Somatic aging pathways regulate reproductive plasticity in Caenorhabditis elegans
Figures
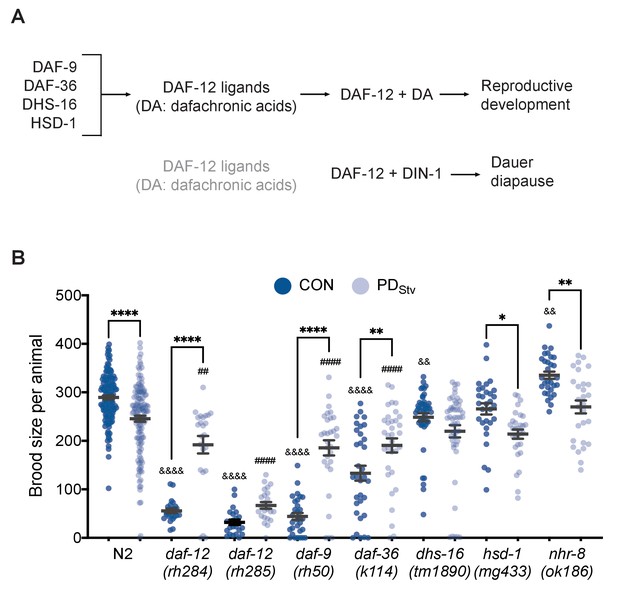
Adult reproductive plasticity is dependent on DAF-12 steroid signaling.
(A) Model of DAF-12 regulation of development. See text for details. (B) Brood size of CON and PDStv in wild-type N2 and mutant strains. * p < 0.05, ** p < 0.01, and **** p < 0.0001 compare CON and PDStv within a strain; &&p < 0.01 and &&&&p < 0.0001 compare N2 CON to mutant CON; ##p < 0.01 and ####p < 0.0001 compare N2 PDStv to mutant PDStv; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data are provided in Figure 1—source data 1.
-
Figure 1—source data 1
Dafachronic acid-dependent DAF-12 signaling is required for decreased fecundity after starvation-induced dauer formation.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig1-data1-v2.xlsx
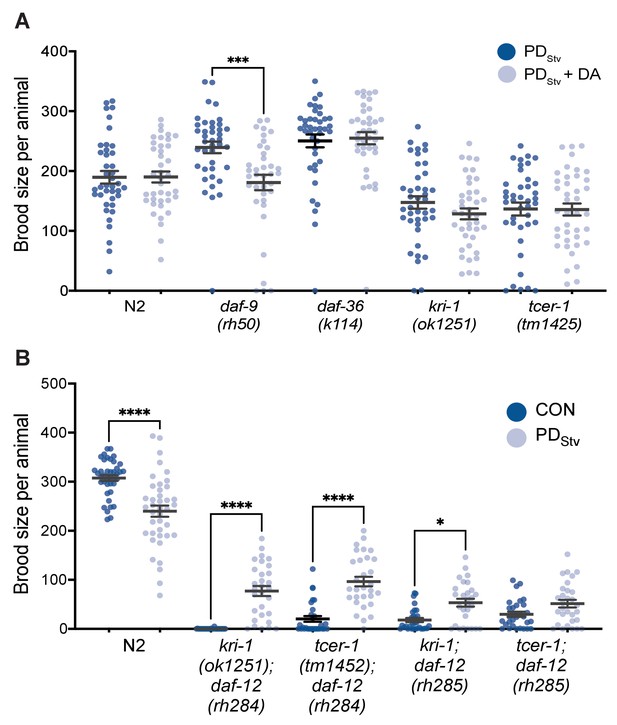
Dafachronic acid affects adult reproductive plasticity.
(A) Brood size comparison between PDStv grown in the presence of Δ7-dafachronic acid (Δ7-DA) (PDStv + DA) and PDStv grown in the absence of Δ7-DA (PDStv - DA) in wild-type N2, daf-9(rh50), daf-36(k114), kri-1(ok1251), and tcer-1(tm1425). *** p < 0.001 compares PDStv and PDStv + DA within a given genotype; one-way ANOVA with Sidak’s multiple comparison test. Additional brood size data are provided in Figure 1—figure supplement 1—source data 1. (B) Brood size comparison of PDStv relative to CON for wild-type N2, kri-1(ok1251); daf-12(rh284), tcer-1(tm1425); daf-12(rh284), kri-1(ok1251); daf-12(rh285), and tcer-1(tm1425); daf-12(rh285). * p < 0.05 and **** p < 0.0001 compare CON and PDStv within a genotype; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional brood size data are provided in Figure 1—figure supplement 1—source data 2.
-
Figure 1—figure supplement 1—source data 1
Brood size comparison between PDStv grown in the presence of delta7-dafachronic acid (PDStv + DA) and PDStv grown in the absence of (delta7-DA (PDStv - DA).
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig1-figsupp1-data1-v2.xlsx
-
Figure 1—figure supplement 1—source data 2
Brood size comparison of PDStv relative to CON.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig1-figsupp1-data2-v2.xlsx
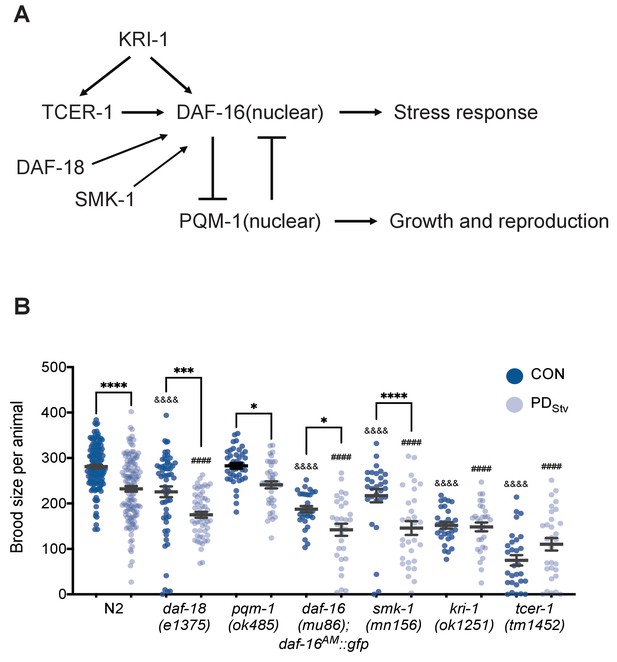
TCER-1 and KRI-1 regulate the decreased fecundity phenotype in PDStv adults.
(A) Model of the regulation of DAF-16 nuclear localization. See text for details. (B) Brood size of CON and PDStv in wild-type N2, daf-18(e1375), pqm-1(ok485), daf-16(mu86); daf-16AM::gfp, smk-1(mn156), kri-1(1251), and tcer-1(tm1452). * p < 0.05, *** p < 0.001, and **** p < 0.0001 compare CON and PDStv within a genotype; &&&&p < 0.0001 compares N2 CON to mutant CON; ####p < 0.0001 compares N2 PDStv to mutant PDStv; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data are provided in Figure 2—source data 1.
-
Figure 2—source data 1
TCER-1 and KRI-1 regulate the decreased fecundity phenotype in PDStv adults.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig2-data1-v2.xlsx
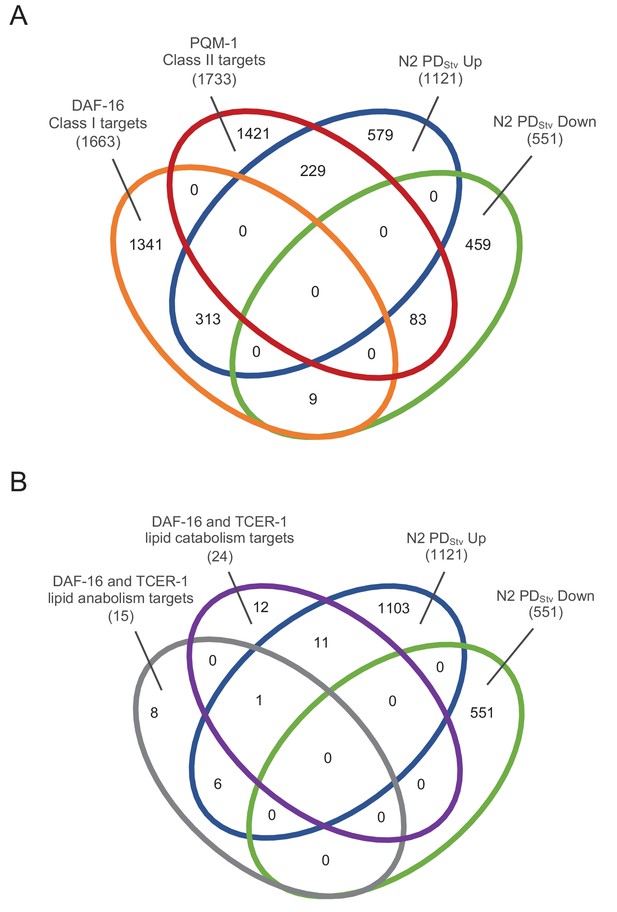
Differentially expressed genes in wild-type N2 PDStv are enriched for DAF-16 targets.
(A) Comparison of DAF-16 and PQM-1 targets (Tepper et al., 2013) with differentially expressed genes in N2 PDStv to N2 CON (Ow et al., 2018). See Figure 2—figure supplement 1—source data 1. (B) Comparison of DAF-16 and TCER-1 targets with functions in lipid metabolism from germ line-less glp-1(e2141ts) animals (Amrit et al., 2016) with differentially expressed genes in N2 PDStv to N2 CON (Ow et al., 2018). See Figure 2—figure supplement 1—source data 2.
-
Figure 2—figure supplement 1—source data 1
Comparison of DAF-16 and PQM-1 targets (Tepper et al., 2013) with differentially expressed genes in N2 PDStv to N2 CON (Ow et al., 2018).
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig2-figsupp1-data1-v2.xlsx
-
Figure 2—figure supplement 1—source data 2
Comparison of DAF-16 and TCER-1 targets with functions in lipid metabolism from germ line-less glp-1(e2141ts) animals (Amrit et al., 2016) with differentially expressed genes in N2 PDStv to N2 CON (Ow et al., 2018).
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig2-figsupp1-data2-v2.xlsx
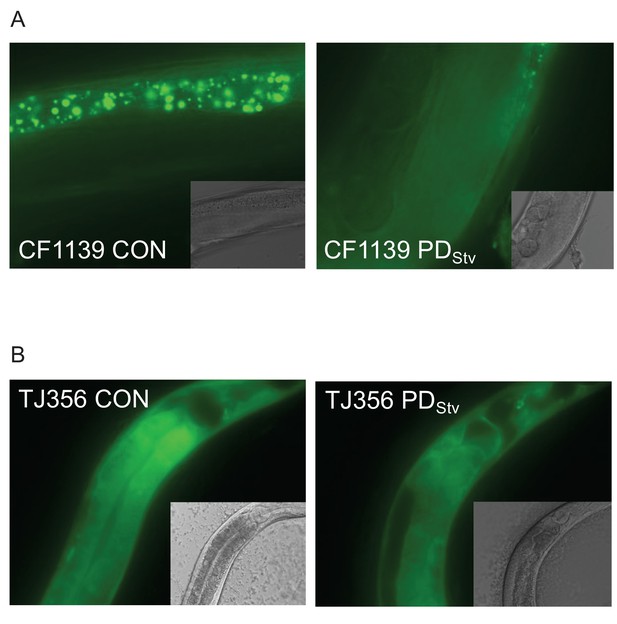
Localization of DAF-16::GFP in CON and PDStv one-day-old adults in (A) CF1139 (daf-16(mu86) I; muIs61 [(pKL78) daf-16::gfp + rol-6(su1006)]) and (B) TJ356 (zIs356 [daf-16p::daf-16a/b::gfp + rol-6(su1006)]).
The GPF observed in CF1139 CON is intestinal background signal.
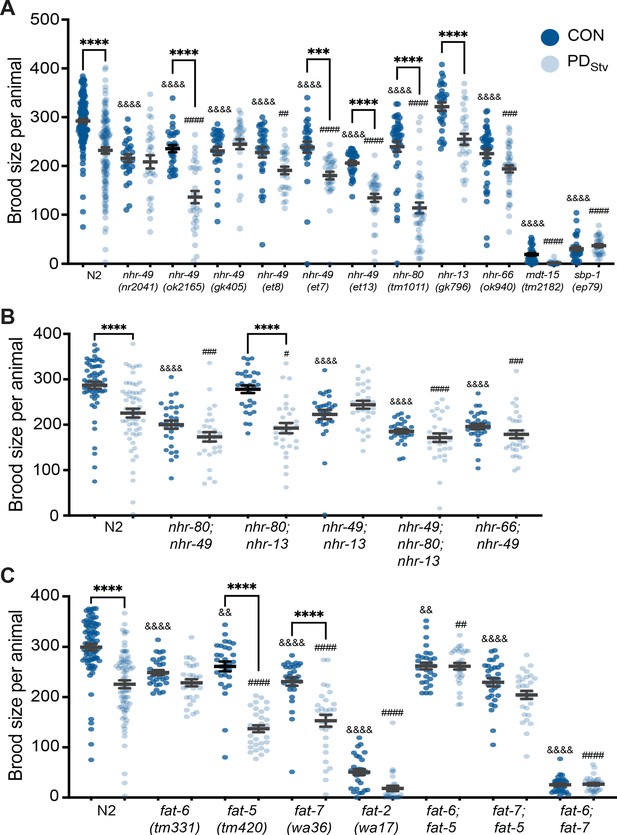
Fatty acid metabolism pathways modulate adult reproductive plasticity.
(A, B, C) Brood sizes of CON and PDStv in wild-type N2 and mutant strains. *** p < 0.001 and ****p < 0.0001 compare CON and PDStv within a genotype; &&p < 0.01 and &&&& p < 0.0001 compare N2 CON to mutant CON; ## p < 0.01, ### p < 0.001, and #### p < 0.0001 compare N2 PDStv to mutant PDStv; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data are provided in Figure 3—source data 1, Figure 3—source data 2, and Figure 3—source data 3.
-
Figure 3—source data 1
Fatty acid metabolism pathways modulate adult reproductive plasticity.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Fatty acid metabolism pathways modulate adult reproductive plasticity.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig3-data2-v2.xlsx
-
Figure 3—source data 3
Fatty acid metabolism pathways modulate adult reproductive plasticity.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig3-data3-v2.xlsx
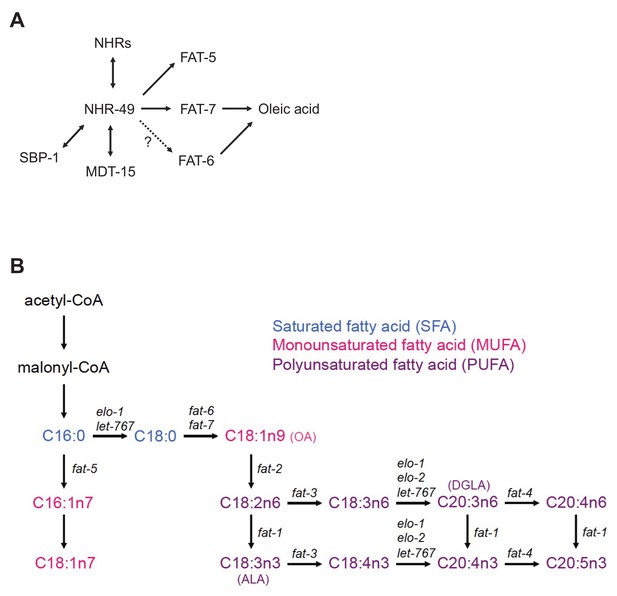
Fatty acid metabolism in C. elegans.
(A) Model of oleic acid synthesis. NHRs refer to NHR-80, NHR-13, and NHR-66. (B) Model of fat metabolism. Saturated fatty acids, SFAs (blue): C16:0 (palmitic), C18:0 (stearic); monounsaturated fatty acids, MUFAs (pink): C18:1n9 (oleic, OA), C16:1n7 (palmitoleic), C18:1n7 (vaccenic); polyunsaturated fatty acids, PUFAs (purple): C18:2n6 (linoleic), C18:3n6 (γ-linolenic), C20:3n6 (dihomo-γ-linolenic, DGLA), C20:4n6 (arachidonic), C18:3n3 (α-linolenic, ALA), C18:4n3 (stearidonic), C20:3n3 (eicosatrienoic), C20:5n3 (eicosapentaenoic).
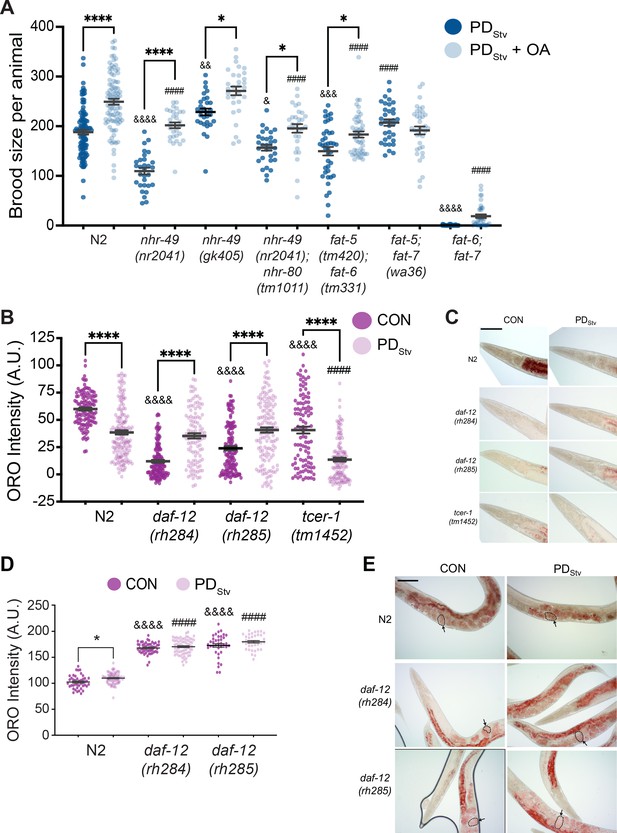
Lipid metabolism is affected in wild-type postdauer adults that experienced starvation-induced dauer.
(A) Brood size of wild-type N2 PDStv and mutant PDStv with or without oleic acid (OA) supplementation. * p < 0.05 and **** p < 0.0001 compare PDStv to PDStv + OA within a genotype; & p < 0.05, &&p < 0.01, &&& p < 0.001, and &&&& p < 0.0001 compare N2 PDStv to mutant PDStv; #### p < 0.0001 compares of N2 PDStv + OA to mutant PDStv + OA; one-way ANOVA with Sidak’s multiple comparison test. Additional data are provided in Figure 4—source data 1. (B) Oil Red O (ORO) intensity in CON and PDStv one-day-old adults. **** p < 0.0001 compares CON and PDStv of the same genotype; &&&&p < 0.0001 compares N2 CON to mutant CON; #### p < 0.0001 compares N2 PDStv to mutant PDStv; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. A.U.: arbitrary units. Additional data are provided in Figure 4—source data 2. (C) Representative micrographs of one-day-old adults stained with ORO. Scale bar: 100 μM. (D) ORO intensity of embryos measured in utero in one-day-old adults. * p < 0.05 compares CON and PDStv within a genotype; &&&& p < 0.0001 compares N2 CON to mutant CON; #### p < 0.0001 compares PDStv of N2 to mutant strains; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data is provided in Figure 4—source data 3. (E) Representative micrographs of ORO-stained adults. Dotted outlines and arrows are representative of ORO-stained in utero embryos quantified in (D). Scale bar: 100 μM.
-
Figure 4—source data 1
Brood size of wild-type N2 PDStv and mutant PDStv with or without oleic acid (OA) supplementation.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig4-data1-v2.xlsx
-
Figure 4—source data 2
Oil Red O (ORO) intensity in CON and PDStv one-day-old adults.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig4-data2-v2.xlsx
-
Figure 4—source data 3
ORO intensity of embryos measured in utero in one-day-adults.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig4-data3-v2.xlsx
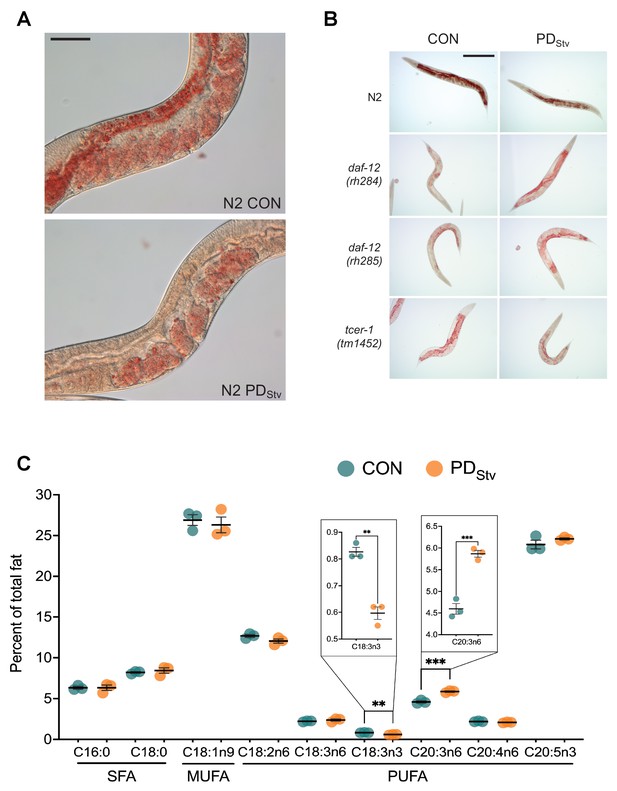
Postdauer one-day-old wild-type N2 adults exhibit fat-stained embryos despite the absence of intestinal fat stores.
(A) Representative micrographs of ORO-stained wild-type N2 CON and PDStv. Scale bar: 50 μM. (B) Representative micrographs of ORO-stained wild-type N2, daf-12(rh284), daf-12(rh285), and tcer-1(tm1452) CON and PDStv quantitated in Figure 4B. Scale bar: 250 μM. (C) Fatty acid composition of N2 CON and PDStv one-day-old adults. ** p < 0.01, ***p < 0.001; Student’s t-test. Error bars represent S.E.M. SFAs (saturated fatty acids): C16:0 (palmitic), C18:0 (stearic); MUFA (monounsaturated fatty acid): C18:1n9 (oleic); PUFAs (polyunsaturated fatty acids): C18:2n6 (linoleic), C18:3n6 (γ-linolenic), C18:3n3 (α-linolenic), C20:3n6 (dihomo-γ-linolenic), C20:4n6 (arachidonic), C20:5n3 (eicosapentaenoic). Additional data are provided in Figure 4—figure supplement 1—source data 1.
-
Figure 4—figure supplement 1—source data 1
Fatty acid composition of N2 CON and PDStv one-day-old adults.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig4-figsupp1-data1-v2.xlsx
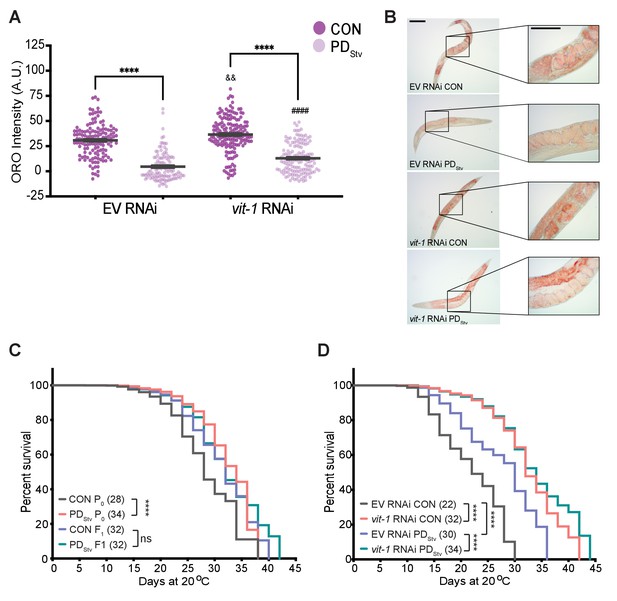
Vitellogenesis and adult lifespan are affected in postdauer animals.
(A) ORO staining in N2 CON and PDStv one-day-old adults fed with vit-1 RNAi or control empty RNAi vector (EV). **** p < 0.0001 compares CON and PDStv of the same RNAi condition; &&p < 0.01 and ####p < 0.0001 compare vit-1 to EV RNAi in N2 CON and PDStv, respectively; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data are provided in Figure 5—source data 1. (B) Representative micrographs of ORO-stained adults (anterior pharynx as in Figure 4C) quantified in (A). Insets show the presence of in utero ORO stained embryos following RNAi knockdown. Scale bars: 100 μM. (C) Adult lifespan assay of N2 CON and PDStv P0 and F1 generations. **** p < 0.0001, ns (not significant); log-rank (Mantel-Cox) test. Median survival (days) is indicated in parenthesis. Additional data are provided in Figure 5—source data 2. (D) Adult lifespan assay of N2 CON and PDStv fed with vit-1 or control empty vector (EV) RNAi. ****p < 0.0001; log-rank (Mantel-Cox) test. Median survival (days) is indicated in parenthesis. Additional data are provided in Figure 5—source data 3.
-
Figure 5—source data 1
ORO staining in N2 CON and PDStv one-day-adults fed with vit-1 RNAi or control empty RNAi vector (EV).
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig5-data1-v2.xlsx
-
Figure 5—source data 2
Adult lifespan assay of N2 CON and PDStv P0 and F1 generations.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig5-data2-v2.xlsx
-
Figure 5—source data 3
Adult lifespan assay of N2 CON and PDStv fed with vit-1 or control empty vector RNAi.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig5-data3-v2.xlsx
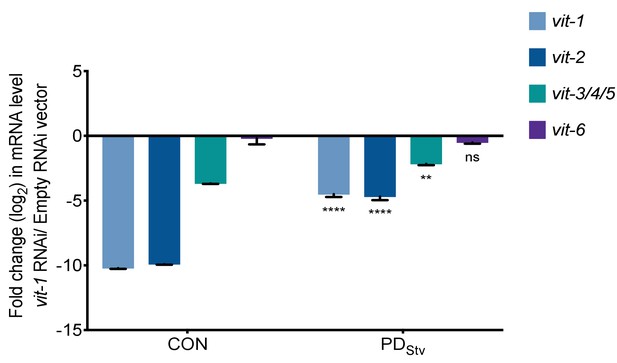
mRNA level of vit-1, vit-2, vit-3/4/5, and vit-6 in one-day-old N2 CON and PDStv adults as measured by reverse transcription quantitative PCR (RT-qPCR) following RNAi-by-feeding with k09f5.2 (vit-1) or control empty RNAi vector.
act-1 was used as the normalization control. **** p < 0.0001 and ns (no significance) refer to the comparison of PDStv to CON for a given RNAi knockdown; two-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data is provided in Figure 5—figure supplement 1—source data 1.
-
Figure 5—figure supplement 1—source data 1
mRNA level of vit-1, vit-2, vit-3/4/5, and vit-6 in one-day-old N2 CON and PDStv adults following RNAi-by-feeding with vit-1 or control empty RNAi vector.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig5-figsupp1-data1-v2.xlsx
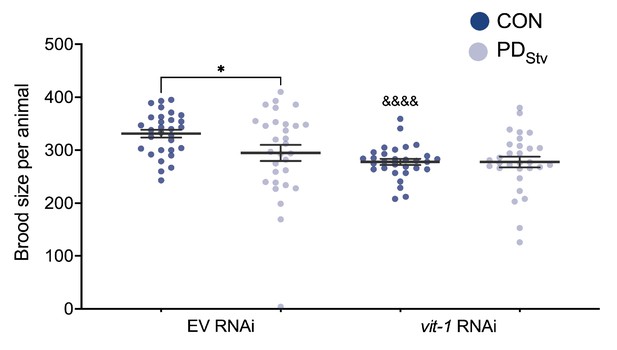
Altered vitellogenesis does not affect fecundity in postdauer animals.
Brood size of N2 CON and PDStv fed with vit-1 or control empty vector (EV) RNAi. ****p < 0.0001 compares CON and PDStv of the same RNAi condition; and p < 0.05 compares N2 CON vit-1 RNAi to CON EV RNAi; Student’s t-test. Error bars represent S.E.M. Additional data is provided in Figure 5—figure supplement 2—source data 1.
-
Figure 5—figure supplement 2—source data 1
Altered vitellogenesis does not affect fecundity in postdauer animals.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig5-figsupp2-data1-v2.xlsx
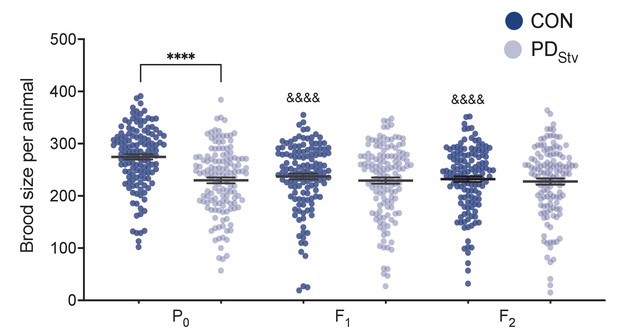
Reproductive plasticity in postdauer animals is not subject to transgenerational inheritance.
(A) Transgenerational brood size (P0, F1, and F2 generations) of wild-type N2 CON and PDStv. ****p < 0.0001 compares CON and PDStv of the same generation; &&&&p < 0.0001 compares the P0 with the F1, and F2 generations of N2 controls; one-way ANOVA with Dunnett’s multiple comparison test. Error bars represent S.E.M. Figure 5—figure supplement 3—source data 1.
-
Figure 5—figure supplement 3—source data 1
Reproductive plasticity in postdauer animals is not subject to transgenerational inheritance.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig5-figsupp3-data1-v2.xlsx
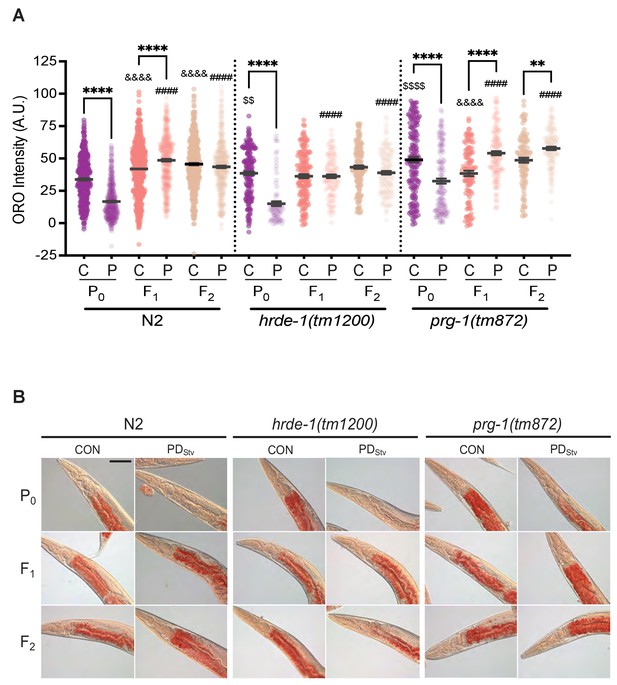
Generational inheritance of starvation memory is dependent upon germline-specific RNAi pathways.
(A) ORO staining in wild-type N2, hrde-1(tm1200), and prg-1(tm872) CON and PDStv one-day-old adults from P0, F1, and F2 generations. *p < 0.05 and ****p < 0.0001 compares CON (C) and PDStv (P) within a genotype and generation; &&&&p < 0.0001 compares controls between generations within a strain; ####p < 0.0001 compares PDStv between generations within a strain; $$p < 0.01 and $$$$p < 0.0001 compares N2 CON to mutant CON of the P0 generation; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data are provided in Figure 6—source data 1. (B) Representative micrographs of N2, hrde-1(tm1200), and prg-1(tm872) CON and PDStv from P0, F1, and F2 generations stained with ORO quantified in (A). Scale bar: 50 μM.
-
Figure 6—source data 1
ORO staining in wild-type N2, hrde-1(tm1200), and prg-1(tm872) CON and PDStv one-day-old adults from P0, F1, and F2 generations.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig6-data1-v2.xlsx
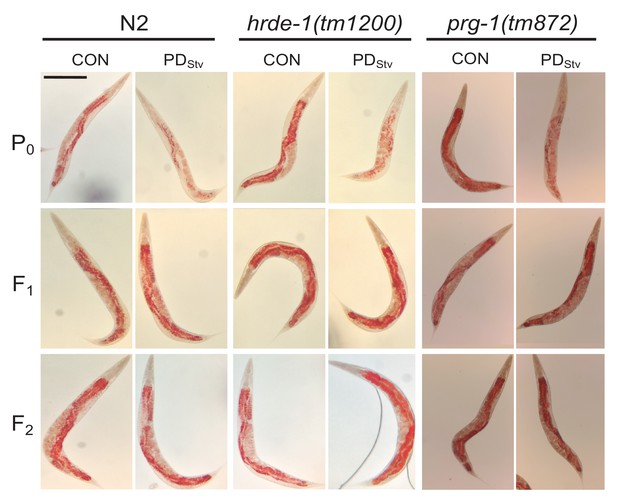
Representative micrographs of N2, hrde-1(tm1200), and prg-1(tm872) CON and PDStv from P0, F1, and F2 generations stained with ORO and quantified in Figure 6A.
Scale bar: 250 μM.
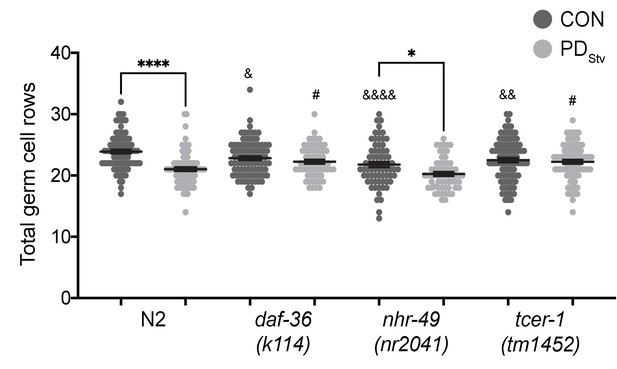
DAF-36 and TCER-1 regulate the onset of germline proliferation.
Total germ cell rows in CON and PDStv wild-type N2 and mutant larva exhibiting L3 vulva morphology (see Materials and Methods). * p < 0.05 and **** p < 0.0001 compare CON and PDStv within a genotype; & p < 0.05, &&p < 0.01, and &&&&p < 0.0001 compare N2 CON with mutant CON of mutants; #p < 0.05 compares PDStv of N2 to PDStv of mutants; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data are provided in Figure 7—source data 1.
-
Figure 7—source data 1
Total germ cell rows in CON and PDStv wild-type N2 and mutant larva.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig7-data1-v2.xlsx
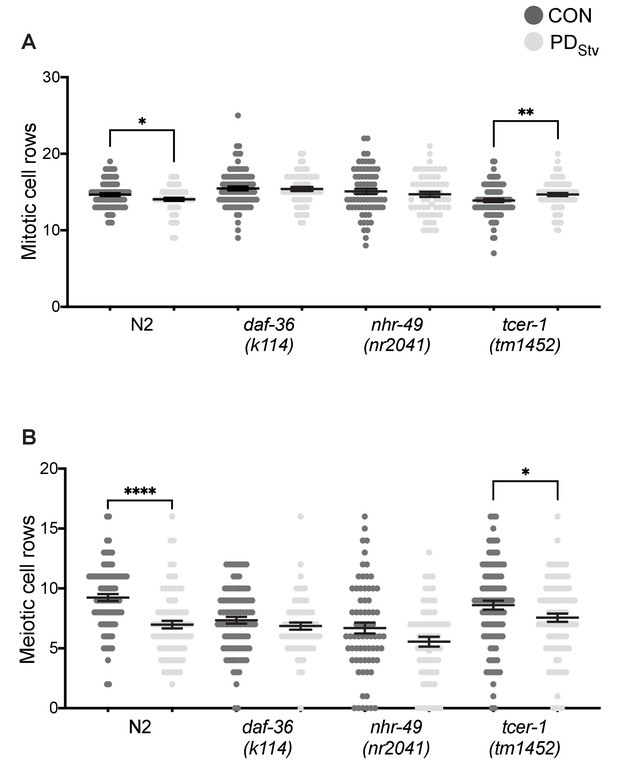
DAF-36 and TCER-1 are required for the delay in the onset of germline proliferation in postdauer animals.
(A) Mitotic cell rows at the L3 stage in CON and PDStv wild-type N2, daf-36(k114), nhr-49(nr2041), and tcer-1(tm1452) was determined by subtracting the meiotic cell rows from the total cell rows for each animal. *p < 0.05 and ** p < 0.01 compares CON and PDStv of the same genotype; Student’s t-test. (B) Meiotic (transition zone) cell rows at the L3 stage in CON and PDStv N2, daf-36(k114), nhr-49(nr2041), and tcer-1(tm1452). * p < 0.05 and **** p < 0.0001 compares CON and PDStv of the genotype; Student’s t-test. Error bars represent S.E.M. Additional data are provided in Figure 7—figure supplement 1—source data 1.
-
Figure 7—figure supplement 1—source data 1
DAF-36 and TCER-1 are required for the delay in the onset of germline proliferation in postdauer animals.
- https://cdn.elifesciences.org/articles/61459/elife-61459-fig7-figsupp1-data1-v2.xlsx
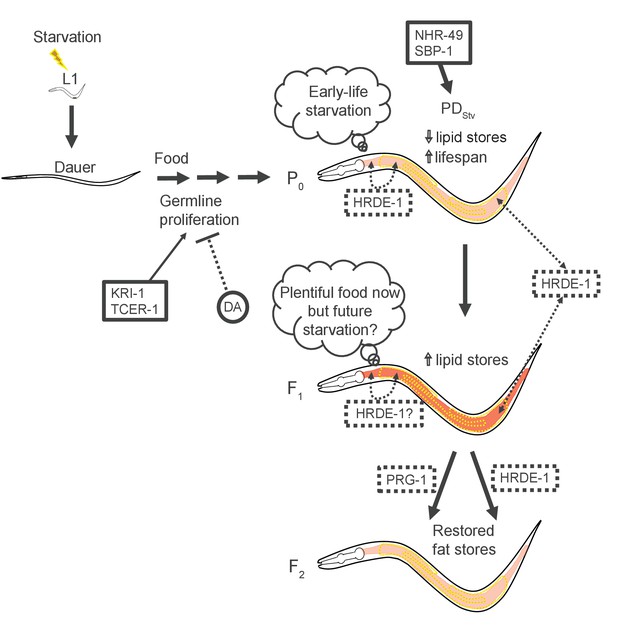
Model of endocrine signaling pathways acting as contributors to fatty acid metabolism of PDStv animals.
Signaling between the soma and the germ line establishing metabolic plasticity and the transmission of ancestral starvation memory are dependent on the germline nuclear RNAi pathway.
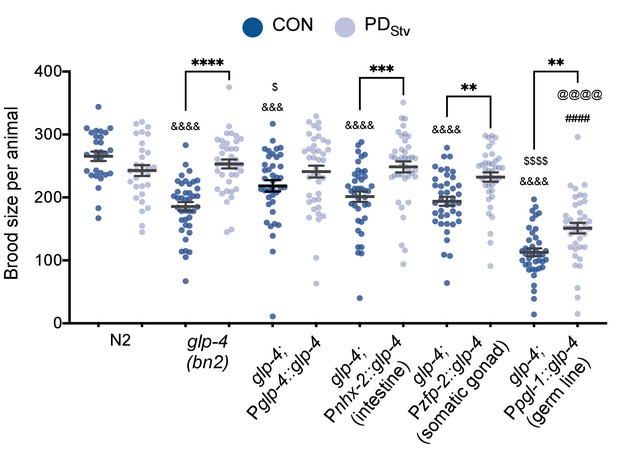
GLP-4 is required in multiple tissues to regulate adult reproductive plasticity.
Brood size comparison of PDStv relative to CON in wild-type N2, glp-4(bn2), and tissue-specific MosSCI rescue strains of glp-4. Assays were done at 15°C. ** p < 0.01, *** p < 0.001, and **** p < 0.0001 compare PDStv to CON within a genotype; &&& p < 0.001 and &&&& p < 0.0001 compare CON of N2, mutant and glp-4 tissue-specific rescues; ####p < 0.0001 compares PDStv of N2 to mutant and rescue strains; $p < 0.05 and $$$$p < 0.0001 compare CON of glp-4 and rescue strains; @@@@p < 0.0001 compares PDStv of glp-4 and rescue strains; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional brood size data are provided in Appendix 1—figure 1—source data 1.
-
Appendix 1—figure 1—source data 1
GLP-4 is required in multiple tissues to regulate adult reproductive plasticity.
- https://cdn.elifesciences.org/articles/61459/elife-61459-app1-fig1-data1-v2.xlsx
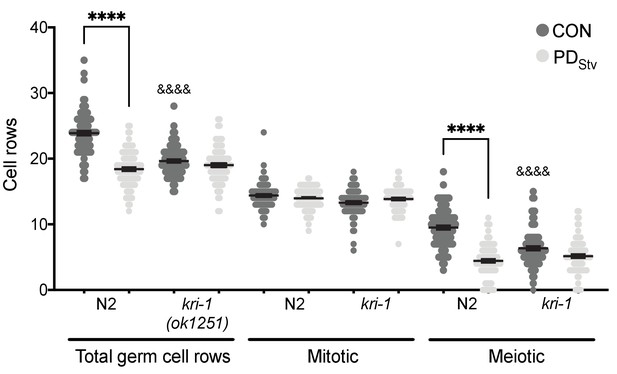
KRI-1 regulates the onset of germline proliferation.
Total, mitotic, and meiotic cell rows in control and PDStv animals exhibiting L3 vulva morphology in wild-type N2 and kri-1(ok1251). **** p < 0.0001 compares of CON and PDStv within a genotype; &&&&p < 0.0001 compares N2 CON to mutant CON within a type of cell row count; one-way ANOVA with Sidak’s multiple comparison test. Error bars represent S.E.M. Additional data are provided in Appendix 1—figure 2—source data 1.
-
Appendix 1—figure 2—source data 1
Total, mitotic, and meiotic cell rows in control and PDStv animals in wild type and kri-1(ok1254).
- https://cdn.elifesciences.org/articles/61459/elife-61459-app1-fig2-data1-v2.xlsx
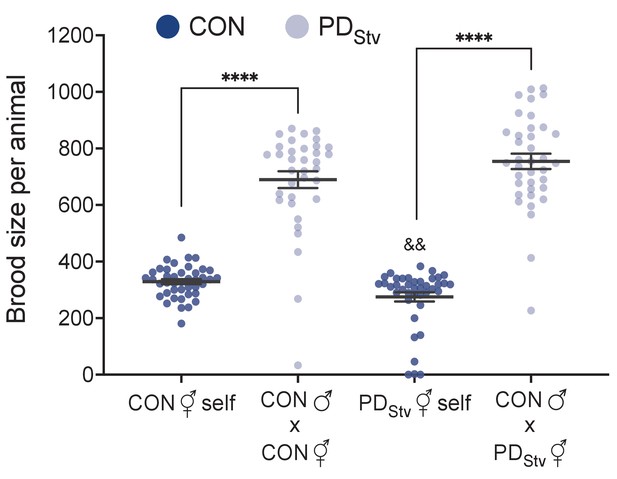
Decreased fecundity in postdauer animals results from a reduction of sperm available for self-fertilization.
Brood size of wild-type N2 CON and PDStv self-fertilized hermaphrodites (⚥) and CON or PDStv hermaphrodites mated with CON males (♂). **** p < 0.0001 compares self-fertilized CON or PDStv hermaphrodites to CON or PDStv hermaphrodites mated to CON males; &&p < 0.01 compares self-fertilized CON to self-fertilized PDStv hermaphrodites; Student’s t-test. Error bars represent S.E.M. Additional data are provided in Appendix 1—figure 3—source data 1.
-
Appendix 1—figure 3—source data 1
Decreased fecundity in postdauer animals results from a reduction of sperm available for self-fertilization.
- https://cdn.elifesciences.org/articles/61459/elife-61459-app1-fig3-data1-v2.xlsx
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (include species here) | ||||
| Strain, strain background (Caenorhabditis elegans) | N2 | Caenorhabditis Genetics Center | Wild type | |
| Strain, strain background (Caenorhabditis elegans) | AA82 | Caenorhabditis Genetics Center | daf-12(rh284) X | |
| Strain, strain background (Caenorhabditis elegans) | AA85 | Caenorhabditis Genetics Center | daf-12(rh285) X | |
| Strain, strain background (Caenorhabditis elegans) | AA292 | Caenorhabditis Genetics Center | daf-36(k114) V | |
| Strain, strain background (Caenorhabditis elegans) | AA1052 | Adam Antebi, Max Planck Institute | dhs-16(tm1890) V | |
| Strain, strain background (Caenorhabditis elegans) | AE501 | Caenorhabditis Genetics Center | nhr-8(ok186) IV | |
| Strain, strain background (Caenorhabditis elegans) | BS1080 | Tim Schedl, Washington University | gld-1::gfp::3xflag | |
| Strain, strain background (Caenorhabditis elegans) | BX26 | Caenorhabditis Genetics Center | fat-2(wa17) IV | |
| Strain, strain background (Caenorhabditis elegans) | BX106 | Caenorhabditis Genetics Center | fat-6(tm331) IV | |
| Strain, strain background (Caenorhabditis elegans) | BX107 | Caenorhabditis Genetics Center | fat-5(tm420) V | |
| Strain, strain background (Caenorhabditis elegans) | BX110 | Caenorhabditis Genetics Center | fat-6(tm331) IV; fat-5(tm420) V | |
| Strain, strain background (Caenorhabditis elegans) | BX153 | Caenorhabditis Genetics Center | fat-7(wa36) V | |
| Strain, strain background (Caenorhabditis elegans) | BX156 | Caenorhabditis Genetics Center | fat-6(tm331) IV; fat-7(wa36) V | |
| Strain, strain background (Caenorhabditis elegans) | BX160 | Caenorhabditis Genetics Center | fat-7(wa36) fat-5(tm420) V | |
| Strain, strain background (Caenorhabditis elegans) | CB1375 | Caenorhabditis Genetics Center | daf-18(e1375) IV | |
| Strain, strain background (Caenorhabditis elegans) | CE541 | Caenorhabditis Genetics Center | sbp-1(ep79) III | |
| Strain, strain background (Caenorhabditis elegans) | CF1139 | Caenorhabditis Genetics Center | daf-16(mu86) I; muIs61 [(pKL78) daf16::gfp + rol-6(su1006)] | |
| Strain, strain background (Caenorhabditis elegans) | CF2052 | Caenorhabditis Genetics Center | kri-1(ok1251) I | |
| Strain, strain background (Caenorhabditis elegans) | CF2167 | Caenorhabditis Genetics Center | tcer-1(tm1452) II | |
| Strain, strain background (Caenorhabditis elegans) | EG6699 | Caenorhabditis Genetics Center | ttTi5605 II; unc-119(ed3) III; oxEx1578 [eft-3p::gfp + Cbr-unc-119] | |
| Strain, strain background (Caenorhabditis elegans) | GR2063 | Caenorhabditis Genetics Center | hsd-1(mg433) I | |
| Strain, strain background (Caenorhabditis elegans) | RG1228 | Caenorhabditis Genetics Center | daf-9(rh50) X | |
| Strain, strain background (Caenorhabditis elegans) | SEH301 | This study | nhr-13(gk796) V backcrossed | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH302 | This study | nhr-49(nr2041) I; nhr-80(tm1011) III | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH303 | This study | nhr-49(nr2041) I; nhr-13(gk796) V | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH304 | This study | nhr-49(nr2041) I; nhr-80(tm1011) III; nhr-13(gk796) V | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH312 | This study | daf-16(mu86) I; muEx158 (daf-16AM::gfp + sur-5p::gfp) | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH319 | This study | nhr-49(et8) I backcrossed | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH326 | This study | nhr-49(et13) I backcrossed | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH327 | This study | nhr-49(et7) I backcrossed | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH342 | This study | nhr-49(nr2041) I; nhr-66(ok940) IV | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH343 | This study | nhr-49(gk405) I backcrossed | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH344 | This study | nhr-49(ok2165) I backcrossed | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH350 | This study | pqm-1(ok485) II backcrossed | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH351 | This study | kri-1(ok1251) I; daf-12(rh284) X | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH352 | This study | kri-1(ok1251) I; daf-12(rh285) X | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH353 | This study | tcer-1(tm1452) II; daf-12(rh284) X | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH354 | This study | tcer-1(tm1452) II; daf-12(rh285) X | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH357 | This study | glp-4(bn2) I; pdrSi1 [Pglp-4::glp-4 cDNA::gfp::glp-4 3'UTR; unc-119(+)] II | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH368 | This study | glp-4(bn2) I; pdrSi2 [Pnhx-2::glp-4 cDNA::gfp::glp-4 3'UTR; unc-119(+)] II | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH369 | This study | glp-4(bn2) I; pdrSi3 [Pzfp-2::glp-4 cDNA::gfp::glp-4 3'UTR; unc-119(+)] II | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH370 | This study | glp-4(bn2) I; pdrSi4 [Ppgl-1::glp-4 cDNA::gfp::glp-4 3'UTR; unc-119(+)] II | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SEH383 | This study | hrde-1(tm1200) III backcrossed | Sarah Hall, Syracuse University |
| Strain, strain background (Caenorhabditis elegans) | SS104 | Caenorhabditis Genetics Center | glp-4(bn2) I | |
| Strain, strain background (Caenorhabditis elegans) | SP488 | Caenorhabditis Genetics Center | smk-1(mn156) V | |
| Strain, strain background (Caenorhabditis elegans) | STE68 | Caenorhabditis Genetics Center | nhr-49(nr2041) I | |
| Strain, strain background (Caenorhabditis elegans) | STE69 | Caenorhabditis Genetics Center | nhr-66(ok940) IV | |
| Strain, strain background (Caenorhabditis elegans) | STE70 | Caenorhabditis Genetics Center | nhr-80(tm1011) III | |
| Strain, strain background (Caenorhabditis elegans) | STE73 | Caenorhabditis Genetics Center | nhr-80(tm1011) III; nhr-13(gk796) V | |
| Strain, strain background (Caenorhabditis elegans) | TJ356 | Caenorhabditis Genetics Center | zIs356 [daf-16p::daf-16a/b::gfp + rol-6(su1006)] | |
| Strain, strain background (Caenorhabditis elegans) | WM161 | Caenorhabditis Genetics Center | prg-1(tm872) II | |
| Strain, strain background (Caenorhabditis elegans) | XA7702 | Caenorhabditis Genetics Center | mdt-15(tm2182) III | |
| Strain, strain background (Escherichia coli) | OP50 | Caenorhabditis Genetics Center | OP50 | |
| Recombinant DNA reagent | k09f5.2 | Kamath et al., 2001 | RNAi | |
| Chemical compound, drug | Oleic acid C18:1 | NuChek Prep, Inc.; Elysian, Minnesota | Cat no. U-46-A | |
| Chemical compound, drug | Oil Red O (ORO) | Sigma Aldrich | Cat no. O0625 | |
| Chemical compound, drug | 5-fluoro-2’-deoxyuridine (FUDR) | Sigma Aldrich | Cat no. F0503 | |
| Chemical compound, drug | IPTG | Sigma Aldrich | Cat no. I5502 | |
| Chemical compound, drug | Carbenicillin | Sigma Aldrich | Cat no. C1389 | |
| Chemical compound, drug | Δ7 form of dafachronic acid | Frank Schroeder, Cornell University | ||
| Chemical compound, drug | DAPI stain | Thermo Scientific | Used at a concentration of 1:1000 | |
| Software, algorithm | Spot 5.2 | Nikon | Nikon Eclipse | |
| Software, algorithm | GraphPad Prism | GraphPad Software | v.9 | |
| Software, algorithm | ImageJ software | ImageJ (http://imagej.nih.gov/ij/) |
Additional files
-
Supplementary file 1
Gene expression changes of selected genes in N2 PDStv compared to N2 CON adults (Ow et al., 2018 PLoS Genet 14: e1007219).
- https://cdn.elifesciences.org/articles/61459/elife-61459-supp1-v2.xlsx
-
Supplementary file 2
Strains used in this study.
- https://cdn.elifesciences.org/articles/61459/elife-61459-supp2-v2.xlsx
-
Supplementary file 3
Primer sequences used in this study.
- https://cdn.elifesciences.org/articles/61459/elife-61459-supp3-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61459/elife-61459-transrepform-v2.docx
-
Appendix 1—figure 1—source data 1
GLP-4 is required in multiple tissues to regulate adult reproductive plasticity.
- https://cdn.elifesciences.org/articles/61459/elife-61459-app1-fig1-data1-v2.xlsx
-
Appendix 1—figure 2—source data 1
Total, mitotic, and meiotic cell rows in control and PDStv animals in wild type and kri-1(ok1254).
- https://cdn.elifesciences.org/articles/61459/elife-61459-app1-fig2-data1-v2.xlsx
-
Appendix 1—figure 3—source data 1
Decreased fecundity in postdauer animals results from a reduction of sperm available for self-fertilization.
- https://cdn.elifesciences.org/articles/61459/elife-61459-app1-fig3-data1-v2.xlsx





