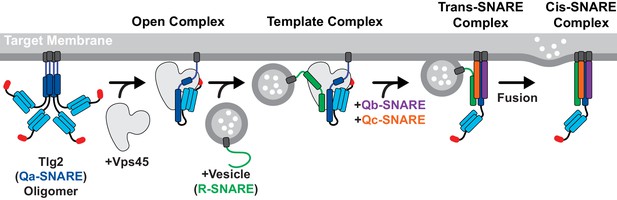
The Sec1/Munc18 protein Vps45 holds the Qa-SNARE Tlg2 in an open conformation
Figures
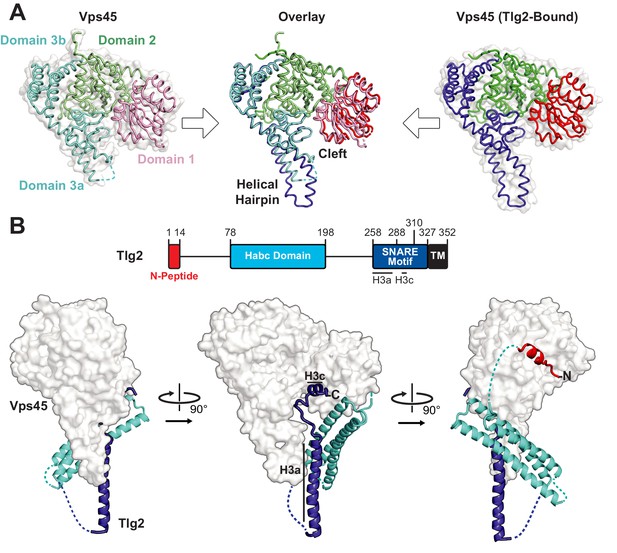
Crystal structures of C. thermophilum Vps45 and Vps45–Tlg2.
(A) Crystal structures of Vps45 (left) and Vps45–Tlg2 (right, showing only Vps45). The comparison shown in the center was generated by aligning domains 2 and 3b. (B) Crystal structure of Vps45–Tlg2. H3a and H3c are helical regions within the SNARE motif that were defined by Misura et al. based on the Munc18–Stx structure (Misura et al., 2000).
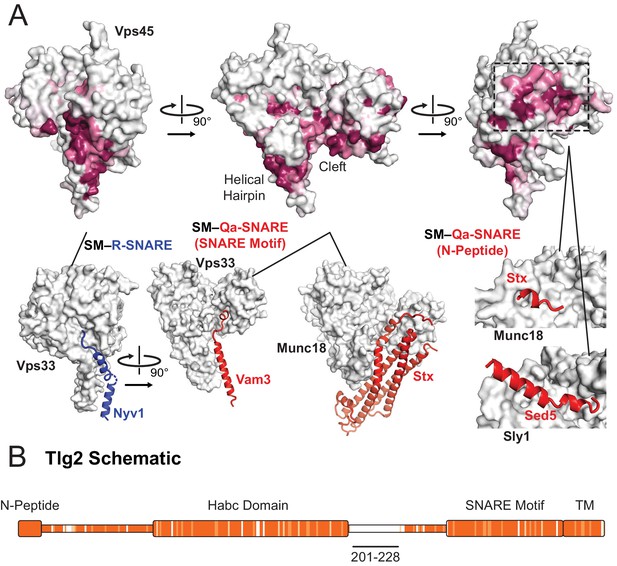
Sequence conservation in Vps45 and Tlg2.
(A) Sequence conservation of Vps45 surface residues as determined by ConSurf (Ashkenazy et al., 2010). Three highly conserved regions on Vps45 have previously been implicated in R-SNARE binding (bottom left, Vps33–Nyv1 (PDB code 5BV0)), Qa-SNARE SNARE motif binding (bottom middle, Vps33–Vam3 (5BUZ) and Munc18–Stx, (3C98)), and N-peptide binding (right Munc18–Stx (3C98) and Sly1-Sed5 (1MQS)). (B) Sequence conservation heat map comparing the Tlg2 sequences from C. thermophilum and C. globosum. The region deleted from Vps45–Tlg2, residues 201–228, is highlighted. The heat map was produced using Clustal Omega (Sievers and Higgins, 2018).
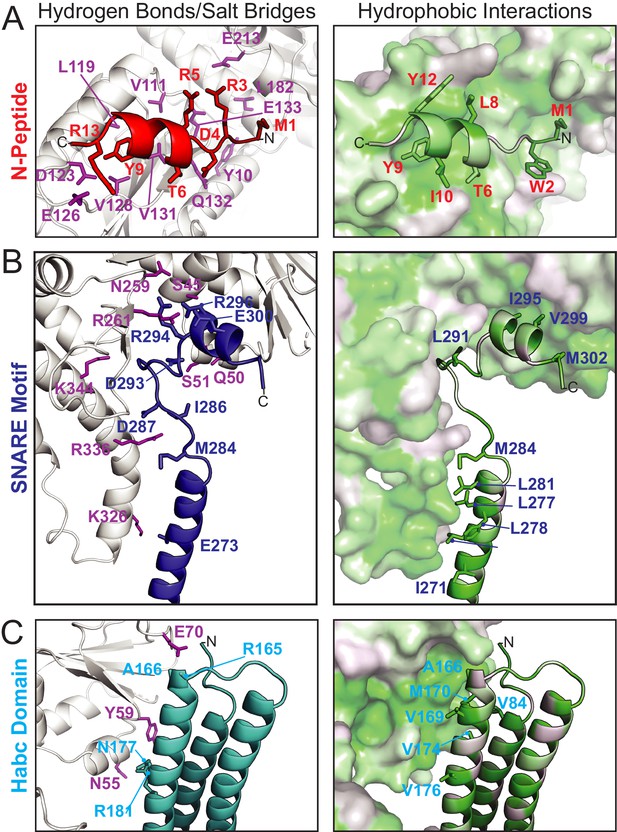
Vps45–Tlg2 interactions.
(A–C) Interactions between Vps45 and the (A) N-peptide, (B) SNARE motif, and (C) Habc domain of Tlg2. The left panels depict residues contributing to hydrogen bonds and/or salt bridges, with Vps45 residues colored purple. The right panels depict hydrophobic interactions, with residues colored from least (white) to most (green) hydrophobic.
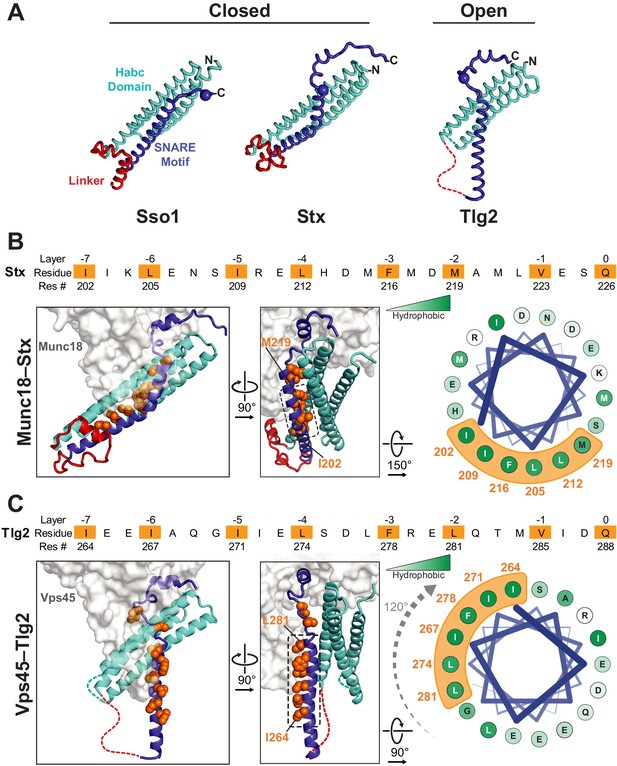
Vps45-bound Tlg2 adopts an open conformation.
(A) Comparison of uncomplexed Sso1 (PDB code 1FIO), Munc18-bound Stx (3C98), and Vps45-bound Tlg2. The locations of the zero-layer Gln residues are indicated with spheres. (B and C) Comparison of the Habc domains and SNARE motifs in the Munc18–Stx and Vps45–Tlg2 complexes. The core residues of the SNARE motifs (layers −7 to 0) are depicted as orange spheres. At the right, helical wheel representations depict the relative rotation of the SNARE motifs (layers −7 to −2, corresponding to helix H3a in Figure 1B) with respect to the rest of the structure.
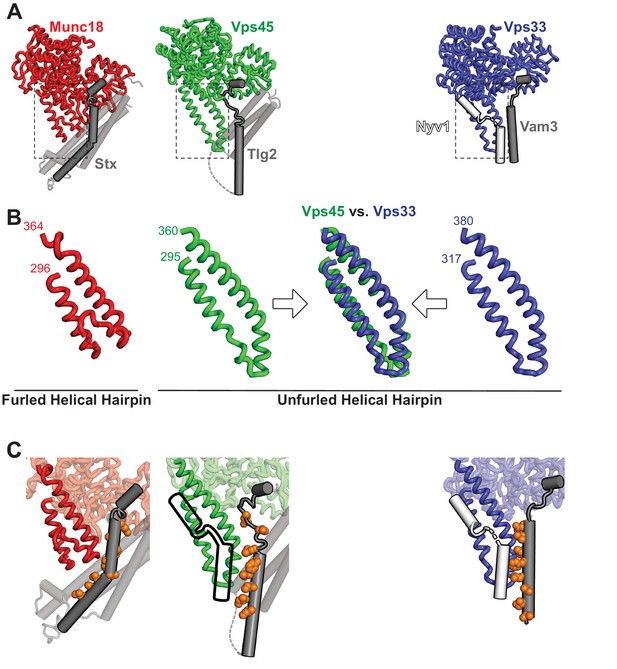
The domain 3a helical hairpin of Tlg2-bound Vps45 is unfurled.
(A) Structures of SM–Qa-SNARE complexes Munc18–Stx and Vps45–Tlg2, as well as a model of the SM–Qa-SNARE–R-SNARE template complex Vps33–Vam3–Nyv1 (Baker et al., 2015). The Qa-SNARE motifs are dark gray, the remainder of the Qa-SNAREs are light gray, and the R-SNARE Nyv1 is white. The model of the template complex was obtained by combining Vps33 and Vam3 from the Vps33–Vps16–Vam3 structure (5BUZ) with Nyv1 from the Vps33–Vps16–Nyv1 structure (5BV0); both of these structures lack SNARE N-terminal regions and contain only the SNARE motifs. (B) Close-up views of the domain 3a helical hairpins (dashed rectangles in panel A). (C) SM–SNARE interactions, emphasizing the domain 3a helical hairpins and the SNARE motifs. For Vps45, a model of the template complex is suggested by superimposing the outline of Vps33-bound Nyv1. As in Figure 3, the core residues of the SNARE motifs (layers −7 to 0) are shown as orange spheres.
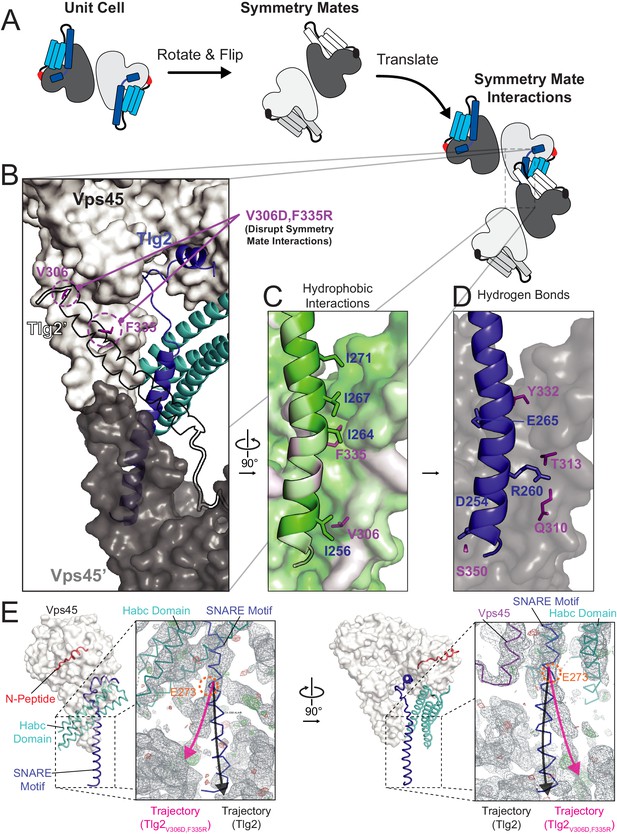
Crystal contacts between the domain 3a helical hairpin and the SNARE motif.
(A) Schematic representation of asymmetric unit and crystal packing. (B–D) Interaction between Tlg2 SNARE motif (blue) with symmetry mate Vps45 (dark gray, named Vps45′). The reciprocal interaction between the symmetry mate Tlg2 (black outline, named Tlg2′) and Vps45 (white) is shown. Residues V306 and F335 were targeted for mutagenesis to disrupt these interactions (purple, V306D and F335R, respectively). Hydrophobic (C) and hydrogen bond/salt bridge (D) stabilizing interactions are shown. Hydrophobicity of each residue is colored on a scale from least (white) to most (green) hydrophobic. (E) Comparison of Vps45–Tlg2 with the electron density map (grey) obtained from Vps45–Tlg2V306D,F335R diffraction data phased using Vps45–Tlg2 (minus the SNARE motif) as the molecular replacement model. The trajectories of the Tlg2 and Tlg2V306D,F335R SNARE motifs (black and pink arrows respectively) diverge near Glu 273 (orange). The unfurled helical hairpin is visible in the right-hand panel.
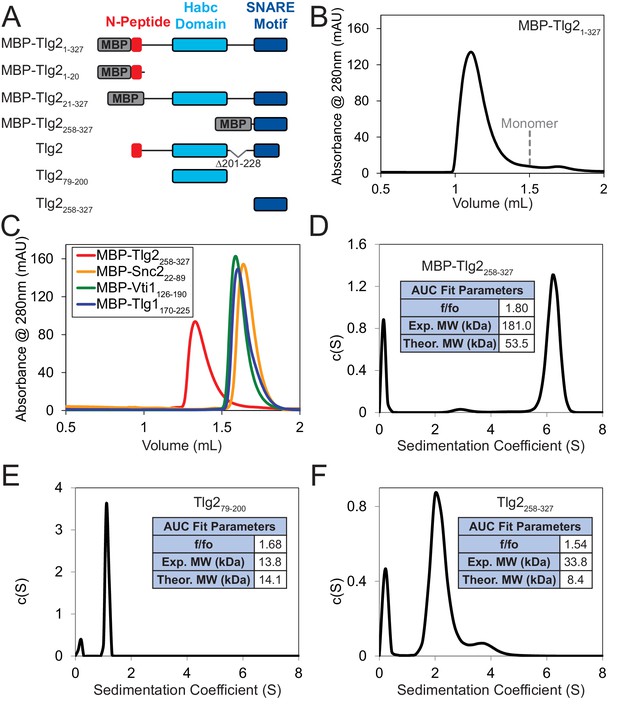
Homo-oligomerization of the Tlg2 SNARE motif.
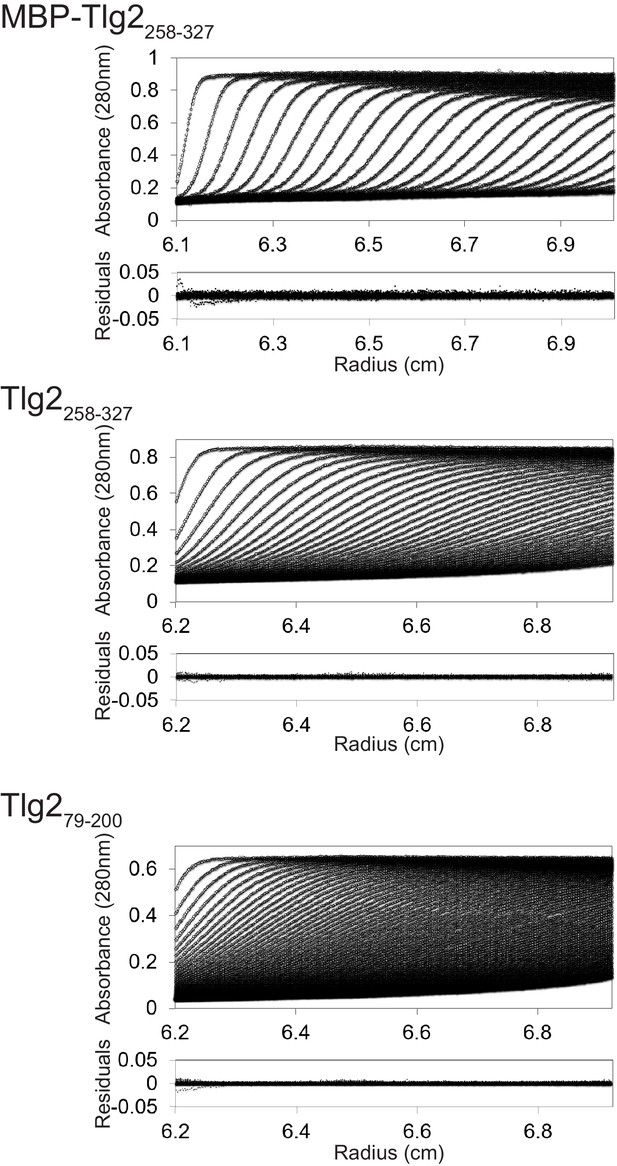
(A) Schematic of the Tlg2 constructs used. (B) Size exclusion chromatography of MBP-Tlg21-327. The predicted position for a monomer, based on size standards, is indicated. (C) Size-exclusion chromatography of the MBP-tagged C. thermophilum SNARE motifs of Tlg2 (Qa-SNARE), Snc2 (R-SNARE), Vti1 (Qb-SNARE), and Tlg1 (Qc-SNARE). (D–F) Sedimentation velocity analytical ultracentrifugation (AUC) and derived parameters (insets). For MBP-Tlg2258-327 (panel D), the experimental molecular weight (181 kDa) falls between those expected for a trimer (161 kDa) and a tetramer (214 kDa); for untagged Tlg2258-327 (panel F), the experimental molecular weight (33.8 kDa) is in excellent agreement with that expected for a tetramer (33.6 kDa). The Habc domain (Tlg279-200) sediments as a monomer (panel E). For data and fits, see Figure 5—figure supplement 1.
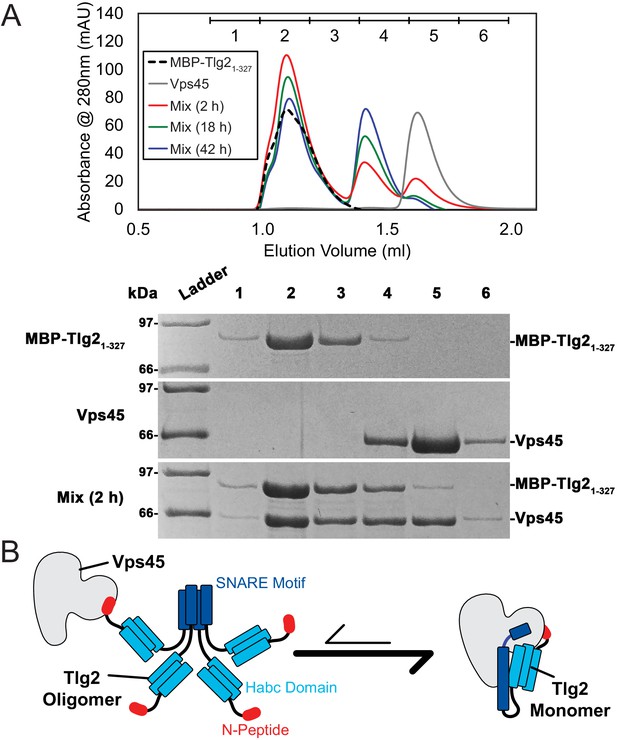
Vps45 rescues Tlg2 from homo-oligomers.
(A) Size-exclusion chromatography (top) of MBP-Tlg21-327, Vps45, and a 1:1 mixture incubated at 20°C for 2, 18, or 42 hr. The indicated fractions were analyzed by SDS-PAGE (bottom). (B) Schematic representation of Vps45-mediated rescue of Tlg2 from homo-oligomers.
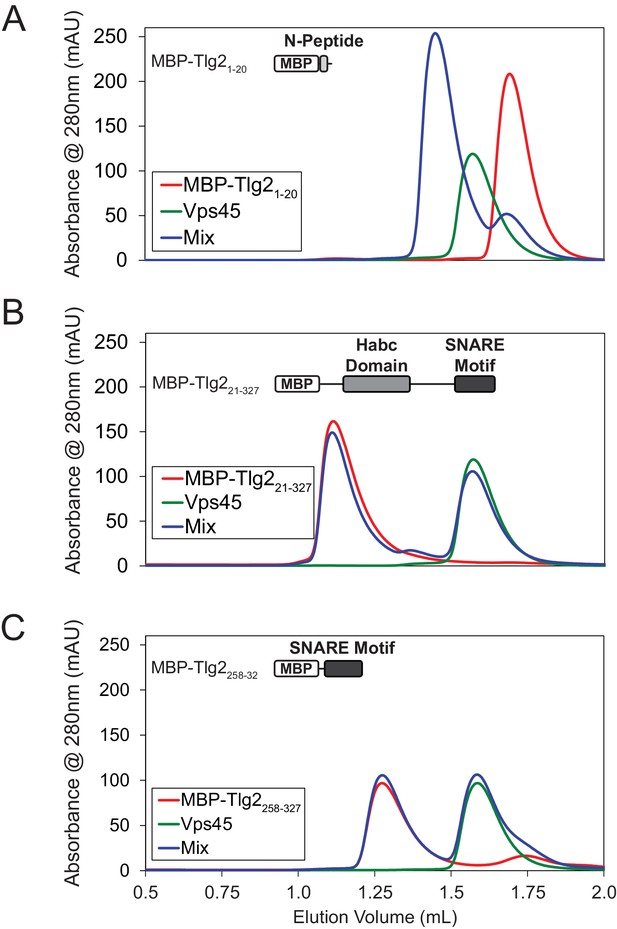
The Tlg2 N-Peptide is required for Vps45 rescue.
Analysis by size-exclusion chromatography of Vps45 binding to (A) MBP-Tlg21-20, (B) MBP-Tlg221-327, and (C) MBP-Tlg2258-327.
Tables
Data Collection and Refinement Statistics.
Values in parenthesis correspond to the highest-resolution shell.
| Vps45 | Vps45–Tlg21-310 | Vps45–Tlg2 | Vps45V306D,F335R–Tlg2 | |
|---|---|---|---|---|
| Data collection | ||||
| Beamline | CHESS (F1) | NSLSII (FMX) | NSLSII (FMX) | NSLSII (FMX) |
| Wavelength (Å) | 0.9782 | 0.9794 | 0.9794 | 0.9793 |
| Space group | P212121 | P21221 | P212121 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 62.49, 93.96, 102.62 | 58.38, 89.43, 209.14 | 58.73, 180.06, 202.09 | 89.14, 58.79, 191.18 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 97.75, 90.0 |
| Resolution (Å) | 35–2.00 (2.03–2.00) | 29–3.88 (4.30–3.88) | 30–2.80 (2.88–2.80) | 30–5.12 (5.73–5.12) |
| Completeness (%) | 99.6 (98.9) | 99.3 (98.1) | 99.8 (98.9) | 98.0 (56.4) |
| Redundancy | 4.6 (4.2) | 13.0 (13.4) | 13.4 (12.3) | 6.2 (5.9) |
| Rmerge | 0.053 (0.564) | 0.246 (2.149) | 0.134 (1.880) | 0.305 (1.222) |
| Rmeas | 0.092 (0.807) | 0.256 (2.234) | 0.144 (1.961) | 0.334 (1.344) |
| <I/σI> | 11.7 (2.0) | 6.9 (1.3) | 13.6 (1.4) | 3.3 (1.1) |
| CC1/2 | 0.940 (0.743) | 0.993 (0.649) | 0.999 (0.731) | 0.980 (0.564) |
| Refinement | ||||
| Resolution (Å) | 35–2.00 (2.04–2.00) | 30–3.90 (4.29–3.90) | 30–2.80 (2.85–2.80) | |
| No. reflections | ||||
| Work | 43025 | 9900 | 51270 | |
| Free | 2268 | 505 | 2676 | |
| Rwork | 0.178 (0.262) | 0.191 (0.291) | 0.194 (0.291) | |
| Rfree | 0.218 (0.300) | 0.242 (0.382) | 0.248 (0.326) | |
| No. atoms | 4551 | 5707 | 11550 | |
| Average B-factor (Å2) | 42.5 | 212.8 | 93.4 | |
| RMSD | ||||
| Bond lengths (Å) | 0.007 | 0.005 | 0.008 | |
| Bond angles (°) | 0.795 | 0.8 | 0.814 | |
| Ramachandran | ||||
| Favored (%) | 98.5 | 94.4 | 95.5 | |
| Outliers (%) | 0.6 | 1.4 | 1.0 | |
| PDB Code | 6XJL | 6XMD | 6XM1 |