Diverse viral proteases activate the NLRP1 inflammasome
Figures
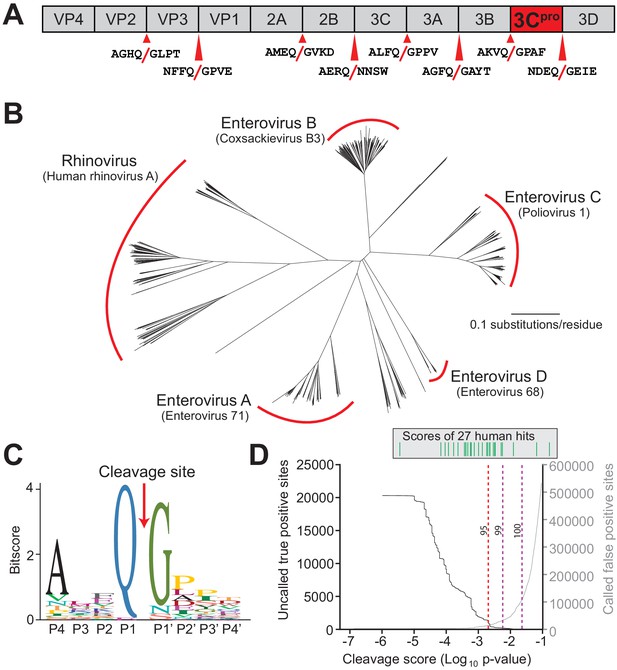
Conserved polyprotein cleavage sites across enteroviruses inform substrate specificity of the enteroviral 3Cpro.
(A) Schematic of 3Cpro cleavage sites (red arrows) within the polyprotein of coxsackievirus B3 Nancy (CVB3), a model enterovirus. Shown are the eight amino acids flanking each cleavage site within the polyprotein. (B) Phylogenetic tree of 796 enteroviral polyprotein coding sequences depicting the major clades of enteroviruses sampled in this study with representative viruses from each clade in parentheses (Supplementary file 2). (C) Eight amino acid polyprotein cleavage motif for enteroviruses (labeled as positions P4 to P4’) generated from the 796 enteroviral polyprotein sequences in (B) using the MEME Suite (Supplementary file 2). (D) Training set data used to determine the motif search threshold for FIMO (Supplementary files 1, 3 and 4). The X-axis represents a log10 of the p-value reported by FIMO as an indicator for the strength of the cleavage motif hit (cleavage score). (Left) The Y-axis depicts the number of uncalled true positives, or motif hits that overlap with the initial set of 8mer polyprotein cleavage sites used to generate the motif, in the training set of enteroviral polyprotein sequences (black). (Right) The Y-axis depicts the number of called false positive sites, or any motif hits found in the polyprotein that are not known to be cleaved by 3Cpro, in the training set of enteroviral polyprotein sequences (gray). (Above) Each line depicts a single, experimentally validated case of enteroviral 3Cpro cleavage site within a human protein as reported in Laitinen et al., 2016 and is ordered along the X-axis by its resulting cleavage score. A vertical dotted line is used to represent the decided threshold that captures 95% of true positive hits and 16 out of 27 reported human hits (Figure 1—figure supplement 1).
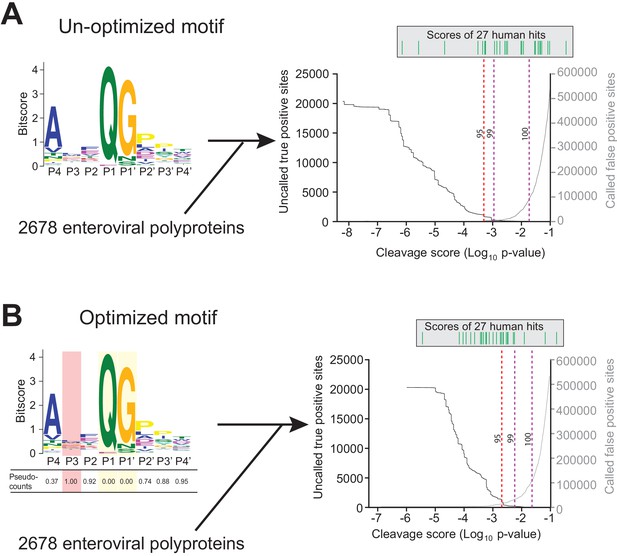
Motif optimization enhances capture of known human targets of enteroviral 3Cpro.
(A) As described in Figure 1B and C and Materials and Methods, the 8mer (P4–P4’) 3Cpro polyprotein cleavage motif was initially generated from unique, concatenated 8mer cleavage sites across 796 enteroviral polyprotein sequences. To assess the capture capability of the motif on both virus and host targets, the motif was then used to conduct a low threshold (p-value=0.1) FIMO (MEME Suite) search across training set of 2678 nonredundant enteroviral polyproteins from ViPR and 27 experimentally validated human targets of 3Cpro (Laitinen et al., 2016). In the graph, the X-axis represents a log10 of the p-value reported by FIMO as an indicator for the strength of the cleavage motif hit, or cleavage score. The left Y-axis depicts the number of uncalled ‘true positives’, or motif hits within the enteroviral polyprotein training set that overlap with the initial set of 8mer polyprotein cleavage sites used to generate the motif (black). The right Y-axis depicts the number of called false positive sites, or any motif hits that are not true positives, in the training set of enteroviral polyprotein sequences (gray). (Above) Each line depicts a single, experimentally validated case of enteroviral 3Cpro cleavage site within a human protein as reported in Laitinen et al., 2016 and is ordered along the x-axis by its corresponding cleavage score. Vertical dotted lines are used to represent the decided thresholds for comparison of capture capability. Capture of human targets at 95%, 99%, or 100% capture of true positives in the polyprotein dataset corresponds to capture of 4, 7, and 16 human hits. (B) Pseudo-counts to the position-specific scoring matrix of the motif shown in (A) were adjusted by total information content where the two most information-dense positions P1 and P1’ are assigned pseudocount = 0 and the least information-dense position P3 pseudocount = 1, and the remaining positions are assigned a pseudocount value relative to the most information-dense position P1. This optimized motif is then used to FIMO search against the same training set as described in (A). Capture of human targets at 95%, 99%, or 100% capture of true positives in the polyprotein dataset corresponds to capture of 16, 23, and 24 human hits.
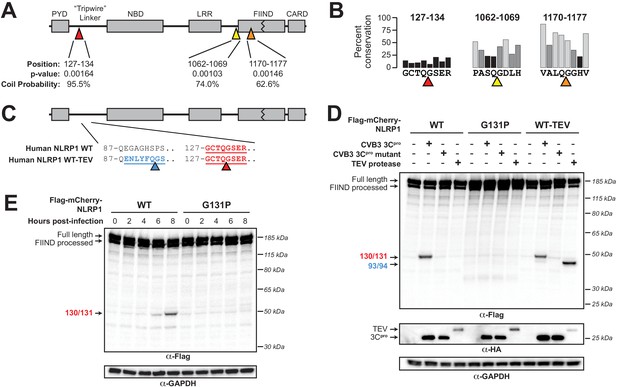
Enterovirus 3Cpro cleaves human NLRP1 at the predicted site of mimicry.
(A) Schematic of the domain structure of NLRP1, with predicted cleavage sites (triangles). FIMO-reported p-values and average NetsurfP-reported coil probabilities (Figure 2—source data 1) are described at the predicted sites. (B) Percent conservation across 100 mammalian species at each position of each predicted 8mer cleavage site within human NLRP1. (C) Schematic of the human NLRP1 sequence used to assess enteroviral cleavage and activation. The predicted enteroviral cleavage site found in the linker region (127-GCTQGSER-134) is shown in red. Human NLRP1 WT-TEV contains an engineered TEV cleavage site between residues 93 and 94 (underlined green) in human NLRP1 WT. (D) Immunoblot depicting human NLRP1 cleavage by CVB3 3Cpro and TEV protease. HEK293T cells were co-transfected using 100 ng of the indicated Flag-tagged mCherry-NLRP1 fusion plasmid constructs with 250 ng of the indicated protease construct and immunoblotted with the indicated antibodies. (E) Immunoblot depicting human NLRP1 cleavage at the indicated timepoints after infection with 250,000 PFU (MOI = ~1) CVB3. HEK293T cells were transfected using 100 ng of either WT NLRP1 or NLRP1 G131P and infected 24–30 hr later. All samples were harvested 32 hr post-transfection and immunoblotted with the indicated antibodies.
-
Figure 2—source data 1
Tabular output of NetSurfP structural predictions for human NLRP1.
NetSurfP output for indicated regions of NLRP1 describing the predicted class (buried or exposed), relative surface accessibility, absolute surface accessibility, probability for alpha-helix, probability for beta-strand, and coil probability for the amino acid positions within each predicted enteroviral 3Cpro cleavage site.
- https://cdn.elifesciences.org/articles/60609/elife-60609-fig2-data1-v2.txt
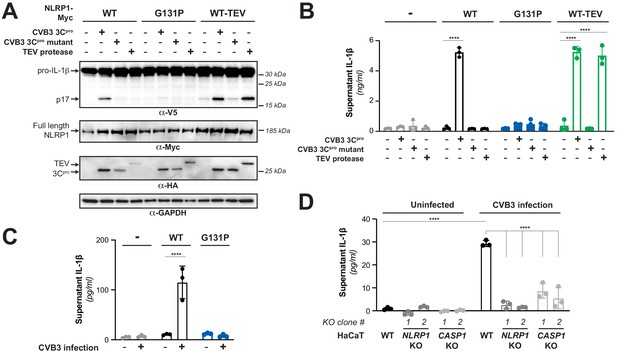
Enterovirus 3Cpro cleavage of human NLRP1 promotes pro-inflammatory cytokine release.
(A) Immunoblot depicting human NLRP1 activation (maturation of IL-1β) by CVB3 3Cpro and TEV protease. HEK293T cells were co-transfected using 100 ng of the indicated protease, 50 ng V5-IL-1β, 100 ng CASP1, 5 ng ASC, and 4 ng of the indicated Myc-tagged NLRP1, and immunoblotted with the indicated antibodies. Appearance of the mature p17 band of IL-1β indicates successful assembly of the NLRP1 inflammasome and activation of CASP1. (B) Bioactive IL-1β in the culture supernatant was measured using HEK-Blue IL-1β reporter cells, which express secreted embryonic alkaline phosphatase (SEAP) in response to extracellular IL-1β. Supernatant from cells transfected as in (A) was added to HEK-Blue IL-1β reporter cells and SEAP levels in the culture supernatant from HEK-Blue IL-1β reporter cells were quantified by the QUANTI-Blue colorimetric substrate. Transfections were performed in triplicate and compared to the standard curve generated from concurrent treatment of HEK-Blue IL-1β reporter cells with purified human IL-1β (Figure 3—figure supplement 2). Data were analyzed using two-way ANOVA with Sidak’s post-test. **** = p<0.0001. (C) CVB3 infection of inflammasome-reconstituted HEK293T cells results in IL-1β release when NLRP1 can be cleaved by 3Cpro. Cells were transfected with the indicated NLRP1 construct and other NLRP1 inflammasome components as in (B). Sixteen hours post-transfection, cells were mock infected or infected with 250,000 PFU (MOI = ~1) CVB3. Eight hours post-infection, culture supernatant was collected and bioactive IL-1β was measured as in (B). (D) CVB3 infection of an immortalized human keratinocyte cell line, HaCaT, activates the NLRP1 inflammasome. WT or knockout (Figure 3—figure supplement 3) HaCaT cell lines were mock infected or infected with 100,000 PFU (MOI = ~0.4) CVB3. Forty-eight hours post-infection, culture supernatant was collected and bioactive IL-1β was measured as in (B).
-
Figure 3—source data 1
Individual data values for Figure 3B, C and D.
- https://cdn.elifesciences.org/articles/60609/elife-60609-fig3-data1-v2.xlsx
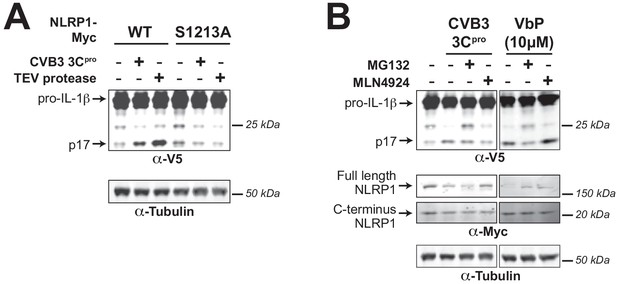
3Cpro-mediated activation of the human NLRP1 inflammasome depends on FIIND autoprocessing and proteosomal degradation.
(A) HEK293T cells were transfected with either WT NLRP1 or a FIIND auto-processed defective mutant (S1213A) along with other components of the NLRP1 inflammasome (CASP1, ASC, and IL-1β) as in Figure 3A. Only cells transfected with WT NLRP1 can produce mature IL-1β upon co-transfection with CVB3 3Cpro or TEV protease as indicated by the appearance of the p17 band. (B) HEK293T cells were transfected as in (A), and then treated with the indicated inhibitors of proteasomal-mediated degradation (0.5 µM MG132) or the N-glycine degron pathway (1.0 µM MLN4924) for 6 hr prior to harvest. Addition of 10 µM VbP, an inhibitor of the NLRP1 inhibitors DPP8/9 (Okondo et al., 2018), was used as a control for protease-independent activation of the inflammasome and is thus unaffected by MLN4924.
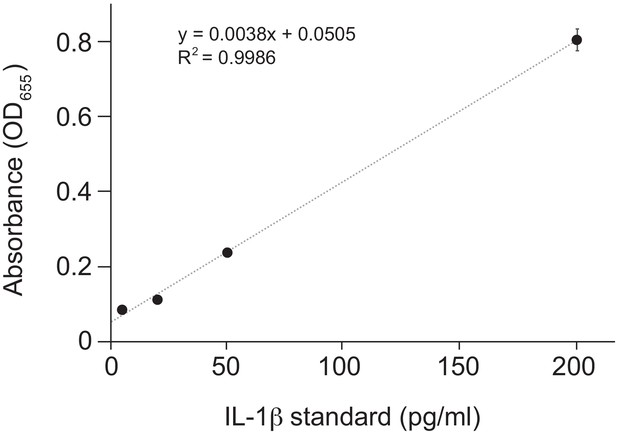
Standard curve for Figure 3B.
Purified human IL-1β was added in duplicate to the indicated final concentration to HEK-Blue IL-1β reporter cells and SEAP activity was measured by increased absorbance at OD655. The indicated linear fit was used to calculate absolute concentrations of bioactive IL-1β from culture supernatants shown in Figure 3B. Note that supernatants from inflammasome-transfected cells was diluted 10-fold before addition to HEK-Blue IL-1β reporter cells to ensure that levels fell within the linear range of the indicated standard curve. Standard curves were generated in an identical manner for each panel of HEK-Blue data shown.
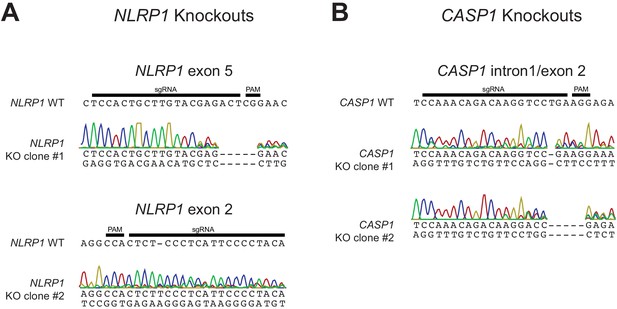
Validation of CRISPR/Cas9-editing of NLRP1 or CASP1 in HaCaT cells by Sanger sequencing.
(A) The WT NLRP1 (exon 5 or exon 2) sequence is shown with CRISPR-targeting sgRNA and PAM sequences indicated. Below each WT sequence is shown the Sanger sequencing chromatogram and associated mutant sequence for each indicated clone. (B) Same as panel A, except indicating WT CASP1 intron 1/exon two sequence and sequencing data from two independently isolated knockout clones.
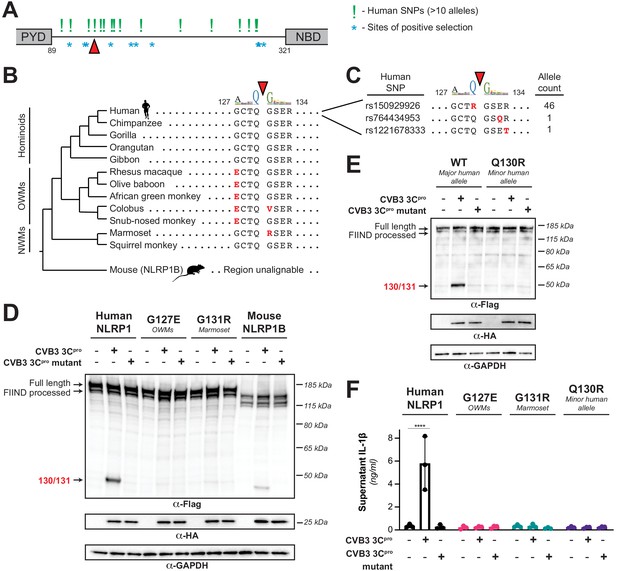
Naturally occurring cleavage site variants alter NLRP1 susceptibility to enteroviral 3Cpro.
(A) Schematic of sites found to be evolving under positive selection (marked as *, from Chavarría-Smith and Vance, 2013) and human SNPs with at least 10 reported instances in the Genome Aggregation Database (GnomAD, Karczewski et al., 2020) (marked as !) within the linker region between the pyrin domain (PYD) and nucleotide binding domain (NBD) of NLRP1. The enteroviral 3Cpro cleavage site between position 130 and 131 is indicated by a red triangle. (B) Phylogenetic tree depicting the enteroviral 3Cpro cleavage site (red triangle) within NLRP1 across three clades of primates – hominoids, Old World monkeys (OWMs), and New World monkeys (NWMs). Mouse NLRP1B lacks any sequence that is alignable to this region of primate NLRP1 (see also Figure 4—figure supplement 1). Amino acid differences to the human NLRP1 reference sequence are highlighted in red. Above the alignment is the enterovirus 3Cpro sequence logo shown in Figure 1. (C) GnomAD-derived allele counts of each missense human SNP (by reference SNP #) within the 8mer of the determined enteroviral 3Cpro cleavage site. (D–E) Immunoblot depicting CVB3 3Cpro cleavage susceptibility of the indicated 8mer site variants introduced into human NLRP1 or full-length wild-type mouse NLRP1B (129 allele) (D) or the cleavage susceptibility of human NLRP1 Q130R, a naturally occurring human population variant (E). (F) Release of bioactive IL-1β into the culture supernatant was measured using HEK-Blue IL-1β reporter cells as in Figure 3B. Data were analyzed using two-way ANOVA with Sidak’s post-test. **** = p<0.0001.
-
Figure 4—source data 1
Individual data values for Figure 4F.
- https://cdn.elifesciences.org/articles/60609/elife-60609-fig4-data1-v2.xlsx
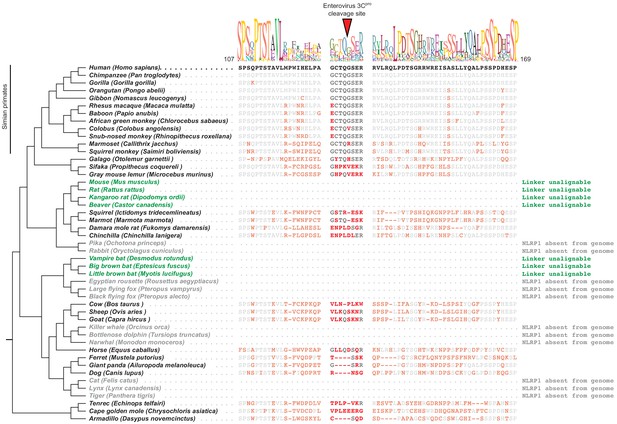
Mammalian NLRP1 phylogenomics and alignment of linker region.
The indicated mammalian NLRP1 sequences were aligned and the region corresponding to residues 107–169 from human NLRP1 were extracted, which is anchored on both ends by well-conserved proline and serine-rich motifs. A consensus sequence generated from alignable sequences in this region is shown above the human sequence. The position of the CVB3 3Cpro cleavage site in human NLRP1 is shown, flanked by four amino acids on both sides (P4->P4’). In other mammals, residues that differ from the human sequence are shown in red. Within the aligned region that corresponds to the CVB3 3Cpro cleavage site, only simian primates have P4, P1 and P1’ residues that would allow cleavage. The only other species that have a plausible cleavage site in this position are sheep and goats (P4 = Val, P1 = Gln, P1’ = Ser), although those residues appear to have evolved independently at those positions. Two clades of species (the ‘mouse-related’ clade of rodents and the microbat clade, marked as green) have NLRP1 protein sequences with N-terminal linkers that are unalignable to human NLRP1 in this region. Four additional clades (lagomorphs, megabats, cetaceans, and felines, marked as gray) lack the NLRP1 gene altogether.
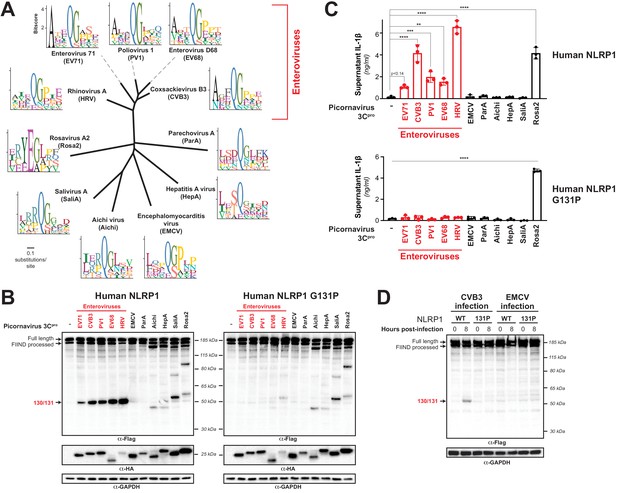
Diverse picornavirus 3Cpros cleave and activate NLRP1 at independently evolved sites.
(A) Phylogenetic tree of 3Cpro protein sequences for the indicated picornaviruses (Figure 5—figure supplement 1; Figure 5—figure supplement 1—source data 1). Shown next to the virus name is the sequence motif generated from the known sites of 3Cpro polyprotein cleavage in that specific virus. (B) Immunoblot depicting human NLRP1 cleavage by the indicated picornaviral 3Cpro. Abbreviations are as in (A). Assays were performed as in Figure 2D. (left) Cleavage assays against WT NLRP1. (right) Human NLRP1 G131P mutant used in Figure 2. (C) Release of bioactive IL-1β into the culture supernatant was measured using HEK-Blue IL-1β reporter cells as in Figure 3B. Data were analyzed using one-way ANOVA with Tukey’s post-test. ** = p<0.01, *** = p<0.001, **** = p<0.0001. (D) Immunoblot depicting human NLRP1 cleavage at the indicated timepoints after infection with 250,000 PFU (MOI = ~1) CVB3 or EMCV. HEK293T cells were transfected using 100 ng of either WT NLRP1 or NLRP1 G131P and, 24 hr later, either mock infected (0 hr timepoint) or infected with CVB3 or EMCV as indicated (8 hr timepoint). All samples were harvested 32 hr post-transfection and immunoblotted with the indicated antibodies.
-
Figure 5—source data 1
Individual data values for Figure 5C and Figure 5—figure supplements 3 and 4.
- https://cdn.elifesciences.org/articles/60609/elife-60609-fig5-data1-v2.xlsx

Alignment of 3Cpros used in this study.
Sequences were aligned using MAFFT (Katoh and Standley, 2013) and used to generate the phylogenetic tree shown in Figure 5A. Asterisks indicate residues 100% conserved in all sequences. The position of the catalytic cysteine, analogous to C147 in CVB3 3Cpro, is highlighted in yellow.
-
Figure 5—figure supplement 1—source data 1
Table of pairwise percent sequence identity of 3Cpros used in this study as determined from the alignment shown in Figure 5—figure supplement 1.
- https://cdn.elifesciences.org/articles/60609/elife-60609-fig5-figsupp1-data1-v2.txt
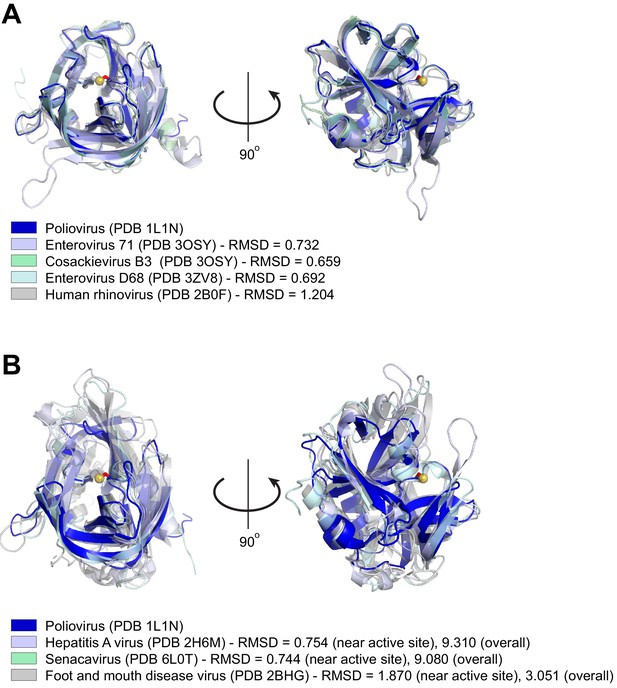
Structural similarity of picornavirus 3Cpros.
(A) Structures of the indicated enterovirus 3Cpros were aligned to poliovirus 3Cpro using Pymol (https://pymol.org/2/ - The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.). For each enterovirus species used in this study, PDB codes are indicated in parentheses and Pymol-calculated RMSD values are shown. The catalytic cysteine, C147, in the poliovirus 3Cpro structure is shown as a ball-and-stick representation (red stick and yellow ball for the catalytic S atom). Despite substantial divergence in amino acid identity (see Figure 5—figure supplement 1—source data 1), the overall fold and active site configuration of these enzymes is similar. (B) Structures of the indicated picornavirus 3Cpros were aligned to poliovirus 3Cpro using Pymol, with a similar representation of the catalytic C147 residue in poliovirus 3Cpro as in (A). RMSD values are shown for alignments to either the whole poliovirus 3Cpro structure (‘overall’ RMSD) or only the 50 amino acids flanking the catalytic cysteine, C147 (‘near active site’ RMSD). The displayed aligned structures are from the ‘near active site’ alignment. Neither senecavirus nor foot and mouth disease virus (FDMV) were used in this study, but are included as additional representatives of non-enterovirus 3Cpro enzymes for which there are experimentally determined molecular structures. Compared to proteases aligned in (A), divergent picornavirus proteases show greater divergence in overall fold as indicated by the larger ‘overall’ RMSD values. However, comparison of the regions around the active site still show high structural similarity, consistent with the constraint on protease evolution and similarity of cleavage specificity shown in Figure 5A.
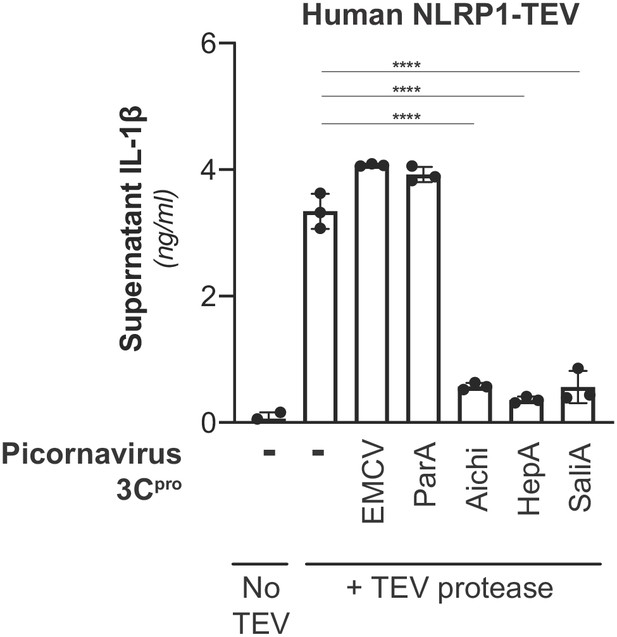
Inhibition of NLRP1 activation by non-enteroviral 3Cpro.
HEK293T cells were transfected with inflammasome components as in Figure 3A using 100 ng TEV protease or pQCXIP empty vector, but with the additional inclusion of a non-enterovirus 3Cpro or empty vector (100 ng). Release of bioactive IL-1β into the culture supernatant was measured using HEK-Blue IL-1β reporter cells as in Figure 3B. Data were analyzed using one-way ANOVA with Tukey’s post-test comparing all conditions containing TEV protease. **** = p<0.0001.
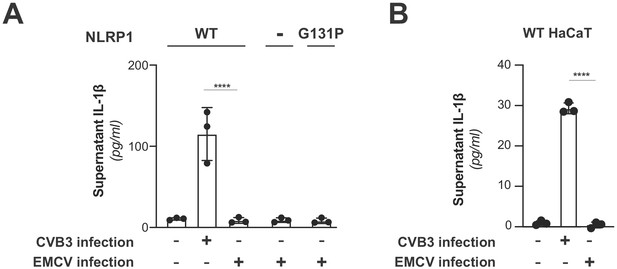
EMCV infection does not activate the NLRP1 inflammasome.
(A) HEK293T cells were transfected with inflammasome components as in Figure 3C and mock infected or infected with 250,000 PFU (MOI = ~1) CVB3 or EMCV. Eight hours post-infection, culture supernatant was collected and bioactive IL-1β was measured as in Figure 3C. Data from the mock and CVB3 infections are reproduced from Figure 3C and included as a point of reference as they were done in parallel. Data were analyzed using two-way ANOVA with Sidak’s post-test. **** = p<0.0001. (B) WT or knockout (Figure 3—figure supplement 3) HaCaT cell lines were mock infected or infected with 100,000 PFU (MOI = ~0.4) CVB3 of EMCV as in Figure 3D. Forty-eight hours post-infection, culture supernatant was collected and bioactive IL-1β was measured as in Figure 3D. Data from the mock and CVB3 infections are reproduced from Figure 3D and included as a point of reference as they were done in parallel. Data were analyzed using two-way ANOVA with Sidak’s post-test. **** = p<0.0001.
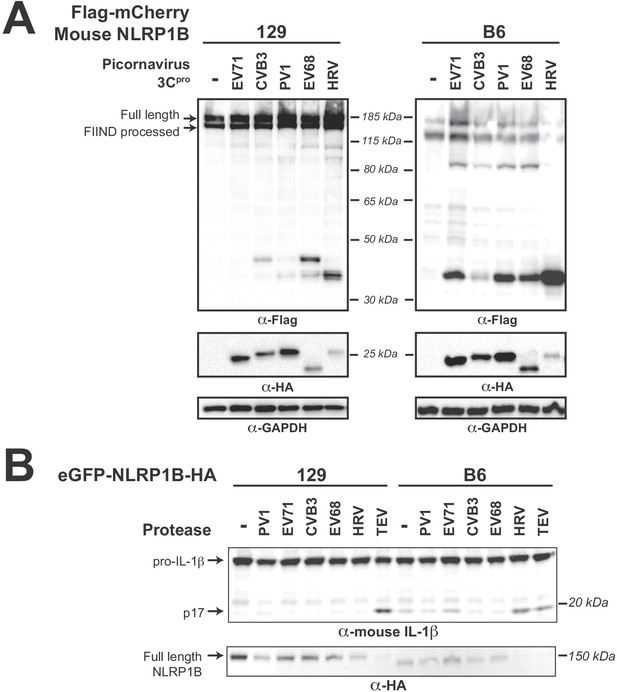
Diverse picornavirus 3Cpros cleave and activate mouse NLRP1B at independently evolved sites.
(A) Immunoblot depicting CVB3 3Cpro cleavage susceptibility of two alleles (129 and B6) of mouse NLRP1B. Assays were performed as in Figure 2D. (B) Immunoblot depicting mouse NLRP1B activation (maturation of IL-1β) by enterovirus 3Cpro and TEV protease. HEK293T cells were co-transfected using 100 ng of the indicated protease, 50 ng mouse IL-1β, 50 ng mouse CASP1, and either 4 ng of 129 NLRP1B or 2.5 ng of B6 NLRP1B constructs, and immunoblotted with the indicated antibodies. Appearance of the mature p17 band of IL-1β indicates successful assembly of the NLRP1B inflammasome and activation of CASP1.

Alignment of N-termini of mouse NLRP1B 129 and B6 alleles.
Sequences were aligned using MAFFT (Katoh and Standley, 2013). Asterisks indicate conserved residues and the start of the nucleotide-binding domain (NBD) is shown.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Homo sapiens) | NLRP1 | NCBI | NCBI: NP_127497.1 | |
| Gene (Mus musculus) | NLRP1B (129) | NCBI | NCBI: AAZ40510.1 | |
| Gene (Mus musculus) | NLRP1B (B6) | NCBI | NCBI: XM_017314698.2 | |
| Cell line (Homo sapiens) | HEK293T | ATCC | Cat# CRL-3216; RRID:CVCL_0063 | |
| Cell line (Homo sapiens) | HEK-Blue IL-1β cells | Invivogen | HKB-IL1B | |
| Cell line (Homo sapiens) | HaCaT (parental) | UC Berkeley Cell Culture Facility | ||
| Cell line (Homo sapiens) | HaCaT Cas9 (WT) | This paper | ||
| Cell line (Homo sapiens) | HaCaT Cas9 ΔNLRP1 #1 (NLRP1 KO clone #1) | This paper | Exon five target (TCCACTGCTTGTACGAGACT) | |
| Cell line (Homo sapiens) | HaCaT Cas9 ΔNLRP1 #2 (NLRP1 KO clone #2) | This paper | Exon two target (TGTAGGGGAATGAGGGAGAG) | |
| Cell line (Homo sapiens) | HaCaT Cas9 ΔCASP1 #1 (CASP1 KO clone #1) | This paper | Exon two target (CCAAACAGACAAGGTCCTGA) | |
| Cell line (Homo sapiens) | HaCaT Cas9 ΔCASP1 #2 (CASP1 KO clone #2) | This paper | Exon two target (CCAAACAGACAAGGTCCTGA) | |
| Recombinant DNA reagent | pcDNA5/FRT/TO (plasmid) | Invitrogen | V652020 | |
| Recombinant DNA reagent | pQCXIP (plasmid) | Takara Bio | 631516 | |
| Recombinant DNA reagent | psPAX2 (plasmid) | Addgene | RRID:Addgene_12260 | Gift from Dr. Didier Trono |
| Recombinant DNA reagent | pMD2.G (plasmid) | Addgene | RRID:Addgene_12259 | Gift from Dr. Didier Trono |
| Recombinant DNA reagent | pLB-Cas9 (plasmid) | Addgene | RRID:Addgene_52962 | Gift from Dr. Feng Zhang |
| Recombinant DNA reagent | pLentiGuide-Puro (plasmid) | Other | Gift from Dr. Mortiz Gaidt | |
| Recombinant DNA reagent | Inflammasome reconstitution plasmids | PMID:27926929 | Gifts from Dr. Russell Vance: human NLRP1 TEV (NLRP1-TEV2), human ASC, human and mouse CASP1, human IL-1B-V5, mouse IL-1B, and TEV protease | |
| Recombinant DNA reagent | CVB3-Nancy infectious clone plasmid | PMID:2410905 | Gift from Dr. Julie Pfeiffer | |
| Recombinant DNA reagent | EMCV-Mengo infectious clone plasmid | PMID:2538661 | Gift from Dr. Julie Pfeiffer | |
| Commercial assay or kit | QUANTI-Blue assay reagent (for HEK-Blue IL-1β cells) | Invivogen | REP-QBS | Includes necessary reagents for measuring IL-1β release from HEK-Blue-IL-1B reporter cell line |
| Chemical compound, drug | TransIT-X2 | Mirus | MIR 6000 | |
| Chemical compound, drug | MG132 | Sigma-Aldrich | M7449 | |
| Chemical compound, drug | MLN4924 | APExBIO | B1036 | |
| Antibody | V5-Tag rabbit monoclonal | Cell Signaling Technology | Cat# 13202; RRID:AB_2687461 | (1:1000) |
| Antibody | Flag-Tag mouse monoclonal | Sigma-Aldrich | Cat# F1804; RRID:AB_262044 | (1:2000) |
| Antibody | Myc-Tag rabbit monoclonal | Cell Signaling Technology | Cat# 2278; RRID:AB_490778 | (1:1000) |
| Antibody | HA-Tag rat monoclonal | Roche | Cat# 11867423001; RRID:AB_390918 | (1:1000) |
| Antibody | GAPDH rabbit monoclonal | Cell Signaling Technology | Cat# 2118; RRID:AB_561053 | (1:2000) |
| Antibody | Goat anti-Rat IgG (H+L) Secondary Antibody, HRP | Thermo Fisher Scientific | Cat# 31470; RRID:AB_228356 | (1:10000) |
| Antibody | Goat anti-Rabbit IgG (H+L) Secondary Antibody, HRP | Biorad | Cat# 170–6515; RRID:AB_11125142 | (1:10000) |
| Antibody | Goat anti-Mouse IgG (H+L) Secondary Antibody, HRP | Biorad | Cat# 170–6516; RRID:AB_11125547 | (1:10000) |
| Antibody | β-Tubulin mouse monoclonal | Sigma-Aldrich | Cat# T4026; RRID:AB_477577 | (1:2000) |
| Antibody | mouse IL-1β goat polyclonal | R and D Systems | Cat# AF-401-NA; RRID:AB_416684 | (1:1000) |
| Sequence-based reagent | Oligonucleotides | Other | See Supplementary file 5 for list of oligonucleotides used in this study | |
| Software, algorithm | MEME v5.0.3 | PMID:25953851 | RRID:SCR_001783 | Motif finder (FIMO) |
| Software, algorithm | MAFFT 7.309 | PMID:23329690 | RRID:SCR_011811 | |
| Software, algorithm | NetSurfP | PMID:30785653 | RRID:SCR_018781 | http://www.cbs.dtu.dk/services/NetSurfP/ (Original); https://services.healthtech.dtu.dk/service.php?NetSurfP-2.0 (Alternate) |
| Software, algorithm | Geneious | PMID:22543367 | RRID:SCR_010519 | Neighbor-joining phylogenetic tree |
| Software, algorithm | BLASTp | PMID:9254694 | RRID:SCR_001010 |
Additional files
-
Supplementary file 1
Training set of enteroviral polyproteins.
Accession IDs are listed for all polyproteins used to benchmark the motif search described in Figure 1D and Figure 1—figure supplement 1. The 8mer cleavage sites and concatenated 8mer sequences are included.
- https://cdn.elifesciences.org/articles/60609/elife-60609-supp1-v2.xlsx
-
Supplementary file 2
Enteroviral polyproteins with unique 8mer 3Cpro cleavage site concatenations.
Accession IDs are listed for all polyproteins used to create the search motif shown in Figure 1C and Figure 1—figure supplement 1 and the enteroviral phylogenetic tree in Figure 1B. The 8mer cleavage sites and concatenated 8mer sequences are included.
- https://cdn.elifesciences.org/articles/60609/elife-60609-supp2-v2.xlsx
-
Supplementary file 3
Un-optimized 3Cpro cleavage motif scores for true positive, false positive and human sites within the enteroviral polyprotein and human training sets.
FIMO-generated p-values and log10(p-value) represent the cleavage score at the provided matched sequence, where (A) is the un-optimized 3Cpro cleavage motif scores for true positive hits within enteroviral polyprotein dataset, (B) is the un-optimized 3Cpro cleavage motif scores for false positive hits within enteroviral polyprotein dataset where unique site matches are shown (26062), and (C) is the un-optimized 3Cpro cleavage motif scores for reported human cleavage sites from the Laitinen et al., 2016 dataset.
- https://cdn.elifesciences.org/articles/60609/elife-60609-supp3-v2.xlsx
-
Supplementary file 4
Optimized 3Cpro cleavage motif scores for true positive, false positive and human sites within the enteroviral polyprotein and human training sets.
FIMO-generated p-values and log10(p-value) represent the cleavage score at the provided matched sequence, where (A) is the optimized 3Cpro cleavage motif scores for true positive hits within enteroviral polyprotein dataset, (B) is the optimized 3Cpro cleavage motif scores for false positive hits within enteroviral polyprotein dataset where unique site matches are shown (24437), and (C) is the optimized 3Cpro cleavage motif scores for reported human cleavage sites from the Laitinen et al., 2016 dataset.
- https://cdn.elifesciences.org/articles/60609/elife-60609-supp4-v2.xlsx
-
Supplementary file 5
List of primers and gBlocks used.
Names and notes contain details on the restriction enzyme sites or point mutations encoded.
- https://cdn.elifesciences.org/articles/60609/elife-60609-supp5-v2.xlsx
-
Supplementary file 6
List of antibodies used for immunoblots.
Manufacturer and dilutions used are noted.
- https://cdn.elifesciences.org/articles/60609/elife-60609-supp6-v2.xlsx
-
Supplementary file 7
List of accession numbers used for sequence alignments.
- https://cdn.elifesciences.org/articles/60609/elife-60609-supp7-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/60609/elife-60609-transrepform-v2.doc





