Magnesium efflux from Drosophila Kenyon cells is critical for normal and diet-enhanced long-term memory
Figures
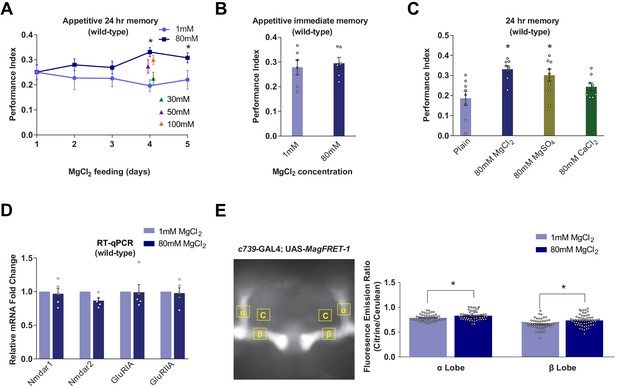
Dietary Mg2+ supplementation enhances Drosophila long-term memory.
(A) Wild-type flies were trained and tested for 24 hr appetitive memory after 1–5 days of ad libitum feeding on food supplemented with Mg2+. Memory was significantly enhanced in flies fed for 4 days with 80 mM MgCl2, as compared to those fed with 1 mM. 80 mM MgCl2 produced marginally higher performance than 50 mM or 100 mM and so was considered optimal (asterisks denote p<0.05, t-test between 1 mM and 80 mM groups for each time point, n = 6–8). (B) 4 days of 80 mM MgCl2 food did not enhance immediate memory. (C) Appetitive 24 hr memory was enhanced by feeding wild-type flies for 4 days with MgCl2 and MgSO4, but not CaCl2. Asterisks denote significant differences (p<0.05, ANOVA, n = 6) between Mg2+ fed and plain groups. (D) RT-qPCR showed no significant differences in glutamate receptor mRNA expression between 1 mM and 80 mM fed flies (t-test, n = 5). (E) c739-GAL4; UAS-MagFRET-1 flies were fed for 4 days on food supplemented with Mg2+. Brains were dissected and fixed and a fluorescence emission ratio measurement (Citrine/Cerulean) was taken as an indicator of [Mg2+]i. The MagFRET signal was significantly greater in the αβ lobes of flies fed with 80 mM MgCl2 than those fed with 1 mM MgCl2 (p<0.05, t-test, n = 52–60). Unless otherwise noted, all data are mean ± standard error of mean (SEM). Asterisks denote significant differences (p<0.05), individual data points displayed as open circles.
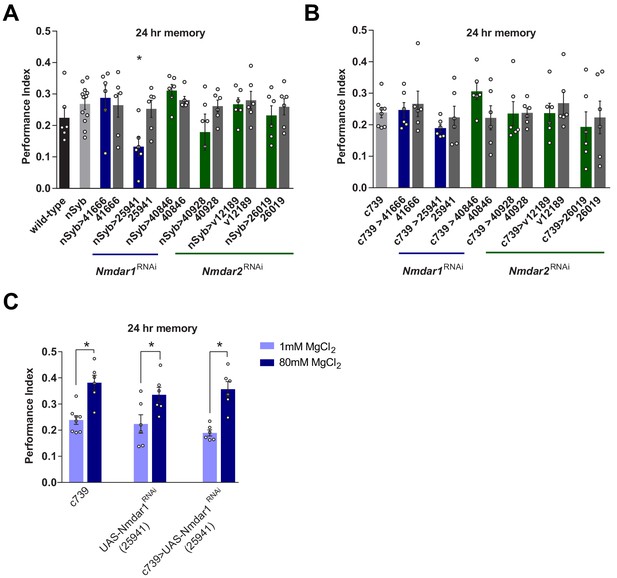
Knockdown of N-methyl-D-aspartate glutamate receptor (NMDAR) in mushroom bodies does not impair Mg2+-enhanced memory.
(A) Expressing UAS-Nmdar1RNAi (BDSC line 25941) in all neurons with nSyb-GAL4 significantly impaired 24 hr memory (p<0.05, ANOVA, n = 6–12). (B) Expressing UAS-Nmdar1RNAi (BDSC line 25941) in αβ Kenyon cells (KCs) with c739-GAL4 did not alter memory (ANOVA, n = 6–8). (F). Mg2+ feeding enhanced LTM in flies expressing UAS-Nmdar1RNAi (BDSC line 25941) in αβ KCs and also in controls (p<0.05, t-test, n = 6–8). Data are mean ± SEM. Individual data points are plotted as circles.
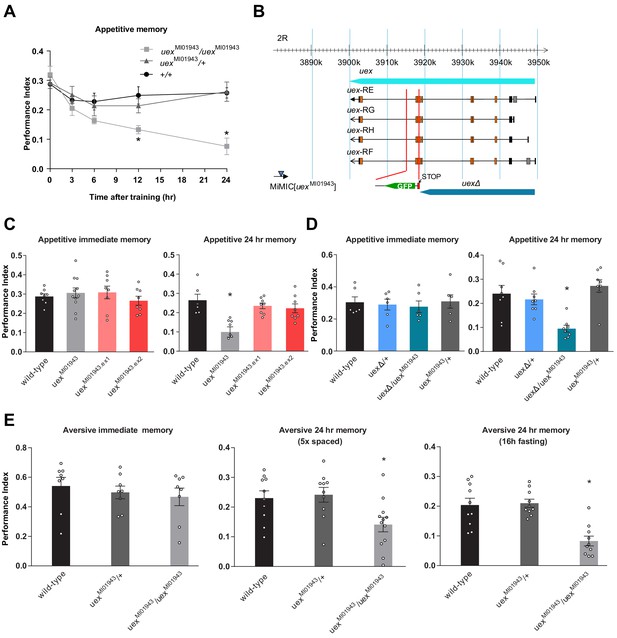
uexMI01943 mutant flies have defective long-term memory (LTM).
(A) Appetitive memory retention was tested at various times after training. Flies homozygous for uexMI01943 showed a significant defect in memory from 12 hr after training, as compared to the performance of heterozygous uexMI01943/+ and wild-type control flies (p<0.05, ANOVA, n = 6–10). (B) The uex locus lies on chromosome 2R between 3,900,285 and 3,949,425 (light blue bar). The four alternate uex transcripts, uex-RE, uex-RG, uex-RH, and uex-RF, all encode the same protein. The uexMI01943 MiMIC (blue triangle) resides ~17 kb downstream of the uex coding region. The CRISPR/Cas9 edited uexΔ allele replaces a 3047 bp fragment, including Exon 7 of uex with a STOP signal (termination codon in all three reading frames) and a GFP cassette, truncating the uex reading frame (dark blue bar). (C) Precise excision of the uexMI01943 MiMIC restores normal 24 hr memory to uexMI01943.ex1 and uexMI01943.ex2 flies (p<0.05, ANOVA, n = 8–11). (D) uexΔ fails to complement the 24 hr memory defect of uexMI01943 (p<0.05, ANOVA, n = 6–8). (E) Flies homozygous for uexMI01943 showed a significant defect in aversive LTM, as compared to the performance of heterozygous uexMI01943/+ and wild-type control flies (p<0.05, ANOVA, n = 8–12). An LTM defect was also observed following five cycles of aversive spaced training and a 16 hr fasting facilitated one-cycle training protocol. Immediate aversive memory was unaffected in uexMI01943 homozygous mutant flies.
-
Figure 2—source data 1
Table of sugar and olfactory sensory acuity controls for all behavioral experiments in this manuscript.
- https://cdn.elifesciences.org/articles/61339/elife-61339-fig2-data1-v2.xlsx
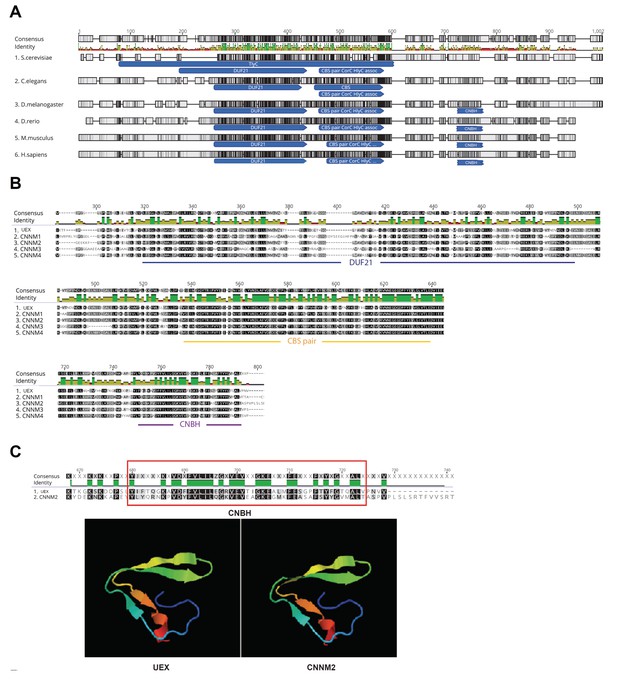
Conservation of UEX with its orthologs.
(A) Alignment of the protein sequences of UEX and its orthologs. The DUF21 and CBS pair domains are highly conserved. Accession numbers for these protein sequences are: 1.NP_014581.1 (Mam3p in Saccharomyces cerevisiae), 2.NP_503052.1 (CNNM-1 in Caenorhabditis elegans), 3.NP_001104391.2 (UEXin Drosophila melanogaster), 4.NP_001138257.1 (CNNM2 in Danio rerio), 5.NPNP_291047.2 (CNNM2 in Mus musculus), 6.NPNP_060119.3 (CNNM2 in Homo sapiens). (B) Focus on the DUF21, CBS pair, and cyclic nucleotide-binding homology (CNBH) domains from UEX and CNNM1-4. Similarity in each aligned sequence is proportional to its gray scale, that is, darker color corresponds to greater similarity. Accession numbers for these protein sequences are: 1.NP_001104391.2 (UEX in Drosophila melanogaster), 2.NP_065081.2 (CNNM1 in Homo sapiens), 3.NP_060119.3 (CNNM2 in Homo sapiens), 4.NP_060093.3 (CNNM3 in Homo sapiens), 5.NP_064569.3 (CNNM4 in Homo sapiens). (C) Protein sequence alignment of the CNBH domains (red rectangle) between UEX and CNNM2 shows high similarity (TM score = 0.98, top panel). The predicted 3D structures of the CNBH domain (lower panel) of UEX (left) and CNNM2 (right) also resemble each other.
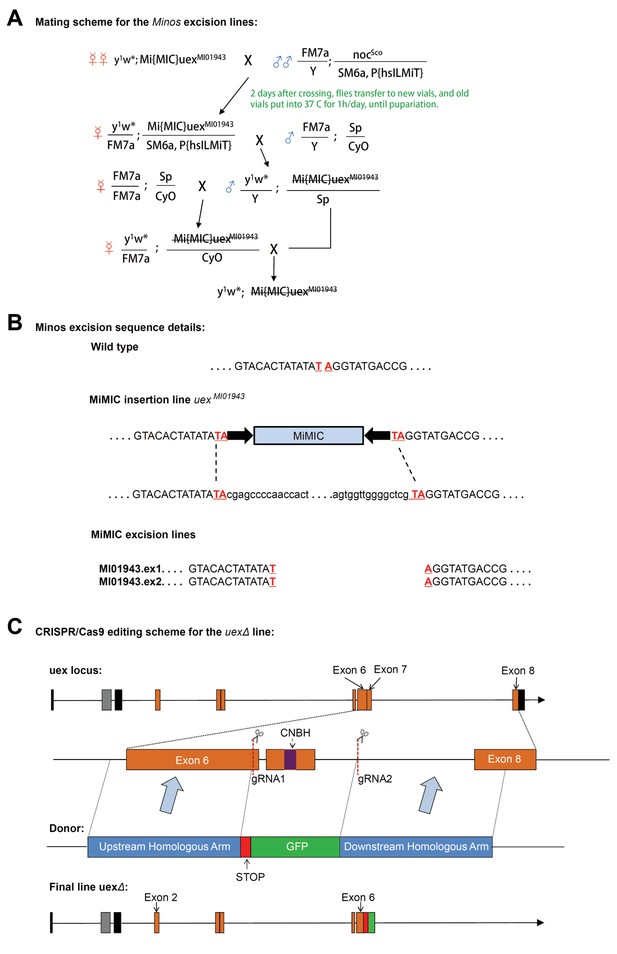
Construction schemes for uex Minos excision and creation of the uexΔ allele.
(A) Mating scheme for Minos excision from uexMI01943 flies. (B) DNA sequence around the MiMIC insertion. Successful excision events were identified in flies with body color reverted to yellow. Genomic DNA from progeny of these flies was extracted, and the uex locus was PCR amplified and sequenced. Two lines (uexMI01943.ex1 and uexMI01943.ex2) were confirmed to be precise excision events, which restored their genomic sequence to the wild-type form. (C) CRISPR-Cas9 editing scheme for the uexΔ lesion. A fragment of 3047 bp spanning part of Exon 6, Exon 7, and part of Exon 8 of the uex locus was replaced by a STOP signal and a GFP cassette. The original sequence was cut by two gRNAs flanking Exon 7, and the new sequence was introduced by homologous recombination using a double-stranded DNA donor. The final uexΔ locus contains a truncated uex reading frame. The GFP cassette facilitates identification of flies with the edited locus.

Knocking down uex expression in αβ Kenyon cells (KCs) impairs LTM.
(A) A uex promoter fragment-GAL4 directs GFP expression in αβc KCs. Anti-GFP immunostained uex-GAL4 (VT23256); UAS-EGFP line. (B) Anti-HA immunostaining of brains harboring the CRISPR/Cas9-edited uex::HA locus shows strong labeling of UEX in all the major subdivisions of the mushroom body (MB). Scale bars 20 µm. (C) RNAi knockdown of uex in all αβ (c739-GAL4) or just αβc (NP7175-GAL4) KCs specifically impaired 24 hr memory. αβs (0770-GAL4) or α′β′ (c305a-GAL4) KC expression had no effect (p<0.05, ANOVA, n = 6–10 for immediate and n = 8–14 for 24 hr memory). (D) Defective LTM was observed if uexRNAi expression was confined to αβ KCs of adult flies using GAL80ts-mediated temporal control. (E) LTM performance was unaffected if the uexRNAi was kept suppressed throughout and (F) LTM performance was restored to normal levels if expression of uexRNAi was re-suppressed for 3 days (p<0.05, ANOVA, n = 6 for immediate and n = 8 for 24 hr memory). (G) Immunostaining shows the effectiveness of uexRNAi. Fluorescence intensity in the αβ and γ lobes of uex::HA flies decreased significantly when UAS-uexRNAi was expressed with MB247-GAL4. Scale bars 20 µm. (H) Quantification of fluorescence intensity in G (p<0.05, t-test, n = 6–8).
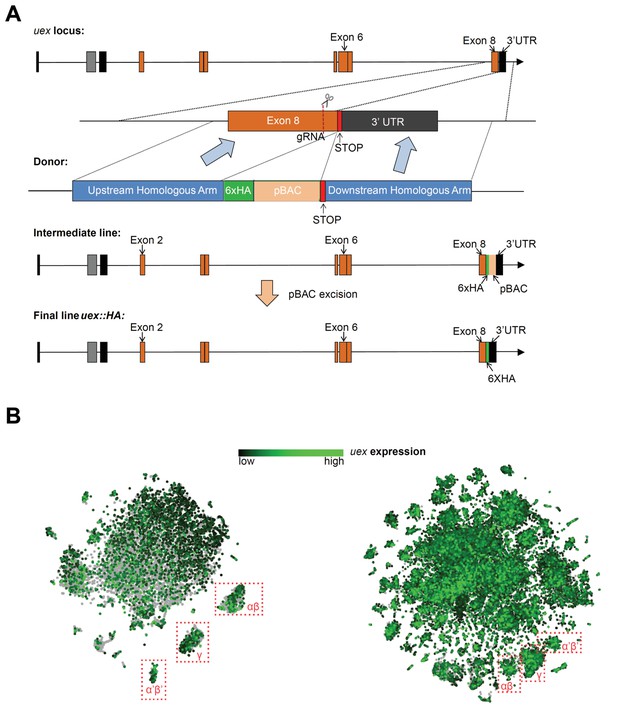
Construction scheme for the uex::HA line and tSNE plots of uex expression.
(A) CRISPR-Cas9 editing scheme for production of the uex::HA locus. A 6xHA tag was inserted immediately before the STOP codon of uex using the 2-step ScarlessDsRed system. In step 1, a pBAC transposon containing a DsRed cassette and a 6xHA tag was inserted in frame with the uex ORF, just prior to the STOP codon, through homologous recombination. In step 2, the pBAC transposon was excised to leave only the 6xHA tag in the final uex::HA locus. (B) tSNE plots show the broad expression of uex in the fly brain, which is consistent with the uex::HA staining result in Figure 3B. Left, data from Croset et al., 2018 and right from Davie et al., 2018 were plotted using the online Scope viewer (http://scope.aertslab.org).
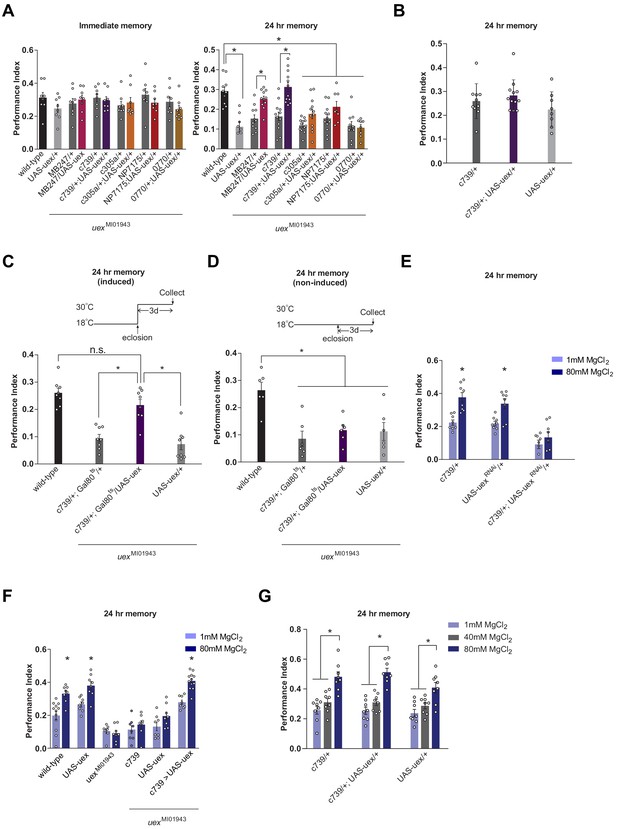
Rescue of the LTM defect in uexMI01943 flies.
Restoring expression of UAS-uex in αβ and γ (MB247-GAL4) or αβ Kenyon cells (KCs) rescued 24 hr memory performance of uexMI01943 flies, whereas expression in αβc, αβs or α′β′ KCs did not (p<0.05, ANOVA and t-test, n = 8–12). (B) Overexpression of UAS-uex in αβ KCs did not enhance 24 hr memory performance in wild-type flies (ANOVA, n = 8–12). (C) Defective LTM was rescued if UAS-uex expression was confined to αβ KCs of adult flies using GAL80ts mediated temporal control (p<0.05, ANOVA, n = 6 for immediate and n = 8 for 24 hr memory) but (D) remained defective if UAS-uex expression was not released. (E) Memory enhancement with dietary Mg2+ is supported by UEX in αβ KCs. Memory of flies expressing UAS-uexRNAi in the αβ KCs cannot be enhanced with Mg2+ feeding (t-test, n = 8). (F) Memory of uexMI01943 mutant flies cannot be enhanced with Mg2+ feeding, but enhancement was restored by expressing UAS-uex in αβ KCs (p<0.05, t-test, n = 8–12). (G) Memory of wild-type flies was not sensitized to Mg2+ enhancement by overexpressing UAS-uex in αβ KCs. Memory was enhanced if the flies were fed with 80 mM MgCl2, but not with suboptimal 40 mM MgCl2 (p<0.05, ANOVA, n = 8).
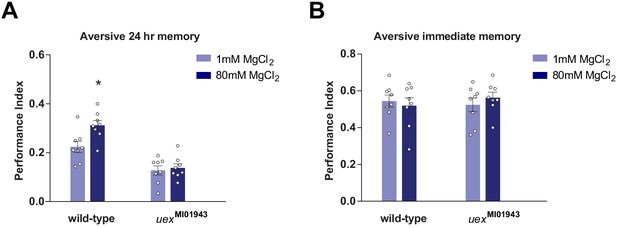
Mg2+ feeding enhanced LTM after aversive spaced training in wild-type but not uexMI01943 mutant flies.
(A) 24 hr memory after five spaced trials of aversive training was enhanced in wild-type, but not uexMI01943 mutant flies, fed for 4 days beforehand with MgCl2 (p<0.05, t-test, n = 8). (B) 4 days of 80 mM MgCl2 food did not enhance immediate aversive memory in either wild-type or mutant flies (t-test, n = 8).
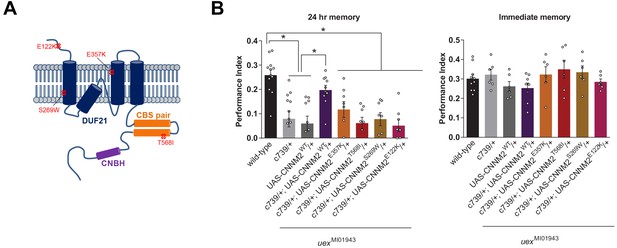
uex encodes an evolutionarily conserved Mg2+ transporter.
(A) Model of CNNM2 protein structure showing clinically relevant point mutations. Adapted and modified from Arjona et al., 2014. (B) Overexpression of wild-type, but not mutant, CNNM2 in αβ Kenyon cells rescues the memory defect of uexMI01943 mutant flies (p<0.05, ANOVA, n = 6–8 for immediate and n = 8–12 for 24 hr memory).
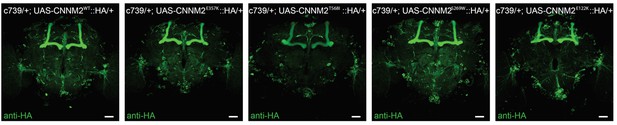
Transgenic expression of mutant variants of CNNM2.
Anti-HA immunostained brains from flies expressing UAS-CNNM2::HA variants driven by c739-GAL4. Genotype from left to right: c739/+; UAS-CNNM2WT::HA/+, c739/+; UAS-CNNM2E357K::HA/+, c739/+; UAS-CNNM2T568I::HA/+, c739/+; UAS-CNNM2S269W::HA/+, c739/+; UAS-CNNM2E122K::HA/+. Scale bar 20 μm.
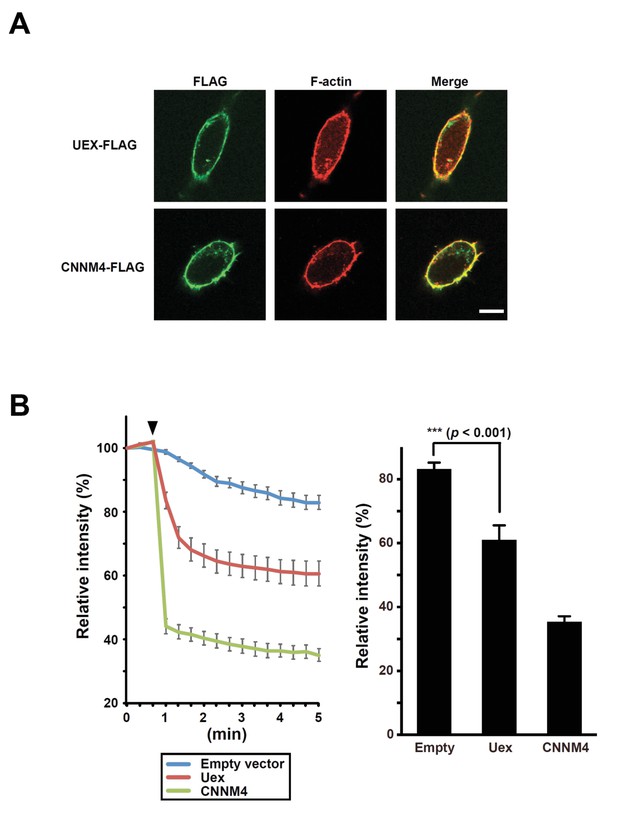
UEX-dependent Mg2+ efflux in HEK293 cells.
(A) HEK293 cells transfected with uex-FLAG or CNNM4-FLAG constructs stained with anti-FLAG antibody (green) and rhodamine-phalloidin (red). Scale bar 10 µm. (B) Mg2+-efflux assay. HEK293 cells transfected with the indicated constructs were loaded with Mg2+ and Magnesium Green, and subjected to Mg2+-depletion at the indicated time point (arrowhead). Line plot indicates the time course of mean relative fluorescence intensities, and bar graph indicates the mean relative fluorescence intensities at 5 min. Data are mean ± SEM (***p<0.001, ANOVA, n = 10).
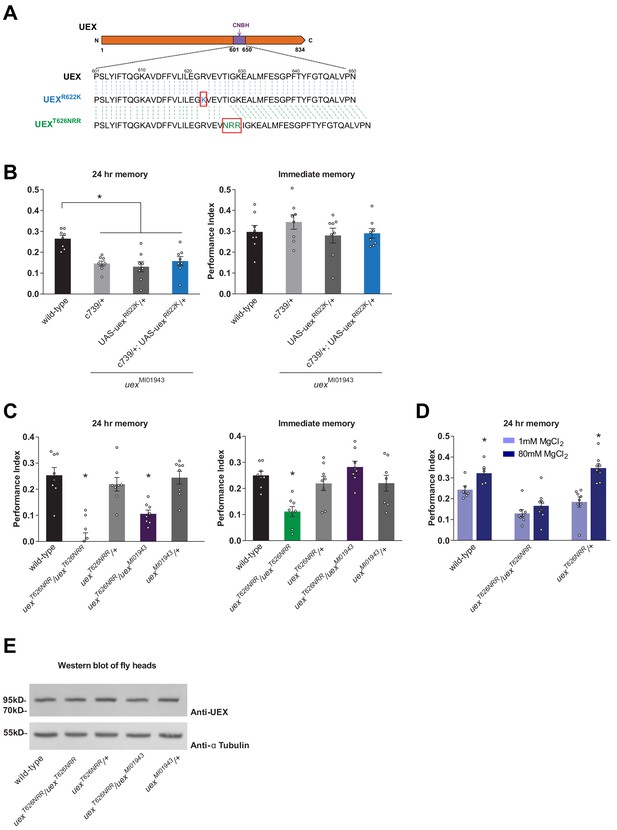
The cyclic nucleotide-binding homology (CNBH) domain of UEX is required for memory.
(A) Schematic showing sequence detail of the CNBH domain in UEX, and the amino acid changes made in uexR622K and uexT626NRR. (B) Expressing a UAS-uexR622K transgene in αβ Kenyon cells did not rescue the LTM defect of uexMI01943 mutant flies (p<0.05, ANOVA, n = 8). Immediate memory was also unaffected. (C) Flies homozygous for uexT626NRR have defective short- and long-term memory, while trans-heterozygous uexT626NRR/uexMI01943 flies only exhibit impaired LTM (*p<0.05, ANOVA, n = 8). (D) Dietary Mg2+ did not enhance memory of homozygous uexT626NRR/ uexT626NRR flies (p<0.05, t-test, n = 8). (E) Western blot analysis of UEX protein expression in fly head extracts. Genotype from left to right: wild-type, uexT626NRR/uexT626NRR, uexT626NRR/+, uexT626NRR/uexMI01943, uexMI01943/+. The blot was first probed with anti-UEX antibody (upper panel), and then stripped and re-probed with anti-Tubulin antibody (lower panel) as a loading control.
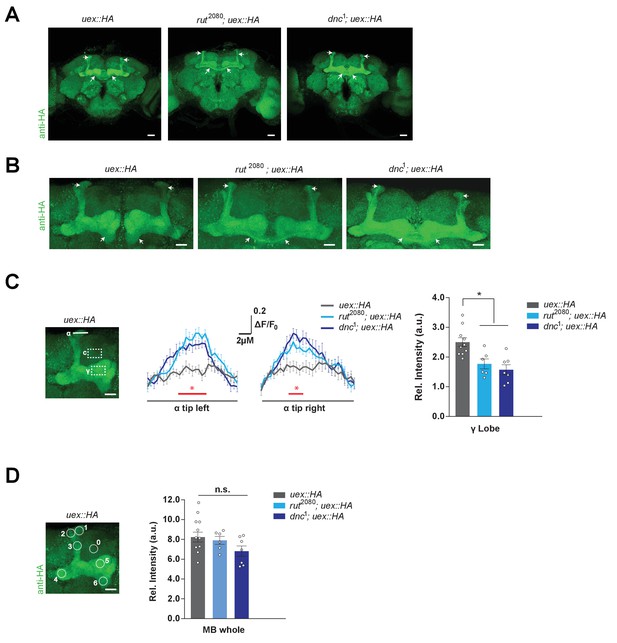
Kenyon cell (KC) uex expression is altered in rutabaga and dunce mutant flies.
(A) Anti-HA stained brains reveal UEX::HA protein localization is altered in rut2080; uex::HA and dnc1; uex::HA flies, becoming more prominent in αβc KCs (arrows). Scale bars 20 µm. (B) Enlarged images of the mushroom bodies (MBs) highlighting αβc KC expression in rut2080 and dnc1 mutant flies, as compared with wild-type uex::HA flies. Scale bars 20 µm. (C) Quantification of fluorescence intensity. Left, micrograph with a measurement line through the α lobe tip and rectangular ROIs for the γ lobe and a control area. Middle, relative fluorescence intensity profiles across the α lobe tip show significantly higher signal in rut2080 and dnc1 mutant flies in the center region occupied by the αβ core KCs (*p<0.05, ANOVA, n = 6–10). Right, the relative intensity in the γ lobe was significantly lower in rut2080 and dnc1 mutant flies, as compared to wild-type controls (*p<0.05, ANOVA, n = 6–10). Scale bars 10 µm. (D) Left, micrograph showing circular ROIs. Right, quantification. Total intensity over all six ROIs on the MBs was not significantly different between the rut2080, dnc1 and wild-type brains (p>0.13; ANOVA, n = 6–10).
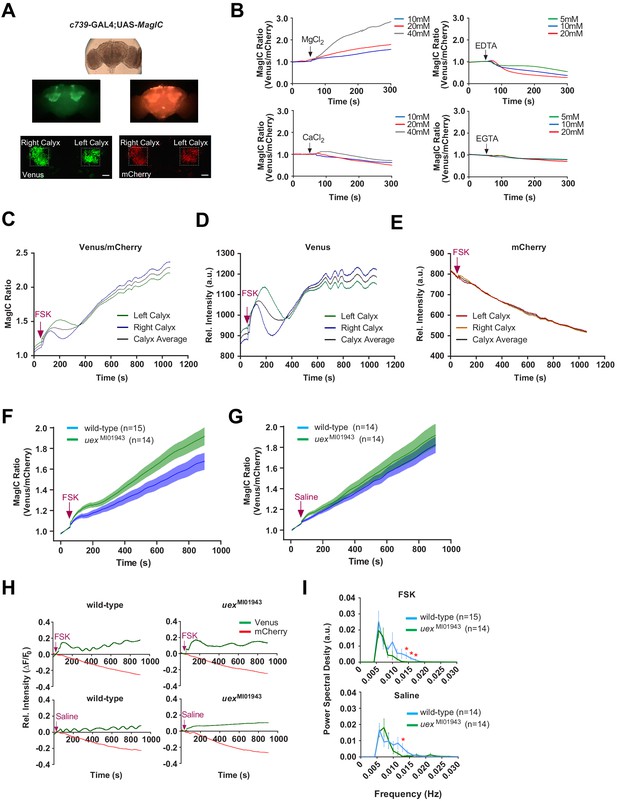
UEX limits a rise in [Mg2+]i and supports a slow oscillation in αβ Kenyon cells (KCs).
(A) Explant fly brain expressing UAS-MagIC driven by c739-GAL4. Upper panel, wide-field phase contrast view; middle panels, fluorescence views of Venus and mCherry channels; lower panel, confocal section at the level of the KC somata showing Venus and mCherry channels. Scale bars 20 µm. (B) MagIC selectively responds to changes in [Mg2+]i in KCs. Traces of MagIC ratio following bath application of 10, 20, or 40 mM MgCl2 or CaCl2; 5, 10, or 20 mM EDTA or EGTA. (C) Representative trace of MagIC ratio following application of FSK shows an initial wave followed by a gradual rise and the development of a slow oscillation. (D) The primary responses result from changes in the Mg2+-sensitive Venus signal. (E) The mCherry signal exhibits a steady decay. (F) FSK-evoked MagIC responses are greater in uex mutant flies. Averaged MagIC responses show that FSK induced a significantly greater increase in uexMI01943 mutant than in wild-type flies. (G). Averaged saline-evoked MagIC responses were not significantly altered in uex mutant flies. (H) Individual Venus (green) and mCherry (red) channel traces showing that the slow oscillation is only evident in the Venus channel of wild-type, but not uex mutant, flies. (I) Power spectral density (PSD) analysis of the time series from 200 to 900 s of all data shows that traces from wild-type flies have significantly more oscillatory activity, centered around 0.015 Hz, than those from uex mutant flies.
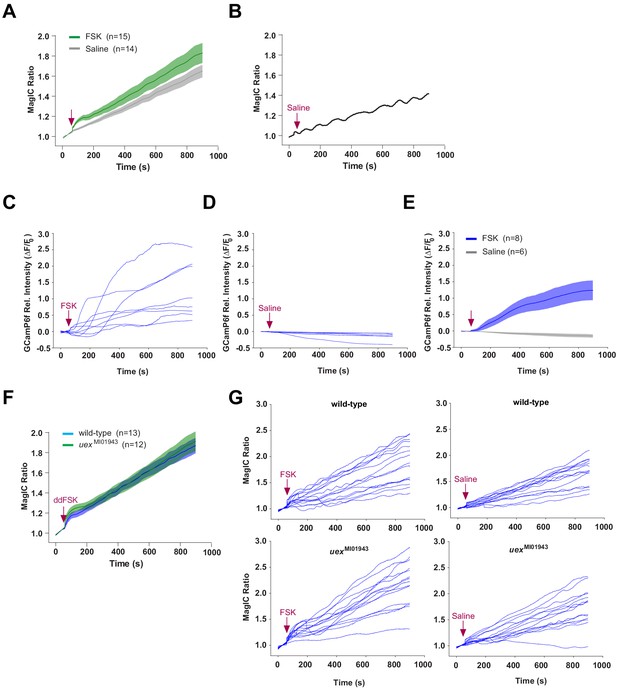
Individual traces for MagIC and GCaMP imaging.
(A) Application of 30 μM FSK induced a MagIC ratio response than saline application in wild-type flies (c739/+; UAS-MagIC/+). (B) A sample trace from (A) showing a fluctuating signal following saline application. (C–E) Traces of relative intensity (ΔF/F0) of GCaMP6f signal from c739/+; UAS-GCaMP6f/+ flies. Slow oscillations were not observed. (C) Individual GCaMP6f traces following FSK application. (D) Individual GCaMP6f traces following saline application. (E) Average GCaMP6f response following FSK application is significantly stronger than that after saline. (F) The average MagIC ratio response following ddFSK application is not sensitive to uex mutation. Responses of wild-type (c739/+; UAS-MagIC/+) and uex mutant flies (c739, uexMI01943/uexMI01943; UAS-MagIC/+) are indistinguishable. (G) Individual MagIC ratio traces from wild-type (c739/+; UAS-MagIC/+, upper panels) and uex mutant (c739, uexMI01943/uexMI01943, lower panels) flies following 30 μM FSK (left panels) or saline (right panels) application.
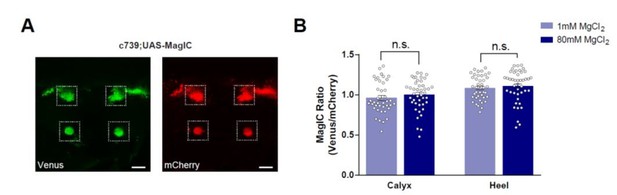
No significant [Mg2+]i changes were detected in the MBs of flies fed with high [Mg2+] food when measured using MagIC in vivo.
(A) Live fly brain expressing UAS-MagIC driven by c739-GAL4. Confocal section at the level of the KC somata and heel region showing Venus (left) and mCherry (right) channels. Scale bars 20μm. (B) c739-GAL4; UAS-MagIC flies were fed for 4 days on food supplemented with Mg2+. Brains were scanned in vivo under confocal microscope. A fluorescence ratio measurement (Venus/mCherry) was taken as an indicator of [Mg2+]i. No significant difference in MagIC ratio was detected between flies fed with 80mM MgCl2 with those fed with 1mM MgCl2 (t-test, n=41-43).
Videos
UEX promotes Mg2+-efflux from HEK293 cells.
Representative movies showing Mg2+-efflux from HEK293 cells transfected with different expression vectors. Imaging protocol is described in Yamazaki et al., 2013. The cells indicated with asterisks in the first frame of each movie are the cells expressing the anti-FLAG immunostained CNNM4 or UEX, which were identified after each live-imaging experiment. Empty vector control is shown in the upper left. The fluorescence signal of CNNM4-FLAG and UEX-FLAG expressing cells decreases rapidly when extracellular Mg2+ is depleted, which was performed between the third and fourth frames in each movie.
Expression of UEX in a wild-type Drosophila brain.
Confocal Z-stack of a uex::HA fly brain stained with anti-HA antibody.
Expression of UEX in a rut2080Drosophila brain.
Confocal Z-stack of a brain from a rut2080; uex::HA fly stained with anti-HA antibody. The αβc Kenyon cells label more prominently than in the wild-type uex::HA brain in Video 2.
Expression of UEX in a dnc1Drosophila brain.
Confocal Z-stack of a brain from a dnc1; uex::HA fly stained with anti-HA antibody. The αβc Kenyon cells label more prominently than in the wild-type uex::HA brain in Video 2.
KC-expressed MagIC responds to Mg2+ application.
Confocal time-series recording from a c739/+; UAS-MagIC/+ fly brain shows an increase in Venus, but not mCherry, fluorescence signal in response to 20 mM MgCl2 application.
KC-expressed MagIC responds to Mg2+ chelation.
Confocal time-series recording from a c739/+; UAS-MagIC/+ fly brain shows a strong decrease in Venus and a weak decrease in mCherry fluorescence signal in response to 10 mM EDTA application.
KC-expressed MagIC reveals slow oscillation of intracellular Mg2+.
Confocal time-series recording from a c739/+; UAS-MagIC/+ fly brain shows a slow oscillation in Venus, but not mCherry, fluorescence signal in response to 30 μM forskolin.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (Drosophila melanogaster) | Canton-S | Originally from W.G.Quinn lab | Canton-S | Waddell Lab stock |
| Genetic reagent (D. melanogaster) | UAS-EGFP | Bloomington Drosophila Stock Center | RRID:BDSC_5431 | |
| Genetic reagent (D. melanogaster) | c739-GAL4 | McGuire et al., 2001 | c739-GAL4 | Lab stock |
| Genetic reagent (D. melanogaster) | c305a-GAL4 | Krashes et al., 2007 | c305a-GAL4 | Lab stock |
| Genetic reagent (D. melanogaster) | NP7175-GAL4 | Tanaka et al., 2004 | NP7175-GAL4 | Lab stock |
| Genetic reagent (D. melanogaster) | 0770-GAL4 | Gohl et al., 2011 | 0770-GAL4 | Lab stock |
| Genetic reagent (D. melanogaster) | MB247-GAL4 | Zars et al., 2000 | MB247-GAL4 | Lab stock |
| Genetic reagent (D. melanogaster) | nSyb-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_51635 | Gift from J. Simpson |
| Genetic reagent (D. melanogaster) | elav-GAL4 | Bloomington Drosophila Stock Center | RRID:BDSC_8765 | |
| Genetic reagent (D. melanogaster) | tubPGAL80ts | McGuire et al., 2003 | tubP-GAL80ts | Lab stock |
| Genetic reagent (D. melanogaster) | UAS-Nmdar1RNAi | Bloomington Drosophila Stock Center | RRID:BDSC_25941 | |
| Genetic reagent (D. melanogaster) | nos-Cas9.P | Bloomington Drosophila Stock Center | RRID:BDSC_54591 | |
| Genetic reagent (D. melanogaster) | nos-Cas9(X) | Fly Stocks of National Institute of Genetics | CAS0002 | |
| Genetic reagent (D. melanogaster) | lig4 KO vasa-Cas9 | Zimmer et al., 2016 | lig4 KO vasa-Cas9 | Gift from C. Zimmer |
| Genetic reagent (D. melanogaster) | PhsILMiT | Bloomington Drosophila Stock Center | RRID:BDSC_24613 | |
| Genetic reagent (D. melanogaster) | rut2080 | Han et al., 1992 | rut2080 | Lab stock |
| Genetic reagent (D. melanogaster) | dnc1 | Dudai et al., 1976 | dnc1 | Lab stock |
| Genetic reagent (D. melanogaster) | uexMI01943 | Bloomington Drosophila Stock Center | RRID:BDSC_32805 | |
| Genetic reagent (D. melanogaster) | uexNC1 | Bloomington Drosophila Stock Center | RRID:BDSC_7176 | |
| Genetic reagent (D. melanogaster) | UAS-uexRNAi | Bloomington Drosophila Stock Center Perkins et al., 2015 | RRID:BDSC_36116 | |
| Genetic reagent (D. melanogaster) | uex-GAL4 | Vienna Drosophila Resource Center | VT23256 | |
| Genetic reagent (D. melanogaster) | UAS-GCaMP6f | Bloomington Drosophila Stock Center | RRID:BDSC_42747 | |
| Genetic reagent (D. melanogaster) | uexMI01943.ex1 | This study | uexMI01943.ex1 | See Methods and Figure 2—figure supplement 2A and B |
| Genetic reagent (D. melanogaster) | uexMI01943.ex2 | This study | uexMI01943.ex2 | See Methods and Figure 2—figure supplement 2A and B |
| Genetic reagent (D. melanogaster) | uexΔ | This study | uexΔ | See Methods and Figure 2—figure supplement 2C |
| Genetic reagent (D. melanogaster) | uex::HA | This study | uex::HA | See Methods and Figure 3—figure supplement 1 |
| Genetic reagent (D. melanogaster) | uexT626NRR | This study | uexT626NRR | See Methods and Figure 6A |
| Genetic reagent (D. melanogaster) | UAS-uex | This study | UAS-uex | See Methods |
| Genetic reagent (D. melanogaster) | UAS-uexR622K | This study | UAS-uexR622K | See Methods and Figure 6A |
| Genetic reagent (D. melanogaster) | UAS-CNNM2WT | This study | UAS-CNNM2WT | See Methods and Figure 5A |
| Genetic reagent (D. melanogaster) | UAS-CNNM2E357K | This study | UAS-CNNM2E357K | See Methods and Figure 5A |
| Genetic reagent (D. melanogaster) | UAS-CNNM2T568I | This study | UAS-CNNM2T568I | See Methods and Figure 5A |
| Genetic reagent (D. melanogaster) | UAS-CNNM2S269W | This study | UAS-CNNM2S269W | See Methods and Figure 5A |
| Genetic reagent (D. melanogaster) | UAS-CNNM2E122K | This study | UAS-CNNM2E122K | See Methods and Figure 5A |
| Genetic reagent (D. melanogaster) | UAS-MagFRET-1 | This study | UAS-MagFRET-1 | See Methods |
| Genetic reagent (D. melanogaster) | UAS-MARIO | This study | UAS-MARIO | See Methods |
| Genetic reagent (D. melanogaster) | UAS-MagIC | This study | UAS-MagIC | See Methods |
| Antibody | Anti-GFP (Rabbit polyclonal) | Invitrogen | Cat# A-11122, RRID:AB_221569 | IF (1:250) |
| Antibody | Anti-HA (Rabbit monoclonal) | New England Biolabs | Cat# 3724T | IF (1:250) |
| Antibody | Anti-FLAG (Rabbit polyclonal) | Sigma-Aldrich | Cat# F-7425, RRID:AB_439687 | IF (1:250) |
| Antibody | Anti-UEX (Rabbit polyclonal) | Eurogentec | Cat# ZGB-15047 | WB (1:2000) |
| Antibody | Anti-Tubulin (Mouse monoclonal) | Sigma-Aldrich | Cat# T-6199, RRID:AB_477583 | WB (1:2000) |
| Antibody | Anti-rabbit IgG (Alexa 488 goat polyclonal) | Invitrogen | Cat# A-11034, RRID:AB_2576217 | IF (1:250) |
| Antibody | Anti-rabbit IgG (HRP-conjugated goat polyclonal) | Thermo Fisher | Cat# 32260, RRID:AB_1965959 | WB (1:5000) |
| Antibody | Anti-mouse IgG (HRP-conjugated goat polyclonal) | Thermo Fisher | Cat# 32230, RRID:AB_1965958 | WB (1:5000) |
| Recombinant DNA reagent | pUAST-uex (plasmid) | This study | pUAST vector containing uex cDNA | |
| Recombinant DNA reagent | pUAST- uexR622K(plasmid) | This study | pUAST vector containing uexR622KcDNA | |
| Recombinant DNA reagent | pUAST- CNNM2WT(plasmid) | This study | pUAST vector containing mouse CNNM2WT cDNA | |
| Recombinant DNA reagent | pUAST- CNNM2E122K(plasmid) | This study | pUAST vector containing mouse CNNM2E122K cDNA | |
| Recombinant DNA reagent | pUAST- CNNM2E357K(plasmid) | This study | pUAST vector containing mouse CNNM2E357K cDNA | |
| Recombinant DNA reagent | pUAST- CNNM2S269W(plasmid) | This study | pUAST vector containing mouse CNNM2S269W cDNA | |
| Recombinant DNA reagent | pUAST- CNNM2T568I(plasmid) | This study | pUAST vector containing mouse CNNM2T568I cDNA | |
| Recombinant DNA reagent | pJFRC-MUH- MagFRET-1 (plasmid) | This paper | pJFRC-MUH vector containing MagFRET-1 CDS | |
| Recombinant DNA reagent | pTW-MARIO (plasmid) | This paper | pTW vector containing MARIO CDS | |
| Recombinant DNA reagent | pTW-MagIC (plasmid) | This paper | pTW vector containing MagIC CDS | |
| Recombinant DNA reagent | pCFD3-dU6:3gRNA vector | Addgene | RRID:Addgene_49410 | |
| Recombinant DNA reagent | pCMVMagFRET-1 | Addgene | RRID:Addgene_50742 | |
| Recombinant DNA reagent | pScarlessHD-2xHA-DsRed | Addgene | 80822 | Gift to Addgene from Kate O’Connor-Giles |
| Recombinant DNA reagent | gRNA constructs for uexΔ | GenetiVision | Y17.C253.Q002 | Generated by GenetiVision for this study |
| Recombinant DNA reagent | Donor construct for uexΔ | GenetiVision | Y17.C253.Q002 | Generated by GenetiVision for this study |
| Recombinant DNA reagent | gRNA construct for uex::HA | WellGenetics | WG-16107 gRNA | Generated by WellGenetics for this study |
| Recombinant DNA reagent | Donor construct for uex::HA | WellGenetics | PWG1521 pUC57-Kan-16107 donor | Generated by WellGenetics for this study |
| Recombinant DNA reagent | gRNA construct for uexT626NRR | This study | ||
| Sequence-based reagent | Gipc1_F | This study | PCR primers | GGGAAAGGACAAAAGGAACCC |
| Sequence-based reagent | uex CDS, Forward | This study | PCR primers | ATCGCCGCGGATGAACACATATTTCATATCATTTATTAC |
| Sequence-based reagent | uex CDS, Reverse | This study | PCR primers | ATCGCTCGAGTTAGGGCTTACTTTGCTTGCTC |
| Sequence-based reagent | uexR622K, fragment 1, Forward | This study | PCR primers | ATGAACACATATTTCATATCATTTATTACAATAATTA |
| Sequence-based reagent | uexR622K, fragment 1, Reverse | This study | PCR primers | GTGACTTCTACTTTACCCTCCAAAATAAG |
| Sequence-based reagent | uexR622K, fragment 2, Forward | This study | PCR primers | GTACTTATTTTGGAGGGTAAAGTAGAAGTCACAATTGGC |
| Sequence-based reagent | uexR622K, fragment 2, Reverse | This study | PCR primers | TTAGGGCTTACTTTGCTTGCTCTCGAATTTG |
| Sequence-based reagent | CNNM2 cDNA, Forward | This study | PCR primers | ATCGCTCGAGATGATTGGCTGTGGCGCTTGTG |
| Sequence-based reagent | CNNM2 cDNA, Reverse | This study | PCR primers | ATCGTCTAGACTATGCGTAGTCTGGCACGTCG |
| Sequence-based reagent | MagFRET-1 CDS, Forward | This study | PCR primers | ATCGCTCGAGGCCACCATGGGCCATATGGTGAGC |
| Sequence-based reagent | MagFRET-1 CDS, Reverse | This study | PCR primers | ATCGTCTAGATTACTTGTACAGCTCGTCCATGCCGAG |
| Sequence-based reagent | MagIC CDS, Forward | This study | PCR primers | CACCAGGATGGCCATCATCAAGGAGTTCATG |
| Sequence-based reagent | MagIC CDS, Reverse | This study | PCR primers | CCGTTACTCGATGTTGTGGCGGATCTTGAA |
| Sequence-based reagent | MARIO CDS, Forward | This study | PCR primers | CACCAGGGCTTGGTACCGAGCTCGGAT |
| Sequence-based reagent | MARIO CDS, Reverse | This study | PCR primers | CCGCCACTGTGCTGGATATCTGCAGAATTCTTA |
| Sequence-based reagent | Inverse PCR of uexMI01943, Set 1, Forward | This study | PCR primers | ATGATAGTAAATCACATTACG3 |
| Sequence-based reagent | Inverse PCR of uexMI01943, Set 1, Reverse | This study | PCR primers | CAATAATTTAATTAATTTCCC3 |
| Sequence-based reagent | Inverse PCR of uexMI01943, Set 2, Forward | This study | PCR primers | CAAAAGCAACTAATGTAACGG |
| Sequence-based reagent | Inverse PCR of uexMI01943, Set 2, Reverse | This study | PCR primers | TTGCTCTTCTTGAGATTAAGGTA |
| Sequence-based reagent | qPCR of Nmdar1, Forward | This study | PCR primers | ATCCCTCGACGTACAACATTGG |
| Sequence-based reagent | qPCR of Nmdar1, Reverse | This study | PCR primers | GAGGTGCTTTATTGTGGTGCTAA |
| Sequence-based reagent | qPCR of Nmdar2, Forward | This study | PCR primers | ACTGCTGGGCAACCTGAG |
| Sequence-based reagent | qPCR of Nmdar2, Reverse | This study | PCR primers | GATTTCCGTCTTGTACGACCA |
| Sequence-based reagent | qPCR of GluRIA, Forward | This study | PCR primers | TTTTCTGGCCGGAATTTAGTT |
| Sequence-based reagent | qPCR of GluRIA, Reverse | This study | PCR primers | CCTGTTCGAAGATTGCACCT |
| Sequence-based reagent | qPCR of GluRIIA, Forward | This study | PCR primers | AACCACCAGATGTCCATCAATG |
| Sequence-based reagent | qPCR of GluRIIA, Reverse | This study | PCR primers | GAAGGTGCGCCACTCATAGT |
| Sequence-based reagent | qPCR of Gapdh, Forward | This study | PCR primers | CTTCTTCAGCGACACCCATT |
| Sequence-based reagent | qPCR of Gapdh, Reverse | This study | PCR primers | ACCGAACTCGTTGTCGTACC |
| Sequence-based reagent | qPCR of Tbp, Forward | This study | PCR primers | ACAGGGGCAAAGAGTGAGG |
| Sequence-based reagent | qPCR of Tbp, Reverse | This study | PCR primers | CTTAAAGTCGAGGAACTTTGCAG |
| Sequence-based reagent | qPCR of Ef1α100E, Forward | This study | PCR primers | GCGTGGGTTTGTGATCAGTT |
| Sequence-based reagent | qPCR of Ef1α100E, Reverse | This study | PCR primers | GATCTTCTCCTTGCCCATCC |
| Sequence-based reagent | uexMI01943 Minos excision, Forward | This study | PCR primers | GTGCCAGACCACTGCACCATC |
| Sequence-based reagent | uexMI01943 Minos excision, Reverse | This study | PCR primers | CCGTACCTATGTCGATTCCCACCTC |
| Sequence- based reagent | uexΔ lesion | This study | CRISPR gRNA1 | ACTTTCCAGTACCTTAGCAC [TGG] |
| Sequence-based reagent | uexΔ lesion | This study | CRISPR gRNA2 | GTCACTCCTCGCGGTACCAC [TGG] |
| Sequence-based reagent | Verification of uexΔ, set 1, Forward | This study | PCR primers | AAGACATGGATTGGCGATTG |
| Sequence-based reagent | Verification of uexΔ, set 1, Reverse | This study | PCR primers | AAGTCGCCATGTTGGATCG |
| Sequence-based reagent | Verification of uexΔ, set 2, Forward | This study | PCR primers | CTGGGCATGGATGAGCTGTA |
| Sequence-based reagent | Verification of uexΔ, set 2, Reverse | This study | PCR primers | CTGGAGCGCAACAATTCTCT |
| Sequence-based reagent | uexT626NRR lesion | This study | CRISPR gRNA | GGTCGTGTAGAAGTCACAAT [TGG] |
| Sequence-based reagent | uexT626NRR lesion | This study | ssODN | GTCTTTATATTTTCACTCAAGGAAAAGCTGTCGACTTTTTTGTACTTATTTTGGAGGGTAAAGTAGAAGTCACAATTGCCAAGGAAGCGCTTATGTTTGAAAGCGGGCCCTTTACTTATT |
| Sequence-based reagent | Screen for uexT626NRR, set 1, Forward | This study | PCR primers | GGTTATTCTCGTATTCCAGTGTACGATGG |
| Sequence-based reagent | Screen for uexT626NRR, set 1, Reverse | This study | PCR primers | GAGATTCAGCATCTAGAGACAAAGACGCAG |
| Sequence-based reagent | Screen for uexT626NRR, set 2, Forward | This study | PCR primers | CGGTCGGGTTAGTTACTCTGGAAGATG |
| Sequence-based reagent | Screen for uexT626NRR, set 2, Reverse | This study | PCR primers | CGCGTAAGCATTCACACTAGCTGAGTAAC |
| Sequence-based reagent | Screen for uexT626NRR, set 3, Forward | This study | PCR primers | GGCTACTTTCCAGTACCTTAGCACTGG |
| Sequence-based reagent | Screen for uexT626NRR, set 3, Reverse | This study | PCR primers | CGCGTAAGCATTCACACTAGCTGAGTAAC |
| Sequence-based reagent | Screen for uexT626NRR, set 4, Forward | This study | PCR primers | CGGAGGTTACTCAATCAAGACGTGTTTC |
| Sequence-based reagent | Screen for uexT626NRR, set 4, Reverse | This study | PCR primers | CGCGTAAGCATTCACACTAGCTGAGTAAC |
| Commercial assay or kit | Direct-zol RNA MiniPrep | Cambridge Bioscience | R2050 | |
| Commercial assay or kit | SuperScript III First-Strand Synthesis SuperMix | Invitrogen | 18080400 | |
| Commercial assay or kit | LightCycler 480 SYBR Green I Master | Roche | 04707516001 | |
| Commercial assay or kit | pENTR/D-TOPO cloning kit | Invitrogen | K240020 | |
| Commercial assay or kit | Gateway LR ClonaseTM II Enzyme mix | Invitrogen | 11791020 | |
| Commercial assay or kit | NEBuilder HiFi DNA Assembly Master Mix | New England Biolabs | E2621S | |
| Commercial assay or kit | ExoSAP-IT PCR Product Cleanup Reagent | Thermo Fisher | 78201 | |
| Chemical compound, drug | MgCl2 | Sigma-Aldrich | M1028 | |
| Chemical compound, drug | MgSO4 | Sigma-Aldrich | M3409 | |
| Chemical compound, drug | CaCl2 | Sigma-Aldrich | 21115 | |
| Chemical compound, drug | KCl | Sigma-Aldrich | 60142 | |
| Chemical compound, drug | EDTA | Sigma-Aldrich | 324504 | |
| Chemical compound, drug | Forskolin | Sigma-Aldrich | F6886 | |
| Chemical compound, drug | 1,9-Dideoxyforskolin | Sigma-Aldrich | D3658 | |
| Chemical compound, drug | Magnesium Green | Invitrogen | M3733 | |
| Chemical compound, drug | Sucrose | Sigma-Aldrich | S0389 | |
| Chemical compound, drug | Mineral oil | Sigma-Aldrich | M5904 | |
| Chemical compound, drug | 3-Octanol | Sigma-Aldrich | 218405 | |
| Chemical compound, drug | 4-Methyl-Cyclohexanol | Sigma-Aldrich | 66360 | |
| Chemical compound, drug | Paraformaldehyde | Fisher Scientific | 15713 | |
| Chemical compound, drug | Phosphate buffered saline tablets | Fisher Scientific | 1282–1680 | |
| Chemical compound, drug | Triton X-100 | Sigma-Aldrich | T9284 | |
| Chemical compound, drug | Vectashield antifade mounting medium | Vector Laboratories | H1000 | |
| Chemical compound, drug | TRIzol RNA isolation reagents | Thermo Fisher | 15596018 | |
| Software, algorithm | Prism 6.0 | GraphPad | RRID:SCR_002798 | https://www.graphpad.com |
| Software, algorithm | SnapGene Viewer 4.1 | SnapGene | RRID:SCR_015052 | https://www.snapgene.com |
| Software, algorithm | Geneious R10.2 | Geneious | RRID:SCR_010519 | https://www.geneious.com |
| Software, algorithm | Fiji/ImageJ 1.4 | NIH | RRID:SCR_002285 | https://imagej.nih.gov |
| Software, algorithm | MATLAB R2017b | Mathworks | RRID:SCR_013499 | https://www.mathworks.com |
| Software, algorithm | Python 3.7 | Python Software Foundation | RRID:SCR_008394 | https://www.python.org |
| Software, algorithm | Visual Studio Code 1.42 | Microsoft | https://code.visualstudio.com | |
| Software, algorithm | Adobe Illustrator CC | Adobe Systems | RRID:SCR_010279 | https://www.adobe.com |
| Software, algorithm | InterPro | EMBL-EBI | RRID:SCR_005829 | https://www.ebi.ac.uk/interpro |
| Software, algorithm | Phyre2 | Genome3D | http://www.sbg.bio.ic.ac.uk/~phyre2 | |
| Software, algorithm | TM-align | Zhang Lab | https://zhanglab.ccmb.med.umich.edu/TM-align/ | |
| Software, algorithm | Chimera 1.11 | UCSF | RRID:SCR_004097 | https://www.cgl.ucsf.edu/chimera/ |





