Structure of the human core transcription-export complex reveals a hub for multivalent interactions
Figures
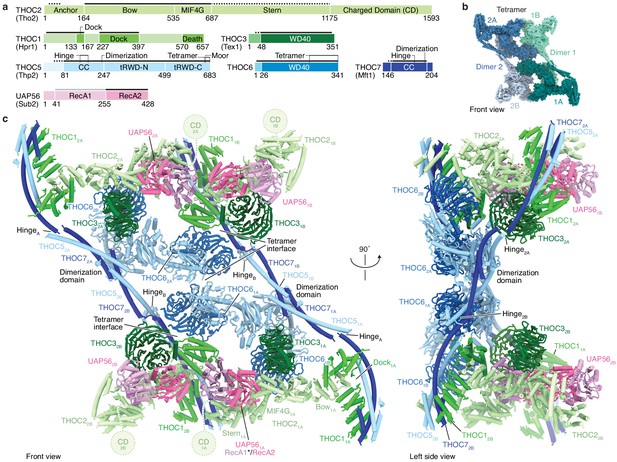
Human THO–UAP56 complex structure.
(a) Domain organization of human THO–UAP56 complex subunits. Solid and dashed lines indicate atomic and backbone regions of the THO–UAP56 structure, respectively. The UAP56 RecA1 lobe is not observed in the structure and indicated with a light gray line. Orthologous yeast protein names are in parenthesis. Color code used throughout. CC, coiled coil; tRWD-N and -C, N- and C-terminal tandem RWD. (b) Surface representation of the THO–UAP56 tetramer structure. Dimer 1 monomers 1A (dark turquoise) and 1B (light turquoise), and dimer 2 monomers 2A (dark blue), 2B (light blue) are indicated. (c) Human THO–UAP56 structure front and left side views. The UAP56 RecA1 lobe was not visible in the cryo-EM density, but is modeled here by superimposing a UAP56 (yeast Sub2) homology model based on a low-resolution THO–Sub2 crystal structure (Ren et al., 2017) (PDB ID 5SUQ) and is indicated with an asterisk. The THOC2 CD domain is disordered (Peña et al., 2012), was truncated for recombinant THO–UAP56 production, and is shown as a dashed circle.
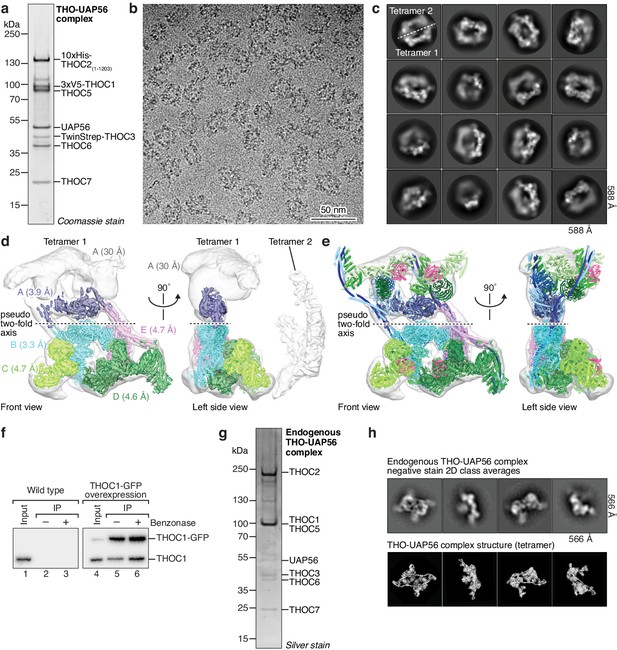
Biochemical characterization and EM of recombinant and endogenous THO–UAP56 complexes.
(a) Protein analysis of the recombinant human THO–UAP56 complex (SDS-PAGE stained with Coomassie blue). (b) Cryo-EM micrograph of the recombinant THO–UAP56 complex. Scale bar, 50 nm. (c) Representative cryo-EM 2D class averages of THO–UAP56 reveal flexibility between and within two adjacent tetramers. (d) Composite cryo-EM density of the THO–UAP56 complex shown in two orthogonal views, front and left side. Colors indicate the respective cryo-EM densities used for modeling (A, dark violet; B, cyan; C, light green; D, dark green; E, magenta). Map A was low-pass filtered to 30 Å (gray, transparent) to show the full tetramer density (front view) and the location of the second tetramer (left side view), which is highly mobile relative to the first. The sharpened densities are shown and were aligned using overlapping regions. See Materials and methods, and Figure 1—figure supplements 2 and 3 for details on image processing. (e) Composite cryo-EM density of THO–UAP56 superimposed on a ribbon model of the final THO–UAP56 model, colored as in Figure 1. (f) The endogenous THO complex forms an oligomer in vivo that does not depend on RNA. THOC1–GFP was expressed ectopically in K562 cells using a lentivirus (lanes 4–6) and purified via the GFP-tag in a single step in absence (lane 5) or presence of Benzonase to degrade nucleic acid (lane 6). Untagged THOC1 did not bind the GFP-beads (lanes 1–3). Proteins where visualized by western blot using an anti-THOC1 antibody (see Materials and methods). (g) Protein analysis of the endogenous human THO–UAP56 complex purified in two steps (SDS-PAGE, Silver stain). See Materials and methods for details. (h) Comparison of representative negative stain EM 2D class averages of the endogenous THO–UAP56 complex (top) to corresponding views of the THO–UAP56 tetramer structure (bottom).
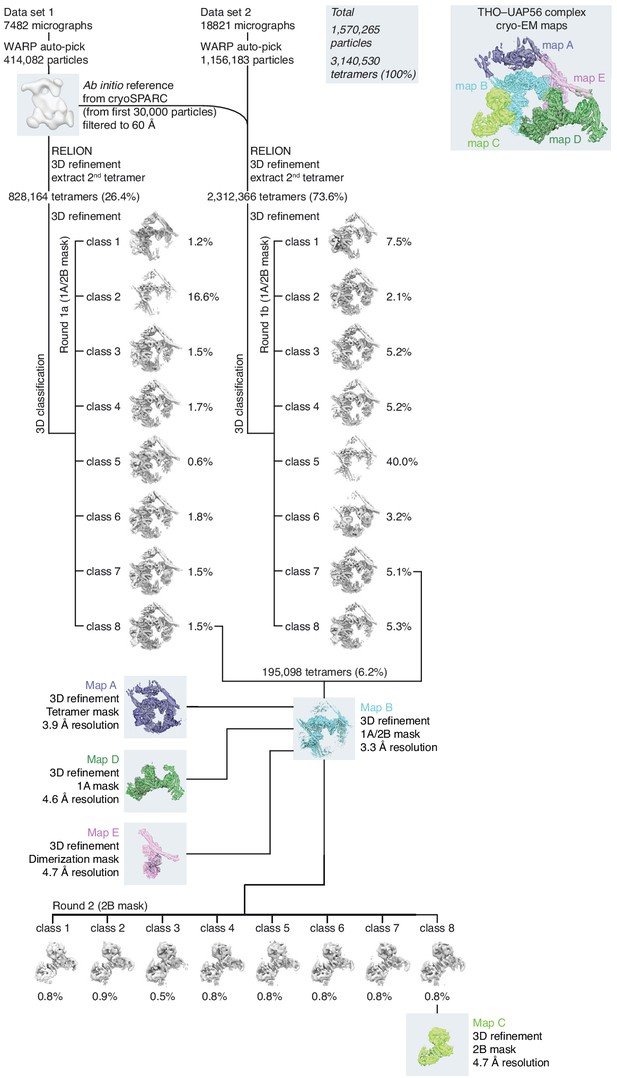
THO–UAP56 cryo-EM image classification.
Three-dimensional image classification tree of the THO–UAP56 cryo-EM data set using an initial cryoSPARC 2.0 (Punjani et al., 2017) model determined from 30,000 particles (Materials and methods). The combined two data sets yielded 1,570,265 THO–UAP56 picked octamer particles using Warp 1.0.7 (Tegunov and Cramer, 2019), from which a total of 3,140,530 tetramer units (100%) were extracted and subsequently analyzed using RELION 3.1 (Scheres, 2012). Classes are colored in gray and visualized at the same density threshold. The percentage of tetramer units contributing to each class is provided. The type of mask and overall resolution is indicated for each 3D refinement subsequent to classification and maps are colored as in Figure 1—figure supplement 1d. For additional details see the Materials and methods.
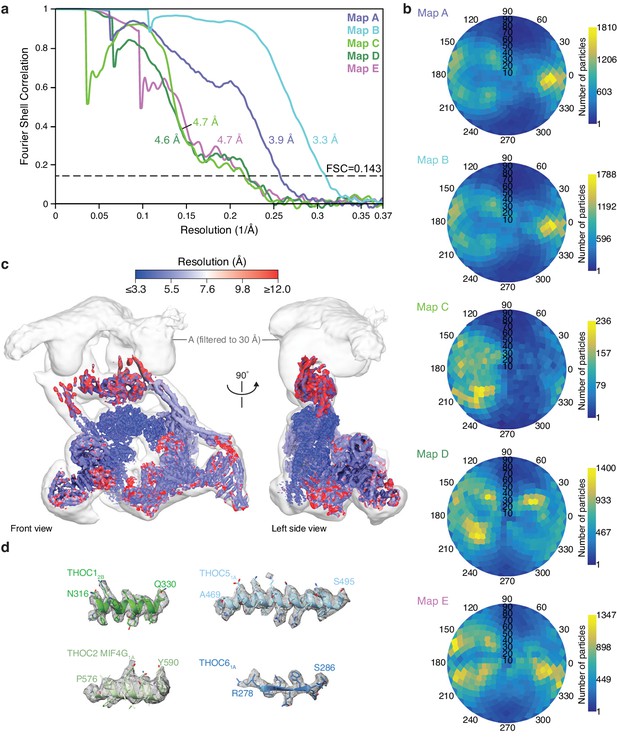
THO–UAP56 cryo-EM reconstructions.
(a) Gold-standard Fourier shell correlation (FSC = 0.143) of the respective A, B, C, D, and E cryo-EM reconstructions. (b) Orientation distribution plots for all particles contributing to the respective A, B, C, D, and E cryo-EM reconstructions. (c) Front and left side views of the composite THO–UAP56 complex cryo-EM density (maps A, B, C, D, E) colored by local resolution as determined by ResMap (Kucukelbir et al., 2014). (d) Representative regions of sharpened THO–UAP56 densities are superimposed on the refined coordinate model. Segments from THOC1 (residues 316–330, helix α13; map B), THOC2 (residues 576–590, α31; map D), THOC5 tRWD-C (residues 469–495, helix α14; map B), and THOC6 (residues 278–286, blade 6 β3; map B) are shown.
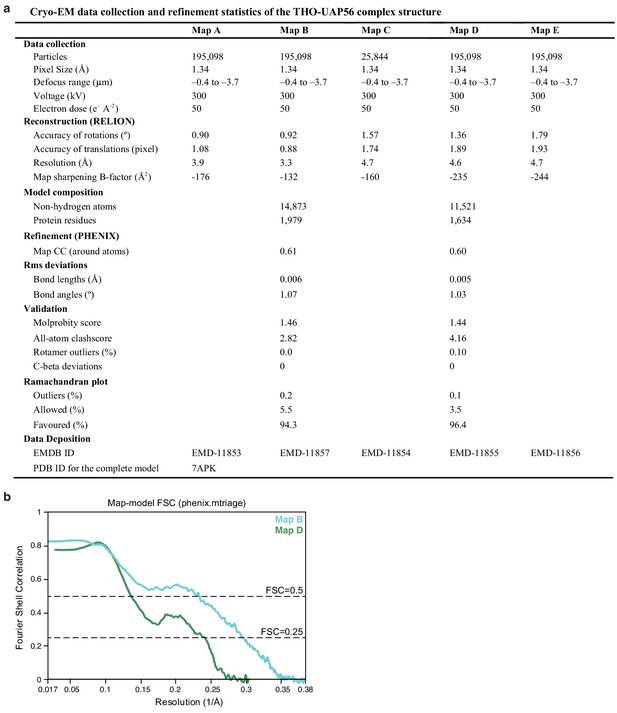
THO–UAP56 cryo-EM data collection and refinement statistics.
(a) Cryo-EM data collection and refinement statistics of the THO–UAP56 structure. Maps A and E were used to position THO–UAP56 monomers 1B and 2A, and THOC5 tRWD–THOC6 from monomer 1B, respectively. (b) Fourier shell correlation between the map B and D cryo-EM densities and the respective refined coordinate model parts, using phenix.mtriage (Afonine et al., 2018).
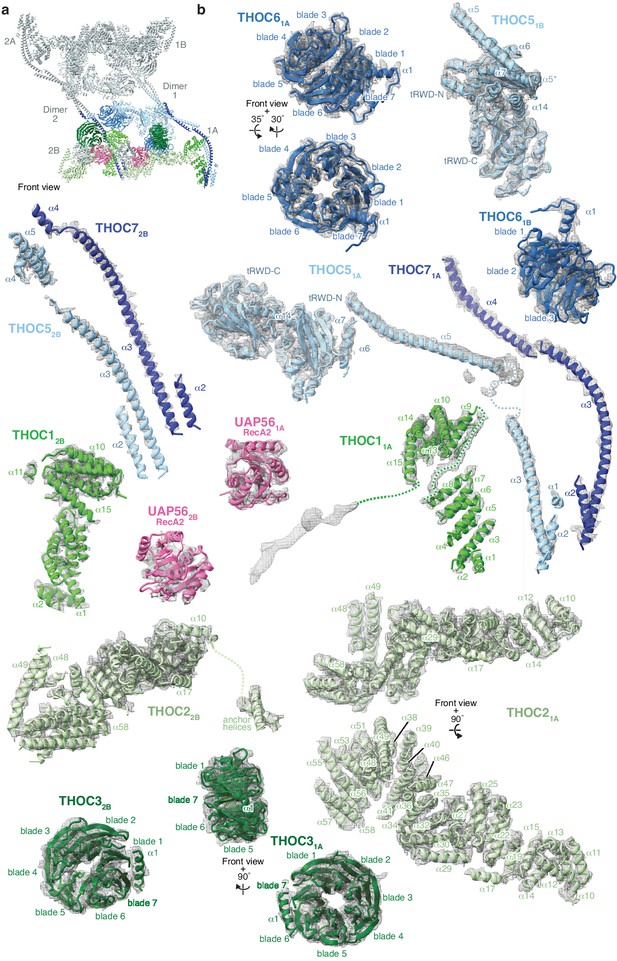
Gallery of THO–UAP56 subunits and their cryo-EM densities.
(a) Front view of the THO–UAP56 complex structure, indicating monomers 1A and 2B that are shown individually in panel b. (b) Gallery of THO–UAP56 complex subunits shown superimposed on their respective cryo-EM densities (maps A-E). Unmodeled densities are shown and selected domains and secondary structure elements are labelled.
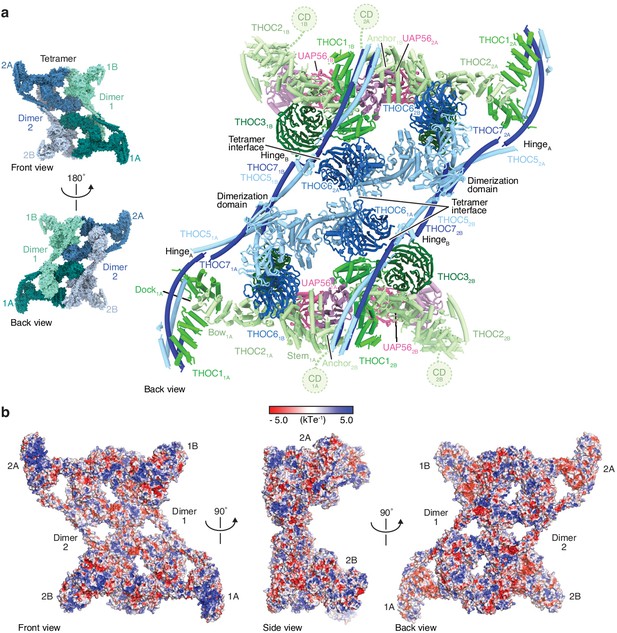
Back view and surface charge distribution of THO–UAP56.
(a) Human THO–UAP56 structure viewed from the back. The insets show surface representations of the THO–UAP56 tetramer structure (front and back view) and are colored as in Figure 1b. (b) Surface representations of the THO–UAP56 tetramer structure (front, left side, and back view), colored according to electrostatic surface potential. This analysis indicates a positive patch at the THOC1–THOC5–THOC7 interface, which is distant from the THOC2–UAP56 interface, and may interact with nucleic acid.
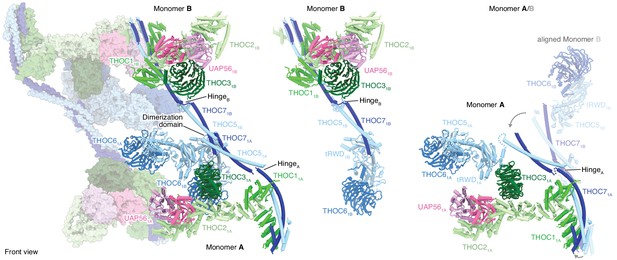
THO–UAP56 tetramer, dimer, and monomer organization.
Overview of the THO–UAP56 architecture from the front, showing the asymmetric dimers 1 and 2 as ribbons and surfaces, respectively on the left. THO–UAP56 monomers 1B (middle) and 1A (right) are shown as ribbons. THOC5, −6, and −7 assume different positions in monomers A and B, shown in the right panel by superposing monomer 1B (transparent ribbons) on monomer 1A via their THOC2 subunits. This movement is indicated with gray arrows. Colors as in Figure 1.
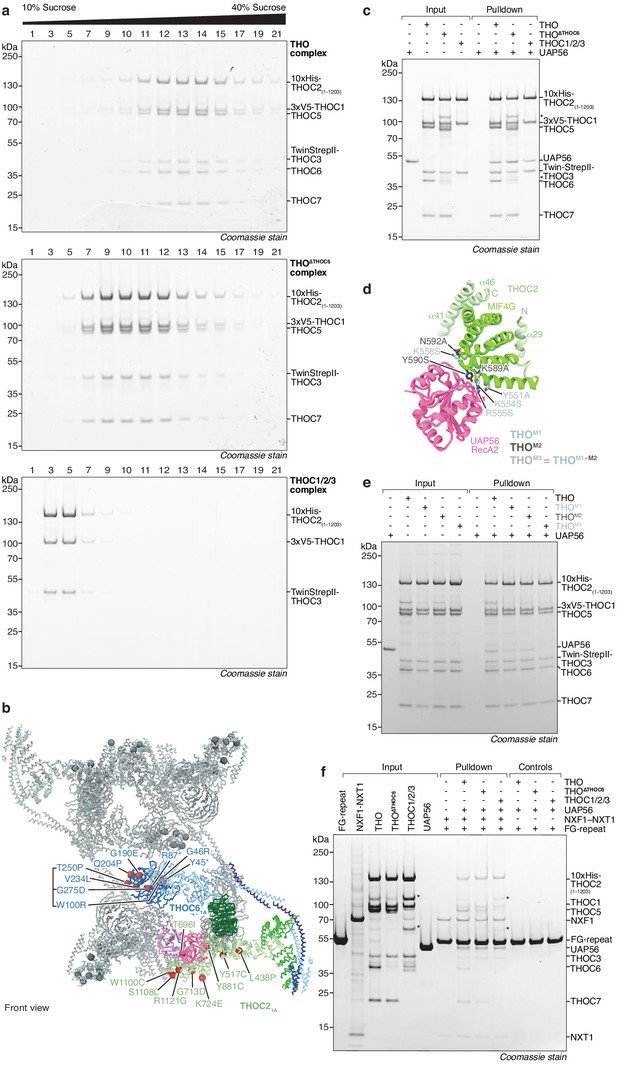
Biochemical probing of THO complex oligomerization, the THO–UAP56 interface, and mapping of disease mutations.
(a) Recombinant human THO (top), THO∆THOC6 (middle), THOC1/2/3 (bottom) complex migration in sucrose density gradients (SDS-PAGE stained with Coomassie blue) indicates an oligomeric state consistent with tetramer, dimer, and monomer architectures, respectively. (b) Location of human disease-associated mutations in THOC2 (Kumar et al., 2020; Kumar et al., 2018; Kumar et al., 2015) and THOC6 (FORGE Canada Consortium et al., 2013; Casey et al., 2016; Anazi et al., 2016; Care4Rare Canada Consortium et al., 2017; Mattioli et al., 2019). Several THOC6 mutations were shown to affect its stability and lead to the loss of interaction with THO subunits (Mattioli et al., 2019), suggesting that stabilization of the THO tetramer is needed for its full activity. In particular, two THOC6 premature stop codon mutations (at residues Y45 and R87) would truncate the THOC6 regions at the tetramer interface, and therefore disrupt this interaction. The mapped THOC2 mutations disrupt local secondary structure or alter the protein surface, but do not affect interfaces with THOC1, THOC3, and UAP56. The THO–UAP56 monomer 1A is colored as in Figure 1 with mutated residues marked as red spheres at their Cα atoms. Monomers 1B, 2A, 2B and associated mutations are colored in gray. The asterisk indicates mutations that result in premature stop codons. (c) Analysis of the human THO–UAP56 complex interaction in a pulldown experiment using the 6-subunit THO complex, THO∆THOC6, or THOC1/2/3 (SDS-PAGE stained with Coomassie blue). The THO complex and its variants were immobilized on Ni-NTA beads via a 10xhistidine tag on the THOC2 subunit. The asterisk indicates contaminants and degradation products. See Materials and methods for details. (d) Ribbon model showing THOC2 MIF4G domain residues (sticks) mutated in panel e to probe the UAP56 interface in a top view. (e) As in panel c, but comparing the THO complex and three THO variants containing THOC2–UAP56 interface mutants (THOM1: THOC2 residues Y551A, K554S, R555S, K558S; THOM2: THOC2 residues K589A, Y590S, N592A; THOM3: combined THOM1 and THOM2 mutants) for their UAP56 affinity. See Materials and methods for details. (f) The human export factor NXF1–NXT1 binds the core THOC1/2/3–UAP56 complex in a pulldown experiment. NXF1–NXT1 was immobilized on amylose beads via an MBP-tagged NUP214 FG-repeat peptide (Materials and methods) and bound THO, THO∆THOC6, or core THOC1/2/3 complex together with UAP56 (SDS-PAGE stained with Coomassie blue). The asterisk indicates contaminants and degradation products. See Materials and methods for details.
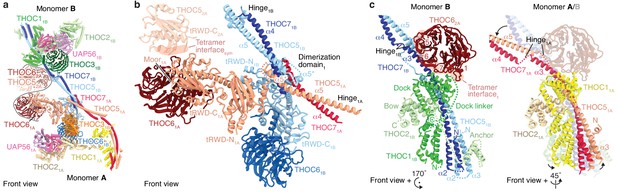
Details of THO–UAP56 tetramer and dimer interfaces.
(a) Ribbon model of the THO–UAP56 complex dimer 1, indicating dimer and tetramer interfaces. Colors for monomer A subunit UAP56 and monomer B are as in Figure 1, monomer A THOC1 (yellow), THOC2 (pale yellow), THOC3 (orange), THOC5 (salmon), THOC6 (maroon), and THOC7 (red). The black line marks the tetramer interface. The THOC5 tRWD-C domain and THOC6 from monomer 2A are shown as transparent ribbons and stabilize the tetramer. (b) THO–UAP56 dimerization domain. Monomer A THOC5 helix α5 and THOC7 α4 make a parallel coiled coil that forms a four-helix bundle with equivalent helices from monomer B. The four-helix bundle is asymmetric (note the α5* position between THOC51A and THOC51B) due to dimerization of the adjacent THOC5 tRWD-N domains. View and colors as in panel a. (c) The THOC5–THOC7 coiled coil binds THOC1 and the THOC2 anchor helices and adopts different positions in presence (left, monomer 1B) or absence (right, monomer 1A) of THOC62A from the neighboring THO–UAP56 monomer 2A. Interacting THOC62A β-propeller blades 1, 5, 6, and seven are labelled. Monomers A and B are superimposed (right) on their THOC2 subunits, and monomer B is transparent. Colors as in panel a.
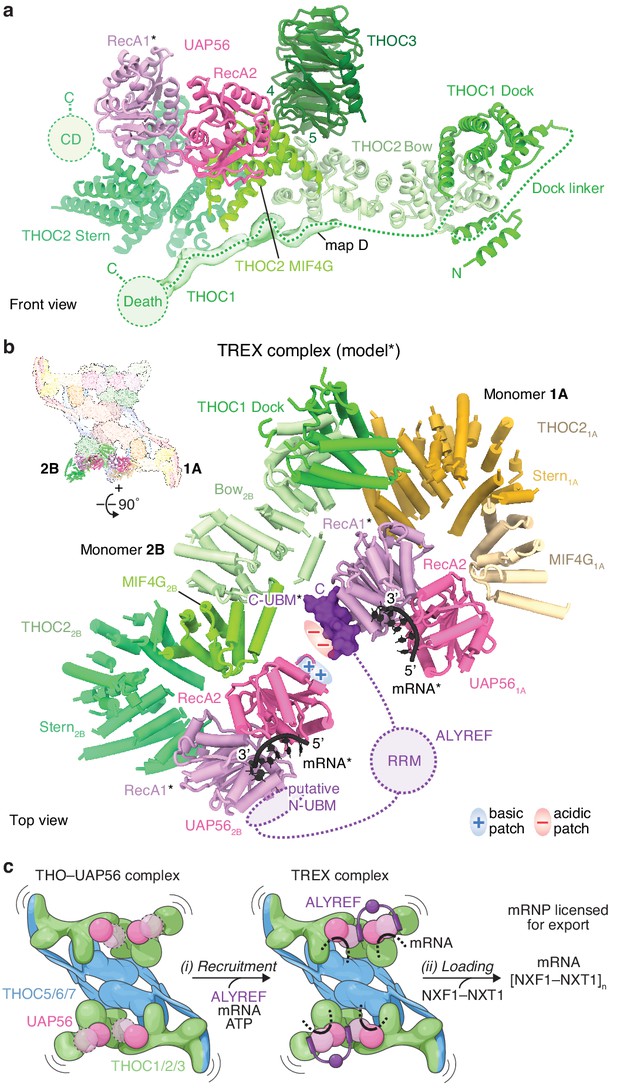
THOC2–UAP56 interface and TREX model.
(a) Details of THOC2 regions (bow, MIF4G, stern, and CD) and their interactions with THOC1, THOC3, and UAP56. THOC3 β-propeller blades 4 and 5 bind the THOC2 bow domain. The UAP56 N-terminal RecA1 lobe is mobile and modeled as for Figure 1, indicated with an asterisk. Unmodeled density (map D) was putatively assigned to THOC1 and binds along the THOC2 bow, MIF4G and stern domains. THOC2 is colored in shades of green, other subunits as in Figure 1. (b) TREX–mRNA complex model. The model suggests that ALYREF may license mRNA export by bridging two adjacent UAP56 helicases through (i) its N- and C-UBMs and (ii) charge complementary between acidic and basic residues of one ALYREF UBM, bound to UAP561A, and the juxtaposed UAP562B. The model was obtained by superimposing the human homology model of the yeast Sub2–Yra1–RNA crystal structure (Ren et al., 2017) (PDB ID 5SUP) on the UAP56 RecA2 lobe, in each monomer. The asterisk indicates modeled elements (UAP56 RecA1, mRNA, ALYREF). THO–UAP56 monomers 1A and 2B are positioned in proximity, according to 3D variability analysis (Figure 4—figure supplement 1d,e). THOC3, −5, –7, the THOC21A bow, THOC11A, and the THOC12B N-terminus were omitted for clarity. Colors as in Figure 3, ALYREF (violet), RNA (black). RRM, RNA-recognition motif; N- and C-UBM, N- and C-terminal UAP56-binding motifs. (c) Cartoon schematic of TREX-dependent mRNA-export licensing. Multivalent interactions between THO–UAP56, mRNA, and mRNP proteins may be required for (i) mRNP recruitment and mRNA binding. ALYREF may bridge two proximal UAP56 helicases in the assembled TREX–mRNA complex, to facilitate (ii) loading of multiple NXF1/NXT1 export factor complexes onto nascent mRNA and license it for export. The curved lines indicate flexibility. The UAP56 RecA1 lobe is mobile in absence of mRNA, indicated by dashed lines and the transparent surface.
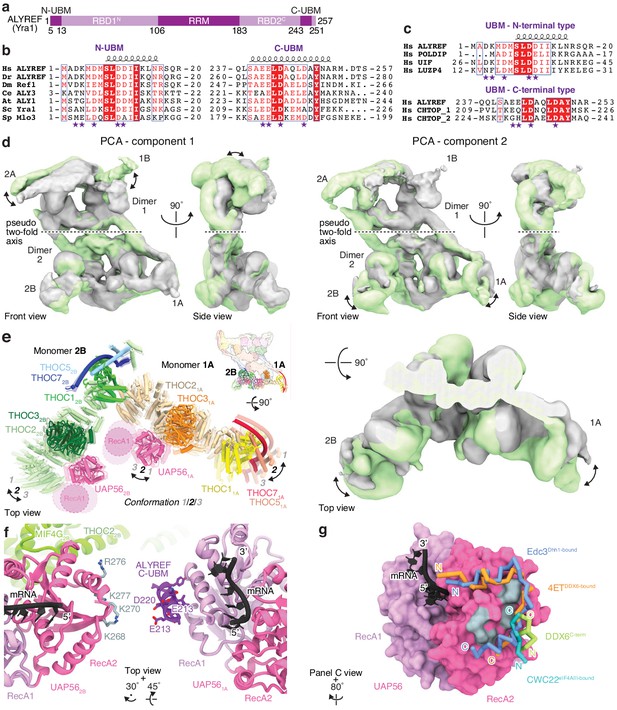
Details of THO–UAP56 flexibility and the UAP56–ALYREF interaction.
(a) Domain organization of human ALYREF (yeast Yra1). UBM, UAP56 Binding Motif; RBD, RNA Binding Domain; RRM, RNA-Recognition Motif. The RBD contains multiple RG- motifs. (b) Amino acid sequence alignment comparing N-terminal (left) and C-terminal (right) UBMs of ALYREF from Homo sapiens (Hs), Danio rerio (Dr), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Arabidopsis thaliana (At), Saccharomyces cerevisiae (Sc), and Schizosaccharomyces pombe (Sp). The alignment was created using Clustal Omega (Sievers et al., 2011) and visualized with ESPript 3 (Robert and Gouet, 2014). A secondary structure assignment, based on PSIPRED (Buchan et al., 2013) for the N-UBM and the crystal structure of Yra1 C-UBM bound to Sub2 (PDB ID 5SUP) for the C-UBM, is shown above the alignment. Purple stars indicate residues predicted to not bind the UAP56 RecA1 lobe. Invariant and conserved residues are indicated with a red background or red font, respectively. (c) Amino acid sequence alignment as in panel b but comparing N-UBMs of human ALYREF, POLDIP3, UIF and LUZP4 on the top, and the C-UBMs of human ALYREF and CHTOP below. (d) Principal component analysis (PCA) of THO–UAP56 cryo-EM data reveals flexibility. Densities for the start (green) and endpoint (gray) of the first two PCA components are shown from front and left side views. Component two is additionally shown from a top view, as in panel e. Dimers 1 and 2, monomers A and B, and the pseudo two-fold symmetry axis are indicated. (e) Modeling of THO–UAP56 monomers 1A and 2B into densities from PCA component two shows that proximal UAP56 helicases approach each other (ribbon model, colored as in Figure 2). Conformation two is the conformation of the THO–UAP56 structure, while conformations 1 and 3 reflect the starting and endpoints for the PCA component two movement shown in panel d. The UAP56 RecA1 lobes are not resolved in our structure and are indicated as dashed circles. The inset shows a surface representation of the THO–UAP tetramer, colored as in Figure 3a, with the subunits shown below depicted as ribbons. THOC5, −6 and −7 were omitted for clarity. (f) Details of the TREX–mRNA complex model shown in Figure 4b (ribbon model, colored as in Figure 4b). Side chains of ALYREF C-UBM residues in stick representation, with conserved acidic residues labelled. Conserved, basic residues on the neighboring UAP562B RecA2 lobe are shown as gray sticks and are labelled. (g) The DEXD-box RNA helicase RecA2 lobe is frequently bound by other proteins. Surface view of RNA-bound UAP56 (homology model based on the Sub2–Yra1–RNA crystal structure [Ren et al., 2017] [PDB ID 5SUP]), with RNA shown as ribbons. RecA1 lobe (purple), RecA2 lobe (pink), RNA (black) and UAP56 RecA2 lobe residues highlighted in panel f (gray). Superimposed on UAP56, and shown as Cα trace, are Edc3 (residues 88–116, Dhh1-bound conformation, PDB ID 4BRU), 4ET (residues 216–238, DDX6-bound conformation, PDB ID 5ANR), DDX6 C-terminus (residues 461–469, PDB ID 4CT4) and CWC22 (residues 131–136, eIF4AIII-bound conformation, PDB ID 4CB9). Other DEXD-box helicases, except for UAP56, were omitted for clarity.
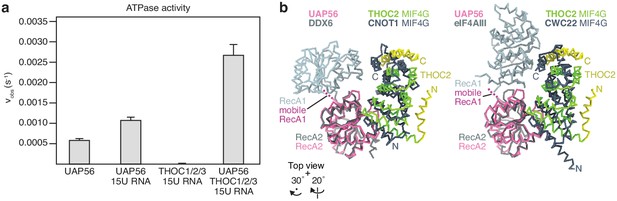
ATPase assay and structural comparisons of the DEXD-box RNA helicase–MIF4G domain interaction.
(a) Human UAP56 ATPase activity is accelerated in vitro by the THOC1/2/3 core complex. Standard deviations are calculated from three independent experiments. For details see the Materials and methods. (b) The DEXD-box helicase RecA2–MIF4G domain interaction is broadly conserved. UAP56 (pink, RecA2) and the interacting THOC2 MIF4G (green, MIF4G α29–36; yellow, α28, α41, α46) were superimposed on DDX6 (light gray) and its interacting CNOT1 MIF4G (dark gray, PDB ID 4CT4) (left) or with eIF4AIII (light gray) and its interacting CWC22 MIF4G (dark gray, PDB ID 4CB9) (right) via their RecA2 domains, respectively. Additional contacts are observed between CNOT1 and CWC22 MIF4G domains and the RecA1 lobe of the respective DEXD-box helicase. Note that a transient interaction between RecA1–THOC2 might not be observed in a cryo-EM experiment, whereas such transient interactions may be trapped in crystal structures.
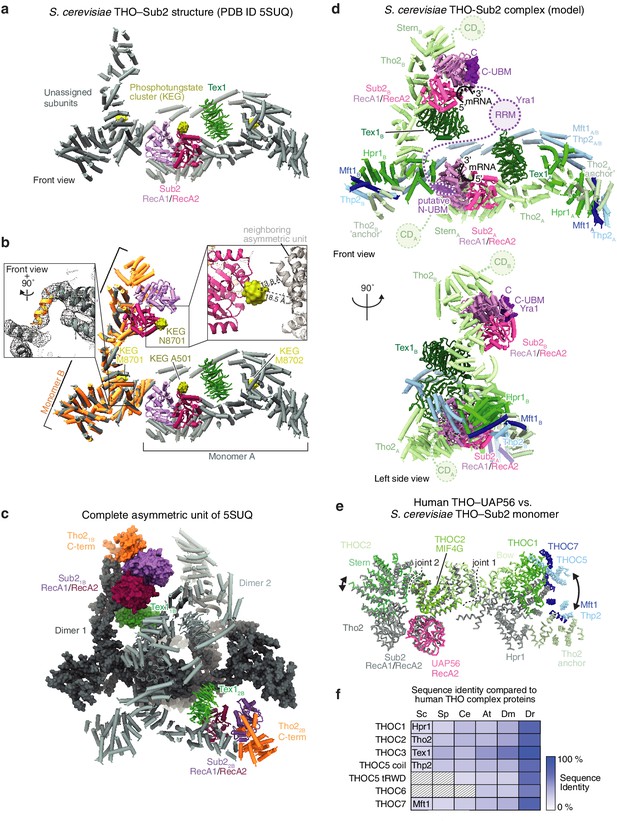
Revised yeast THO–Sub2 model and conservation of the THO architecture.
(a) Ribbon model of the published chimeric yeast THO-Sub2 crystal structure (Ren et al., 2017), which contained Saccharomyces bayanus Tex1, while the other subunits were from Saccharomyces cerevisiae. (Sub2, RecA1, purple; RecA2 pink; unassigned subunits, gray; Tex1, green; PDB ID 5SUQ). Phosphotungsten (KEG) clusters are shown as surfaces (yellow). (b) The published yeast THO–Sub2 crystal structure contained a dimer. Superposition of model residues 8–5289 (corresponding to monomer A, orange) onto model residues 6170–8550 of the unassigned subunits. Related phosphotungsten clusters are shown as surfaces in dark yellow (N8701 and A501) and light yellow (M8701 and M8702). The left inset shows that unmodeled density in the final (2Fo − Fc) map contoured at 1.0σ of monomer B was independently built in the superimposed monomer A (model residues 482–492, orange). The right inset shows a closeup of KEG N8701 (yellow, surface representation) that is located at the previously unmodeled monomer B Sub2 RecA2 lobe, but distant from monomer A. In support of this architecture, the endogenous Saccharomyces cerevisiae THO complex was also reported to form dimers using negative stain EM (Peña et al., 2012). (c) Asymmetric unit of the revised yeast THO–Sub2 crystal structure. The previously built backbone model is colored in gray. The second Sub2 (RecA1, purple; RecA2 pink), Tex1 (green) and Tho2 C-terminus (orange) are shown. The two dimers in the asymmetric unit are shown, dimer one as a surface and dimer two as a ribbon model. Consistent with the model, we observed no clashes in the completed asymmetric unit. (d) Model of the yeast Saccharomyces cerevisiae TREX complex dimer in front and left side views. The model suggests that yeast Yra1, similar to human ALYREF, may bridge two Sub2 helicases that are located ~80 Å apart. The shown homology model is based on the combined revised chimeric yeast THO–Sub2 complex crystal structure (for details see a–c, Materials and methods) and the Sub2–RNA–Yra1 structure (Ren et al., 2017). The Tho2 ‘anchor’ domain is putatively assigned based on secondary structure prediction and homology to the human monomer B THOC2 structure (Figure 1—figure supplement 5b). Colors as in Figure 1, Yra1 (violet), RNA (black). (e) Superposition of the human THO–UAP56 complex monomer 1A (colored as in Figure 4a) with yeast THO–Sub2 monomer A on their THOC2 (yeast Tho2) subunits (gray; Mft1–Thp2, light blue; putative anchor Tho2, light green). Note the good fit of the THOC2 MIF4G domain (between joints 1 and 2), and the structural differences towards the THOC2 Stern (left) and the base of the THOC5–THOC7 coiled coil (right) (indicated by arrows). Tex1, THOC3, and the THOC5 tRWD domain were omitted for clarity. (f) The THO complex architecture was adapted during evolution. The THOC5 tRWD domain and THOC6, which stabilize the tetramer, are found in complex eukaryotes. The heatmap is colored according to sequence identity to the human THO subunits. Shown are Danio rerio (Dr), Drosophila melanogaster (Dm), Caenorhabditis elegans (Ce), Arabidopsis thaliana (At) Saccharomyces cerevisiae (Sc), and Schizosaccharomyces pombe (Sp).
Videos
360 degree rotation of the human THO–UAP56 complex, colored as in Figure 1c.
360 degree rotation of the yeast THO–Sub2 complex, colored as in Figure 4—figure supplement 3d.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (H. sapiens) | THOC1 | Uniprot | Q96FV9 | |
| Gene (H. sapiens) | THOC2 | Uniprot | Q8NI27 | |
| Gene (H. sapiens) | THOC3 | Uniprot | Q96J01 | |
| Gene (H. sapiens) | THOC5 | Uniprot | Q13769 | |
| Gene (H. sapiens) | THOC6 | Uniprot | Q8NI27 | |
| Gene (H. sapiens) | THOC7 | Uniprot | Q6I9Y2 | |
| Gene (H. sapiens) | UAP56/DDX39B | Uniprot | Q13838 | |
| Gene (H. sapiens) | NXF1 | Uniprot | Q9UBU9 | |
| Gene (H. sapiens) | NXT1 | Uniprot | Q9UKK6 | |
| Gene (H. sapiens) | NUP214 | Uniprot | P35658 | |
| Strain, strain background (Escherichia coli) | DH10EmBacY | Geneva Biotech | DH10EmBacY | Strain propagated locally from commercial source stock |
| Strain, strain background (E. coli) | BL21(DE3)RIL | Agilent | 230245 | Strain propagated locally from commercial source stock |
| Cell line (Spodoptera frugiperda) | Sf9 | Thermo Fisher Scientific | IPLB-Sf-21-AE; RRID:CVCL_0549 | Strain propagated locally from commercial source stock |
| Cell line (Trichoplusia ni) | High Five | Thermo Fisher Scientific | BTI-TN-5B1-4; RRID:CVCL_C190 | Strain propagated locally from commercial source stock |
| Cell line (Lenti-X) | Lenti-X | Takara | Cat. #632180 | |
| Cell line (K562) | K562 | DSMZ | ACC 10, RRID:CVCL_0004 | |
| Antibody | anti-THOC1 (Polyclonal rabbit) | Merk | HPA019096 | (dilution 1:500) |
| Antibody | anti-mouse IgG-HRP (Polyclonal goat) | Promega | W4021 | (dilution 1:10000) |
| Recombinant DNA reagent | pACEBac1 | Geneva Biotech | pACEBac1 | |
| Recombinant DNA reagent | pOPINB | Addgene | #41142 | |
| Recombinant DNA reagent | Lentiviral vector | Addgene | #31485 | |
| Software, algorithm | hhpred | doi:10.1093/nar/gki408 | RRID:SCR_010276 | https://toolkit.tuebingen.mpg.de/tools/hhpred |
| Software, algorithm | WARP | doi: 10.1038/s41592-019-0580-y | http://www.warpem.com/warp/ | |
| Software, algorithm | cryoSPARC | doi:10.1038/nmeth.4169 | RRID:SCR_016501 | https://cryosparc.com/ |
| Software, algorithm | Relion | doi:10.7554/eLife.42166 | RRID:SCR_016274 | https://www3.mrc-lmb.cam.ac.uk/relion/ |
| Software, algorithm | ResMap | doi:10.1038/nmeth.2727 | http://resmap.sourceforge.net | |
| Software, algorithm | Modeller | doi:10.1002/cpbi.3 | RRID:SCR_008395 | https://salilab.org/modeller/ |
| Software, algorithm | COOT | doi:10.1107/S0907444910007493 | RRID:SCR_014222 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Software, algorithm | Phenix | doi:10.1107/S2059798318006551 | RRID:SCR_014224 | https://www.phenix-online.org/ |
| Software, algorithm | USCF ChimeraX | doi:10.1002/pro.3235 | RRID:SCR_015872 | https://www.rbvi.ucsf.edu/chimerax/ |
| Software, algorithm | Pymol | Schrödinger LLC, 2020 | RRID:SCR_000305 | https://pymol.org/2/ |
Additional files
-
Supplementary file 1
Coordinate file of the revised yeast THO–Sub2 complex.
- https://cdn.elifesciences.org/articles/61503/elife-61503-supp1-v2.pdb
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61503/elife-61503-transrepform-v2.pdf





