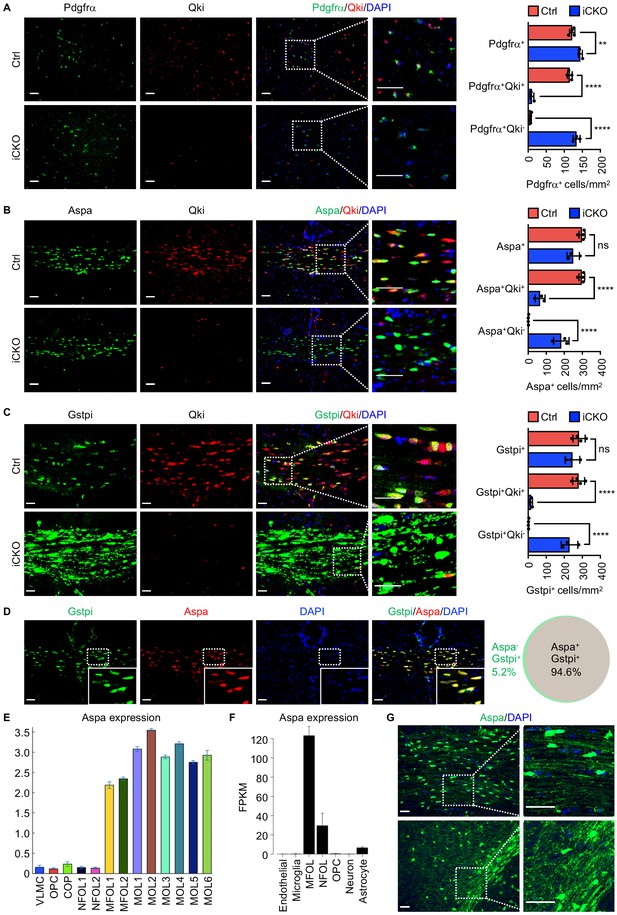
Qki loss in neural stem cells does not impair formation of oligodendrocyte precursor cells or Aspa+Gstpi+myelinating oligodendrocytes.
(A–C) Representative images of and quantification of immunofluorescent staining of Pdgfrα-Qki (A), Aspa-Qki (B), and Gstpi-Qki (C) in the corpus callosum tissues in Qk-Nestin-iCKO mice (n = 3) and control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. (D) Representative images of immunofluorescent staining of Gstpi and Aspa in the corpus callosum tissues in WT mice at 3 weeks of age (n = 4 mice/group). Scale bars, 50 μm. The Venn diagram depicts the overlap of Aspa+ and Gstpi+ oligodendrocytes. (E, F) RNA-seq expression data for Aspa transcripts from the databases of Gonçalo Castelo-Branco’s laboratory (E) and Ben A. Barres’s and Jiaqian Wu’s laboratory (F). VLMC: vascular and leptomeningeal cells; COP: differentiation-committed oligodendrocyte precursors; NFOL: newly formed oligodendrocytes; MFOL: myelin-forming oligodendrocytes; MOL: mature oligodendrocytes. (G) Representative images of immunofluorescent staining of Aspa in the corpus callosum regions in WT mice at 3 weeks of age. Scale bars, 50 μm. Data are shown as mean ± s.d. and were analyzed using one-way ANOVA with Tukey’s multiple comparisons test. **p<0.01; ****p<0.0001; ns: not significant.
-
Figure 2—source data 1
Exact p-values for statistical analysis.
- https://cdn.elifesciences.org/articles/60467/elife-60467-fig2-data1-v2.xlsx
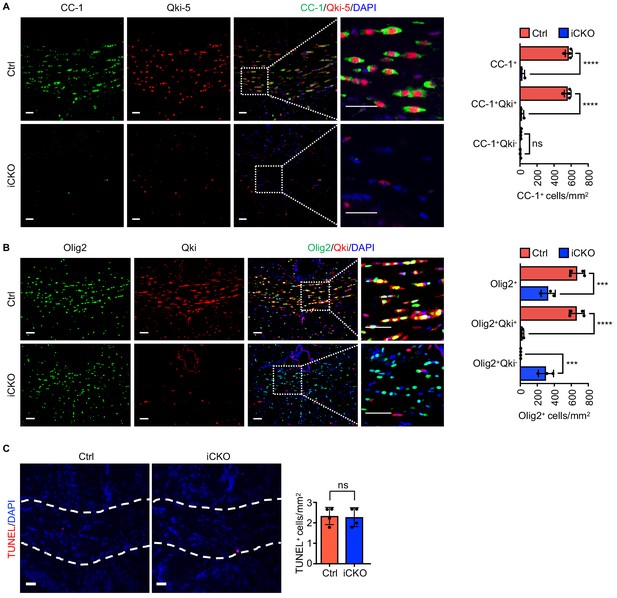
Qki loss in neural stem cells impairs the differentiation of Olig2+AspaGstpi- oligodendroglial lineage cells.
(A, B) Representative images of and quantification of immunofluorescent staining of CC-1–Qki-5 (A) and Olig2-Qki (B) in the corpus callosum tissues in Qk-Nestin-iCKO mice (n = 3) and control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. Data are shown as mean ± s.d. and were analyzed using one-way ANOVA with Tukey’s multiple comparisons test. ***p<0.001; ****p<0.0001; ns: not significant. (C) Representative images of and quantification of TUNEL immunofluorescent staining in the corpus callosum tissues in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection (n = 4 mice/group). Scale bar, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. ns: not significant.
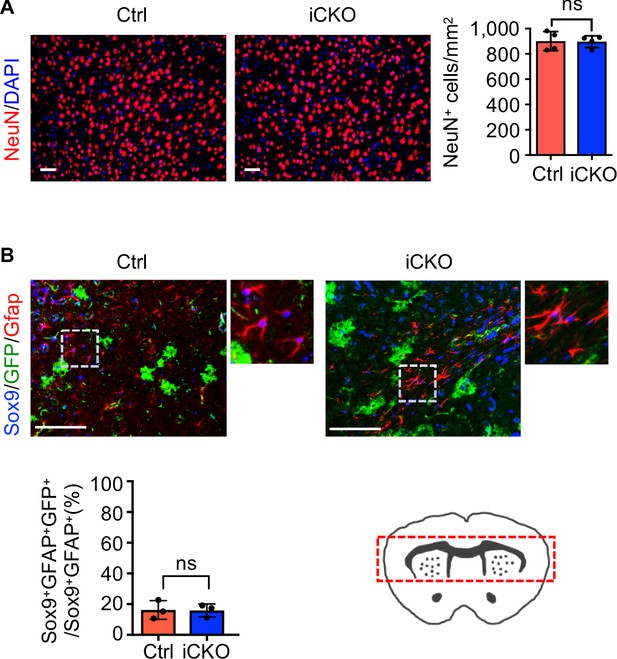
Deletion of Qk in neural stem cells has no effects on neuronal or astrocytic populations.
(A) Representative images of and quantification of immunofluorescent staining of NeuN in the cortex tissues in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection (n = 4 mice/group). Scale bars, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. ns: not significant. (B) Representative images of and quantification of immunofluorescent staining of Sox9, GFP, and Gfap in the brain region within the red dotted box in Qk-Nestin-iCKO and control mice 2 weeks after tamoxifen injection (n = 3 mice/group). Scale bars, 100 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. ns: not significant.
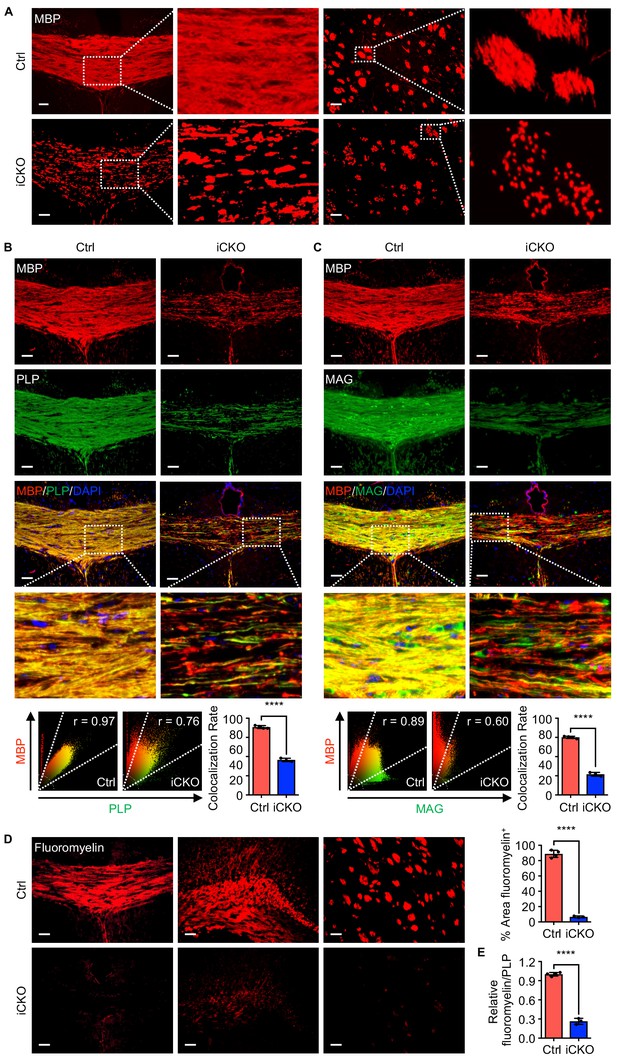
Qki loss leads to defective myelin membrane assembly.
(A) Representative images of immunofluorescent staining of MBP in the corpus callosum tissues in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection. (B, C) Representative images of and quantification of the co-localization rates of immunofluorescent staining of MBP-PLP (B) and MBP-MAG (C) in the corpus callosum tissues in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection. (D) Representative images of and quantification of staining of FluoroMyelin in the corpus callosum tissues in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection. (E) Quantification of the relative ratio of FluoroMyelin to PLP in the corpus callosum tissues in Qk-Nestin-iCKO mice (n = 3) and control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. The r values in the scatter plots (B, C) were calculated using Pearson’s correlation. ****p<0.0001.
-
Figure 3—source data 1
Exact p-values for statistical analysis.
- https://cdn.elifesciences.org/articles/60467/elife-60467-fig3-data1-v2.xlsx
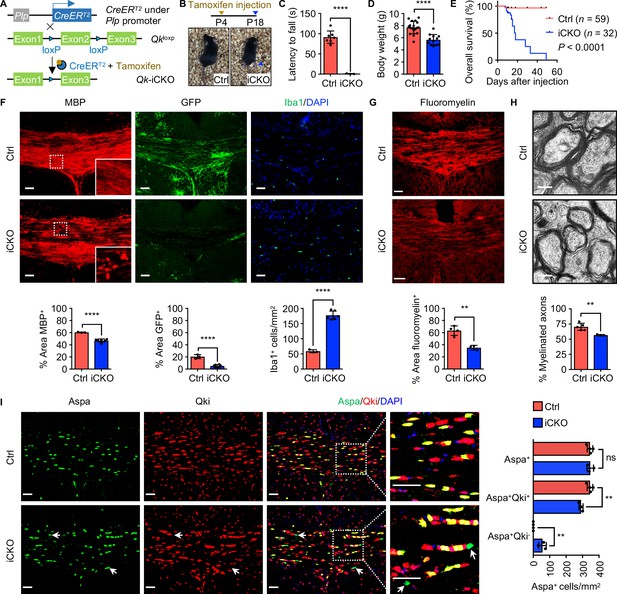
Qk deletion in oligodendrocyte precursor cells leads to defective myelinogenesis without impairing differentiation of Aspa+ myelinating oligodendrocytes.
(A) Schema of the generation of Qk-Plp-iCKO mice. (B) Representative images of severe hind limb paresis in Qk-Plp-iCKO mice 2 weeks after tamoxifen injection. (C) Latency of mice falling off the rotarod at a constant speed (5 rpm). n = 3 mice in the Qk-Plp-iCKO group; n = 7 mice in the control group. (D) Body weights of Qk-Plp-iCKO mice (n = 12) and control mice (n = 18) 2 weeks after tamoxifen injection. (E) Kaplan–Meier curves of and log-rank test results for overall survival in Qk-Plp-iCKO mice (n = 32) and control mice (n = 59). (F) Representative images of and quantification of immunofluorescent staining of MBP, GFP, and Iba1 in the corpus callosum tissues in Qk-Plp-iCKO mice (n = 6) and control mice (n = 3) 2 weeks after tamoxifen injection. Scale bars, 50 μm. (G) Representative images of and quantification of staining of FluoroMyelin in the corpus callosum tissues in Qk-Plp-iCKO mice (n = 3) and control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. (H) Representative electron micrographs of and quantification of the percentage of myelinated axons in the optic nerves in Qk-Plp-iCKO mice (n = 3) and control mice (n = 5) 2 weeks after tamoxifen injection. Scale bars, 500 nm. (I) Representative images of and quantification of immunofluorescent staining of Aspa and Qki in the corpus callosum tissues in Qk-Plp-iCKO mice (n = 3) and control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test (C,D, F–H) or one-way ANOVA with Tukey’s multiple comparisons test (I). **p<0.01; ****p<0.0001; ns: not significant.
-
Figure 4—source data 1
Exact p-values for statistical analysis.
- https://cdn.elifesciences.org/articles/60467/elife-60467-fig4-data1-v2.xlsx
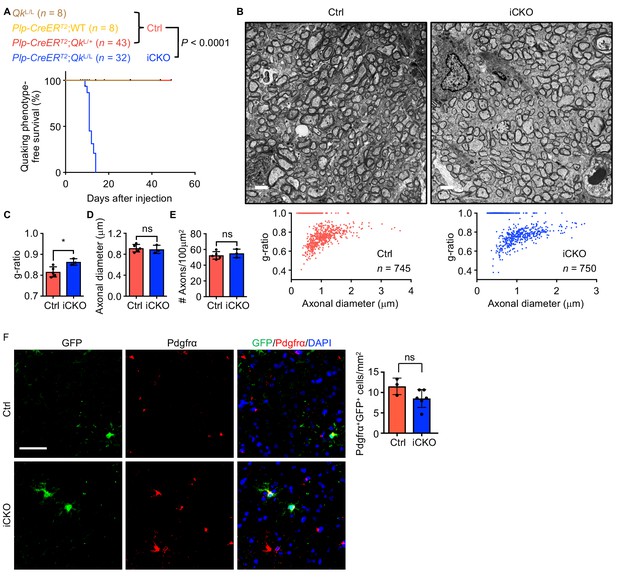
Deletion of Qk in mouse oligodendrocyte precursor cells results in hypomyelination in the central nervous system.
(A) Kaplan–Meier curves of and log-rank test results for quaking phenotype-free survival of QkL/L mice (n = 8), Plp-CreERT2;WT mice (n = 8), Plp-CreERT2;QkL/+ mice (n = 43), and Plp-CreERT2;QkL/L mice (n = 32). (B) Representative electron micrographs of and quantification of the g-ratio in the optic nerves in Qk-Plp-iCKO mice (n = 3) and control mice (n = 5) 2 weeks after tamoxifen injection. Scale bars, 2 μm. (C–E) Quantification of the g-ratio (C) axonal diameter (D) and density of axon (E) in the mice in (B). Data are shown as mean ± s.d. and were analyzed using Student's t test. *p<0.05; ns: not significant. (F) Representative images of and quantification of immunofluorescent staining of GFP and Pdgfrα in the cortex tissues in Qk-Plp-iCKO;mTmG mice (n = 6) and control Plp-CreERT2;mTmG mice (n = 3) 2 weeks after tamoxifen injection. Scale bar, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. ns: not significant.
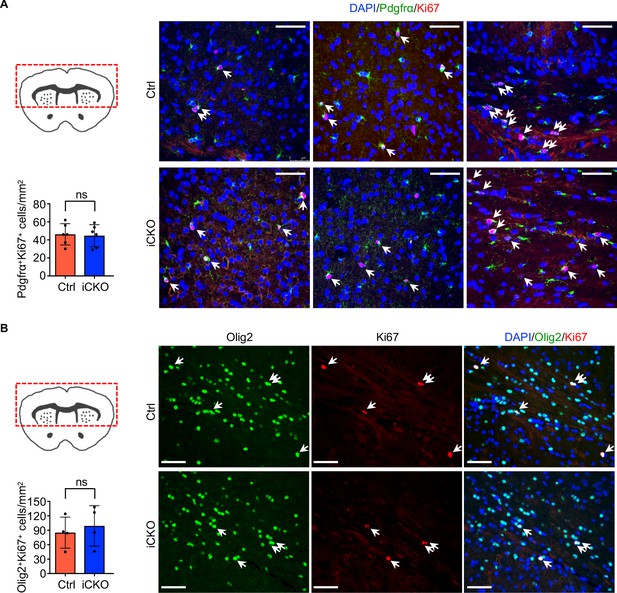
Deletion of Qk does not alter proliferation of oligodendrocyte precursor cells and oligodendroglial lineage cells.
(A, B) Representative images of and quantification of immunofluorescent staining of Pdgfrα and Ki67 (A) and Olig2 and Ki67 (B) in the brain region within the red dotted box in Qk-Plp-iCKO;mTmG mice (n = 6 in A and n = 4 in B) and control Plp-CreERT2;mTmG mice (n = 6 in A and n = 4 in B) 2 weeks after tamoxifen injection. Scale bar, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. ns: not significant. Arrow indicates Pdgfrα+Ki67+ cells (A) and Olig2+Ki67+ cells (B).
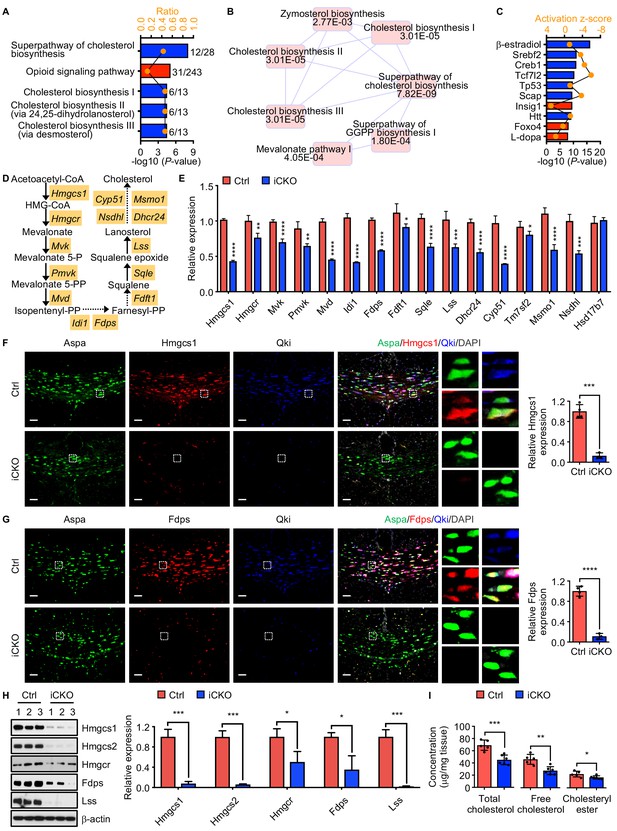
Qki regulates transcription of the genes involved in myelin cholesterol biosynthesis.
(A) Bar graph showing the five canonical pathways most affected by Qki on the basis of differentially expressed genes in Qk-Plp-iCKO mice and control mice (n = 2 mice/group). Blue and red indicate pathways whose activity decreased or increased, respectively, in Qk-Plp-iCKO mice. (B) Overlapping canonical pathway networks for the top 20 canonical pathways with a minimum of three common molecules in different pathways. GGPP: geranylgeranyl diphosphate. (C) Bar graph showing the 10 upstream regulators most enriched in Qk-Plp-iCKO mice. (D) Schema of the cholesterol biosynthesis pathway. (E) Quantification of expression of representative enzymes involved in cholesterol biosynthesis in the corpus callosum tissues in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection according to real-time qPCR (n = 4 mice/group). (F, G) Representative images of and quantification of immunofluorescent staining of Hmgcs1 (F) and Fdps (G) in Aspa+Qki- oligodendrocytes in the corpus callosum of Qk-Nestin-iCKO mice (n = 3) and Aspa+Qki+ oligodendrocytes in the corpus callosum of control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. (H) Immunoblots of and quantification of the levels of expression of the representative enzymes involved in cholesterol biosynthesis in the corpus callosum tissues in Qk-Nestin-iCKO mice and control mice 2 weeks after tamoxifen injection (n = 3 mice/group). (I) Quantification of the cholesterol levels in the corpus callosum tissues in Qk-Nestin-iCKO mice (n = 6) and control mice (n = 5) 2 weeks after tamoxifen injection. Data are shown as mean ± s.d. and were analyzed using Student's t test. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001.
-
Figure 5—source data 1
Exact p-values for statistical analysis.
- https://cdn.elifesciences.org/articles/60467/elife-60467-fig5-data1-v2.xlsx
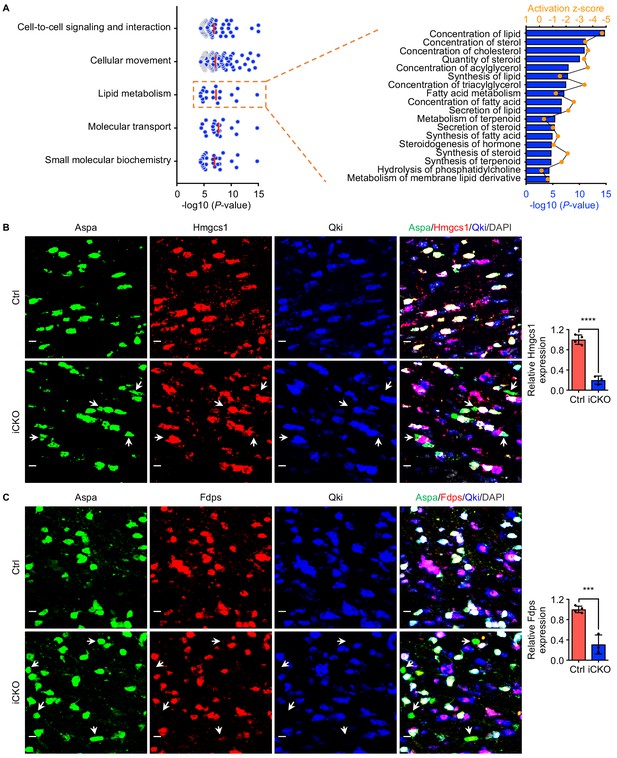
Qki regulates transcription of the genes involved in myelin cholesterol biosynthesis.
(A) Graph of the five enriched molecular and cellular functions most affected by Qki on the basis of differentially expressed genes in Rosa26-CreERT2;QkL/L mice (n = 3 mice) and control mice (n = 5 mice), shown in the left panel, with the individual annotation of lipid metabolism shown in the right panel. (B, C) Representative images of and quantification of immunofluorescent staining of Hmgcs1 (B) and Fdps (C) in Aspa+Qki- oligodendrocytes in the corpus callosum of Qk-Plp-iCKO mice (n = 3) and Aspa+Qki+ oligodendrocytes in the corpus callosum of control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. Data are shown as mean ± s.d. and were analyzed using Student's t test. ***p<0.001; ****p<0.0001.
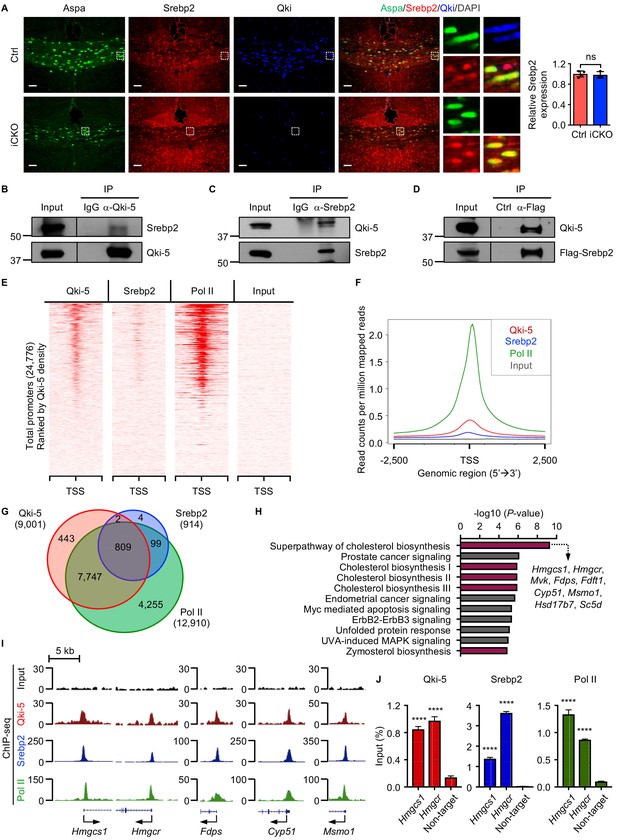
Qki-5 interacts with Srebp2 to regulate transcription of the genes involved in cholesterol biosynthesis.
(A) Representative images of and quantification of immunofluorescent staining of Srebp2 in Aspa+Qki- oligodendrocytes in Qk-Nestin-iCKO mice (n = 3) and Aspa+Qki+ oligodendrocytes in control mice (n = 4) 2 weeks after tamoxifen injection. Scale bars, 50 μm. (B) Results of co-immunoprecipitation (co-IP) using an anti–Qki-5 antibody with differentiated oligodendrocytes followed by detection of Srebp2 via immunoblotting. (C) Results of co-IP using an anti-Srebp2 antibody with differentiated oligodendrocytes followed by detection of Qki-5 via immunoblotting. (D) Results of co-IP using an anti-Flag antibody with differentiated oligodendrocytes having ectopic expression of Flag-Srebp2 followed by detection of Qki-5 via immunoblotting. (E, F) ChIP-seq density heat maps (E) and average genome-wide occupancies (F) of Qki-5, Srebp2, and Pol II in differentiated oligodendrocytes. Regions within 2.5 kb of the transcriptional start site (TSS) are included. All events are rank-ordered from high to low Qki-5 occupancy. (G) Venn diagram of the overlap of Qki-5-, Srebp2-, and Pol II-binding events in the promoter regions in differentiated oligodendrocytes. Promoters are defined as TSS ±2 kb. (H) Canonical pathway analysis of Qki-5-, Srebp2-, and Pol II-co-occupied genes in differentiated oligodendrocytes. Cellular pathways involved in cholesterol biosynthesis are labeled in dark pink. (I) Representative ChIP-seq binding events of Qki-5, Srebp2, and Pol II in the promoter regions of the genes involved in cholesterol biosynthesis. y-axis: normalized reads. (J) ChIP-qPCR results showing the recruitment of Qki-5, Srebp2, and Pol II to the promoter regions of Hmgcs1 and Hmgcr in differentiated oligodendrocytes. Data are shown as mean ± s.d. and were analyzed using Student’s t test. ****p<0.0001; ns: not significant.
-
Figure 6—source data 1
Exact p-values for statistical analysis.
- https://cdn.elifesciences.org/articles/60467/elife-60467-fig6-data1-v2.xlsx
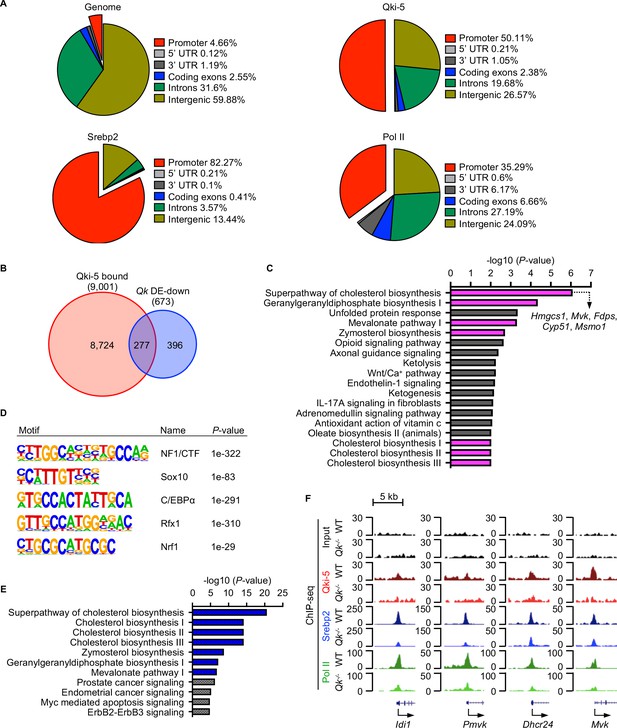
Qki-5 cooperates with Srebp2 to regulate transcription of the genes involved in cholesterol biosynthesis.
(A) Global distribution of the ChIP-seq events of Qki-5, Srebp2, and Pol II in differentiated oligodendrocytes. UTR: untranslated region. (B) Venn diagram of the overlap of Qki-5-bound genes in Qki-5 ChIP-seq from differentiated oligodendrocytes and genes with substantially lower expression in Qk-Plp-iCKO mice than in control mice according to RNA-seq. (C) Canonical pathway analysis of the 277 overlapping genes shown in (B). Cellular pathways involved in cholesterol biosynthesis are labeled in magenta. (D) Sequence motifs enriched in Qki-5 ChIP-seq data from differentiated oligodendrocytes by HOMER motif analysis. (E) Canonical pathway analysis of Srebp2-bound genes in differentiated oligodendrocytes. Cellular pathways involved in cholesterol biosynthesis are labeled in blue. (F) Representative ChIP-seq binding events of Qki-5, Srebp2, and Pol II in the promoter regions of the genes involved in cholesterol biosynthesis in WT and Qk-/- differentiated oligodendrocytes. y-axis: normalized reads.
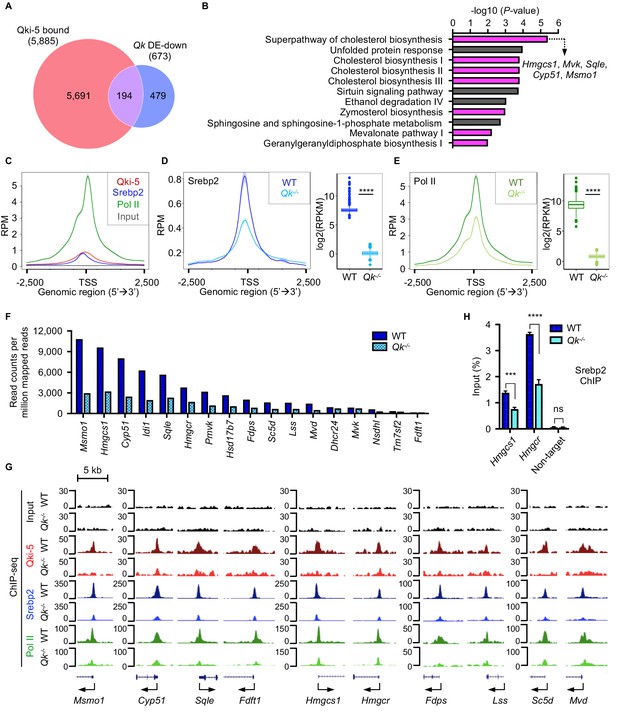
Qki transcriptionally enhances Srebp2-mediated cholesterol biosynthesis.
(A) Venn diagram of the overlap of Qki-5-bound genes in Qki-5 ChIP-seq from freshly isolated mouse oligodendrocytes and the genes with markedly lower expression in Qk-Plp-iCKO mice than in control mice. DE: differentially expressed. (B) Canonical pathway analysis of the 194 overlapping genes shown in (A). Cellular pathways involved in cholesterol biosynthesis are labeled in magenta. (C) Average occupancies of Qki-5, Srebp2, and Pol II in the gene clusters bound by Srebp2 (n = 914) in differentiated oligodendrocytes. Regions within 2.5 kb of the transcriptional start site (TSS) are included. (D, E) Average occupancies of Srebp2 (D) and Pol II (E) in the gene clusters bound by Srebp2 in WT and Qk-/- differentiated oligodendrocytes (left) and comparison of ChIP-seq (right). Regions within 2.5 kb of the TSS are included. RPM: reads counts per million mapped reads; RPKM: reads counts per kilobase per million mapped reads. (F) Bar graphs of the RPM of the Srebp2 ChIP-seq peaks within ± 0.5 kb from the TSS for 17 well-characterized Srebp2 target genes involved in cholesterol biosynthesis in WT and Qk-/- differentiated oligodendrocytes. (G) Representative ChIP-seq binding events of Qki-5, Srebp2, and Pol II in the promoter regions of the genes involved in cholesterol biosynthesis in WT and Qk-/- differentiated oligodendrocytes. y-axis: normalized reads. (H) ChIP-qPCR results showing the recruitment of Srebp2 to the promoter regions of Hmgcs1 and Hmgcr in WT and Qk-/- differentiated oligodendrocytes. Data are shown as mean ± s.d. and were analyzed using Student’s t test. ***p<0.001; ****p<0.0001; ns: not significant.
-
Figure 7—source data 1
Exact p-values for statistical analysis.
- https://cdn.elifesciences.org/articles/60467/elife-60467-fig7-data1-v2.xlsx
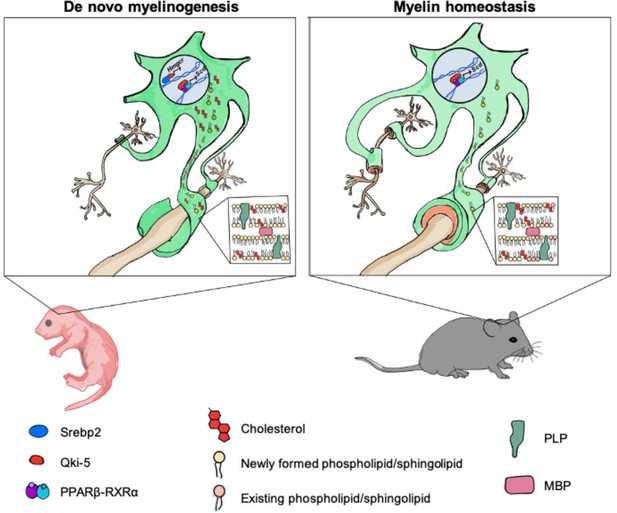
Model of Qki’s roles in regulating cholesterol biosynthesis and fatty acid metabolism during central nervous system myelination and myelin maintenance.
Qki regulates cholesterol biosynthesis in a Srebp2-dependent manner during de novo myelinogenesis but not during myelin maintenance. In contrast, Qki regulates fatty acid metabolism during both de novo myelinogenesis and mature myelin maintenance.
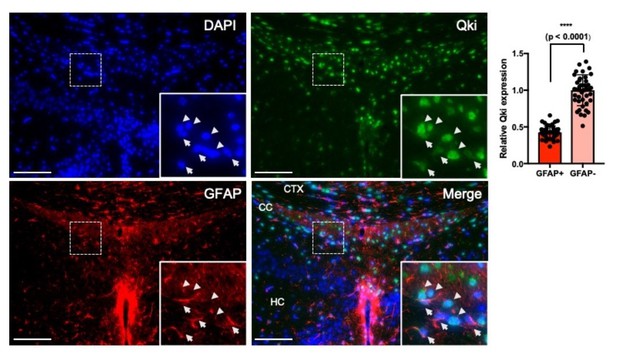
Qki is moderately expressed in GFAP+ astrocytes.
Representative images and quantification of immunofluorescent staining of Qki and GFAP in the cortex/corpus callosum/hippocampus tissues in WT mice at P21. Arrow: Qki+GFAP+ cells. Arrow head: Qki+GFAP- cells. CTX: cortex. CC: corpus callosum. HC: hippocampus. Scale bars, 100 μm.
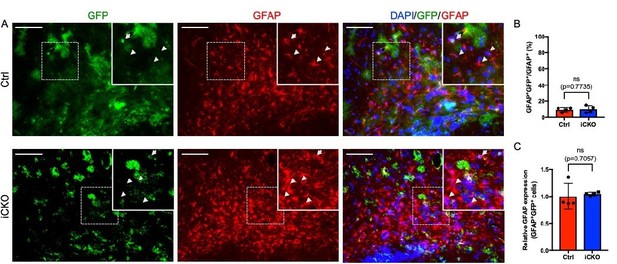
GFP is expressed in a small subpopulation of GFAP+ astrocytes, and Qki loss does not alter GFAP expression.
(A) Representative images of immunofluorescent staining of GFP and GFAP in the corpus callosum tissues in Qk- Nestin-iCKO and control mice two weeks after tamoxifen injection. Arrowhead: GFAP+GFP-cells. Arrow: GFAP+GFP+ cells. Scale bars, 100 μm. (B) Quantification of GFAP+GFP+ cells in Qk-Nestin-iCKO (n = 4) and control (n = 4) mice two weeks after tamoxifen injection shown in A. (C) Quantification of relative GFAP expression in GFAP+GFP+ cells from Qk-Nestin-iCKO (n = 4) and control (n = 4) mice two weeks after tamoxifen injection shown in A.
Videos
Defect in motor coordination displaying tremors and ataxia in Qk-Nestin-iCKO mice.
Defect in motor coordination displaying tremors and ataxia in Qk-Plp-iCKO mice.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Mouse monoclonal anti-MBP | BioLegend | Cat# SMI-94R | |
| Antibody | Mouse monoclonal anti–β-Amyloid | BioLegend | Cat# SIG-39220 | |
| Antibody | Rabbit polyclonal anti-PLP | Abcam | Cat# ab105784 | |
| Antibody | Goat polyclonal anti-Iba1 | Abcam | Cat# ab107159 | |
| Antibody | Rabbit polyclonal anti-Hmgcs1 | Abcam | Cat# ab155787 | |
| Antibody | Rabbit polyclonal anti-Fdps | Abcam | Cat# ab153805 | |
| Antibody | Rabbit polyclonal anti-Lss | Abcam | Cat# ab80364 | |
| Antibody | Mouse monoclonal anti-RNA polymerase II CTD repeat YSPTSPS | Abcam | Cat# ab817 | |
| Antibody | Rabbit monoclonal anti-MAG | Cell Signal Technology | Cat# 9043 | |
| Antibody | Rabbit polyclonal anti-GFP | Cell Signal Technology | Cat# 2555 | |
| Antibody | Rabbit monoclonal anti-Pdgfrα | Cell Signal Technology | Cat# 3174 | |
| Antibody | Mouse monoclonal anti-Ki67 | Cell Signal Technology | Cat# 9449 | |
| Antibody | Rabbit polyclonal anti-Aspa | Millipore | Cat# ABN1698 | |
| Antibody | Rabbit polyclonal anti-Olig2 | Millipore | Cat# AB9610 | |
| Antibody | Mouse monoclonal anti-NeuN | Millipore | Cat# MAB377 | |
| Antibody | Mouse monoclonal anti-Apc (CC-1) | Millipore | Cat# OP80 | |
| Antibody | Rabbit polyclonal anti-Gstpi | MBL International | Cat# 311 | |
| Antibody | Mouse monoclonal anti-Qki (Pan) | Sigma-Aldrich | Cat# SAB5201536 | |
| Antibody | Rabbit polyclonal anti-Hmgcs2 | Sigma-Aldrich | Cat# SAB2107997 | |
| Antibody | Rabbit polyclonal anti-Srebp2 | Sigma-Aldrich | Cat# HPA031962 | |
| Antibody | Mouse monoclonal anti–β-actin | Sigma-Aldrich | Cat# A5441 | |
| Antibody | Mouse monoclonal anti-Flag | Sigma-Aldrich | Cat# F1804 | |
| Antibody | Rabbit polyclonal anti-Hmgcr | Thermo Fisher Scientific | Cat# PA5-37367 | |
| Antibody | Rabbit polyclonal anti-Srebp2 | Cayman Chemical | Cat# 10007663 | |
| Antibody | Mouse monoclonal anti-Gfap | BD Biosciences | Cat# 556330 | |
| Antibody | Normal rabbit IgG | Santa Cruz Technology | Cat# sc-2027 | |
| Antibody | Rabbit polyclonal anti–Qki-5 | This paper | immunized with a short synthetic peptide (CGAVATKVRRHDMRVHPYQRIVTADRAATGN) | |
| Antibody | Mouse monoclonal anti-AnkG | Sigma-Aldrich | MABN466 | |
| Antibody | Rabbit polyclonal anti–Sox9 | Sigma-Aldrich | AB5535 | |
| Antibody | Rabbit polyclonal anti-Caspr | Gift from Dr. Rasband lab | ||
| Antibody | Mouse monoclonal anti-PanNav | Gift from Dr. Rasband lab | (K58/35) | |
| Chemical compound | Tamoxifen | Sigma-Aldrich | Cat# T5648 | |
| Chemical compound | 4-hydroxytamoxifen | Sigma-Aldrich | Cat# H7904 | |
| Chemical compound | Poly-L-ornithine | Sigma-Aldrich | Cat# P3655 | |
| Chemical compound | 3,3',5-Triiodo-L-thyronine | Sigma-Aldrich | Cat# T2877 | |
| Chemical compound | Anti-Flag M2 Magnetic Beads | Sigma-Aldrich | Cat# M8823 | |
| Chemical compound | Citrate buffer | Poly Scientific R&D Corp | Cat# s2506 | |
| Chemical compound | FluoroMyelin Red | Thermo Fisher Scientific | Cat# F34652 | |
| Chemical compound | Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140122 | |
| Recombinant protein | Laminin Mouse Protein, Natural | Thermo Fisher Scientific | Cat# 23017015 | |
| Chemical compound | Neurobasal Medium | Thermo Fisher Scientific | Cat# 21103049 | |
| Chemical compound | B-27 | Thermo Fisher Scientific | Cat# 17504044 | |
| Chemical compound | GlutaMAX Supplement | Thermo Fisher Scientific | Cat# 35050061 | |
| Chemical compound | Puromycin Dihydrochloride | Thermo Fisher Scientific | Cat# A1113802 | |
| Recombinant protein | Dynabeads Protein G | Thermo Fisher Scientific | Cat# 10004D | |
| Chemical compound | HardSet Antifade Mounting Medium with DAPI | Vector Laboratories | Cat# H1500 | |
| Chemical compound | NeuroCult Basal Medium | Stemcell Technologies | Cat# 05700 | |
| Chemical compound | NeuroCult Proliferation Supplement | Stemcell Technologies | Cat# 05701 | |
| Recombinant protein | Recombinant Human EGF | ProteinTech | Cat# AF-100-15 | |
| Recombinant protein | Recombinant Human FGF-basic (146 a.a.) | ProteinTech | Cat# 100-18C | |
| Chemical compound | DNase (RNase-free) | Qiagen | Cat# 79254 | |
| Chemical compound | cOmplete, Mini Protease Inhibitor Cocktail | Roche Diagnostics | Cat# 11836153001 | |
| Chemical compound | RNase A | Thermo Fisher Scientific | Cat# 12091021 | |
| Chemical compound | Proteinase K (PK) Solution | Promega | Cat# MC5005 | |
| Commercial assay | Neural Tissue Dissociation Kit | Miltenyi Biotec | Cat# 130-092-628 | |
| Commercial assay | In-Fusion HD Cloning Plus | Takara Bio | Cat# 638910 | |
| Commercial assay | RNeasy Mini Kit | Qiagen | Cat# 74104 | |
| Commercial assay | QIAquick PCR Purification Kit | Qiagen | Cat# 28104 | |
| Commercial assay | SuperScript III First-Strand Synthesis SuperMix for qRT-PCR | Thermo Fisher Scientific | Cat# 11752250 | |
| Commercial assay | SuperSignal West Pico PLUS Chemiluminescence System | Thermo Fisher Scientific | Cat# 34579 | |
| Commercial assay | iTaq Universal SYBR Green Supermix | Bio-rad Laboratories | Cat# 1725122 | |
| Commercial assay | DC Protein Assay Kit | Bio-rad Laboratories | Cat# 5000121 | |
| Commercial assay | Total Cholesterol Assay Kits | Cell Biolabs | Cat# STA-390 | |
| Commercial assay | KAPA Hyper Prep Kit | Kapa Biosystems | Cat# 004477 | |
| Commercial assay | In Situ Cell Death Detection Kit | Millipore Sigma | 11684795910 | |
| Deposited data | Gene expression profile | This paper | GEO: GSE145116 | |
| Deposited data | Gene expression profile | This paper | GEO: GSE145117 | |
| Deposited data | ChIP sequencing data | This paper | GEO: GSE144756 | |
| Deposited data | ChIP sequencing data | Zhou et al., 2020 | GEO: GSE126577 | |
| Experimental model | Mouse: B6.(Cg)-Nestin-CreERT2 | Imayoshi et al., 2008 | N/A | |
| Experimental model | Mouse: B6.(Cg)-Plp1-CreERT2 | The Jackson Laboratory | Stock No.: 005975 | |
| Experimental model | Mouse: B6.129-Rosa26-CreERT2 | The Jackson Laboratory | Stock No.: 008463 | |
| Experimental model | Mouse: B6.129(Cg)-ROSAmT/mG | The Jackson Laboratory | Stock No.: 007676 | |
| Experimental model | Mouse: B6.(Cg)-Qk-loxP | Shingu et al., 2017 | N/A | |
| Recombinant DNA reagent | pLKO-puro Flag-Srebp2 | Addgene | Cat# 32018 | |
| Recombinant DNA reagent | pLKO-puro 3X Flag-Srebp2 | This paper | N/A | |
| Software and algorithm | ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ | |
| Software and algorithm | HISAT2 | Johns Hopkins University | https://ccb.jhu.edu/software/hisat2/index.shtml | |
| Software and algorithm | StringTie | Johns Hopkins University | https://ccb.jhu.edu/software/stringtie/ | |
| Software and algorithm | DESeq2 | Bioconductor | http://bioconductor.org/packages/DESeq2/ | |
| Software and algorithm | IPA | Qiagen | https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/ | |
| Software and algorithm | Trim Galore | Babraham Institute | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ | |
| Software and algorithm | Bowtie | Johns Hopkins University | http://bowtie-bio.sourceforge.net/tutorial.shtml | |
| Software and algorithm | SAMtools | Li et al., 2009 | https://github.com/samtools/samtools | |
| Software and algorithm | MACS2 | Feng et al., 2012 | https://github.com/taoliu/MACS/ | |
| Software and algorithm | deeptools | Ramírez et al., 2016 | https://github.com/deeptools/deepTools | |
| Software and algorithm | ngsplot | Shen et al., 2014 | https://github.com/shenlab-sinai/ngsplot | |
| Software and algorithm | Prism 8 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ | |
| Software and algorithm | HOMER | UCSD | http://homer.ucsd.edu/homer |
Additional files
-
Supplementary file 1
A complete list of the sequences of the primer pairs used in this study.
- https://cdn.elifesciences.org/articles/60467/elife-60467-supp1-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60467/elife-60467-transrepform-v2.docx





