Pregnancy success in mice requires appropriate cannabinoid receptor signaling for primary decidua formation
Figures
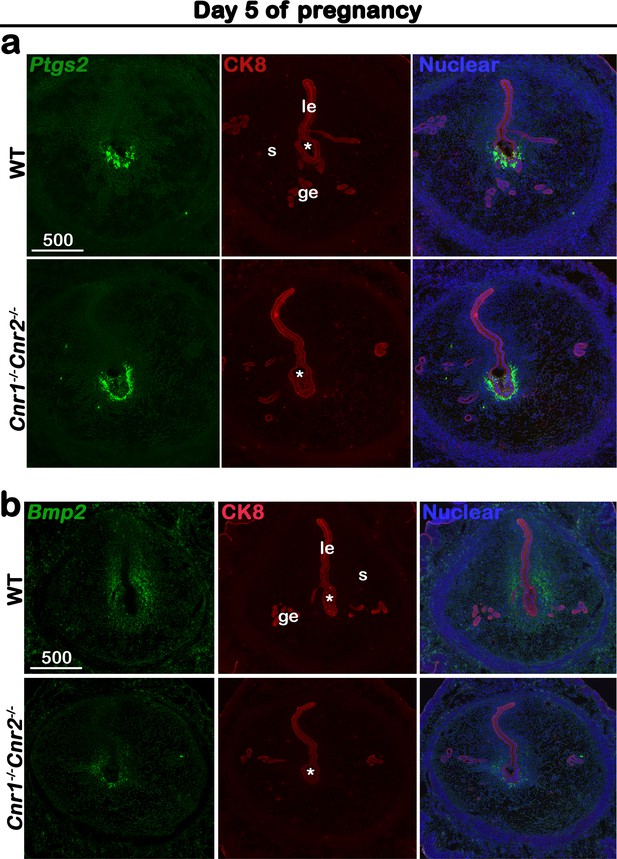
Cnr1-/-Cnr2-/- females show normal responses to embryonic stimulation during implantation, but decidual process is compromised.
(a) In situ hybridization of Ptgs2 in uteri on day 5 of pregnancy. The expression patterns of Ptgs2 critical for implantation show no significant difference in WT and Cnr1-/-Cnr2-/- females. CK8 staining outlines uterine epithelial cells. (b) In situ hybridization of Bmp2 in uteri on day 5 of pregnancy. Decidual responses in Cnr1-/-Cnr2-/- females are much weaker than those in WT females. le, luminal epithelium; s, stroma; ge, glandular epithelium; Asterisks, positions of embryos; Scale bars, 500 μm. All images are representative of three independent experiments.
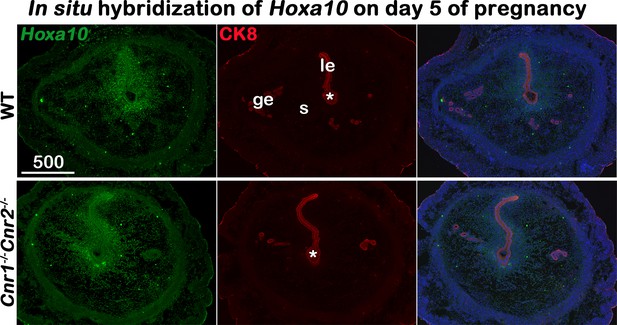
In situ hybridization of Hoxa10 in uteri on day 5 of pregnancy.
Decidual responses in Cnr1-/-Cnr2-/- females are weaker than those in WT females. le, luminal epithelium; s, stroma; ge, glandular epithelium; Asterisks, positions of embryos; Scale bars, 500 μm. All images are representative of three independent experiments.
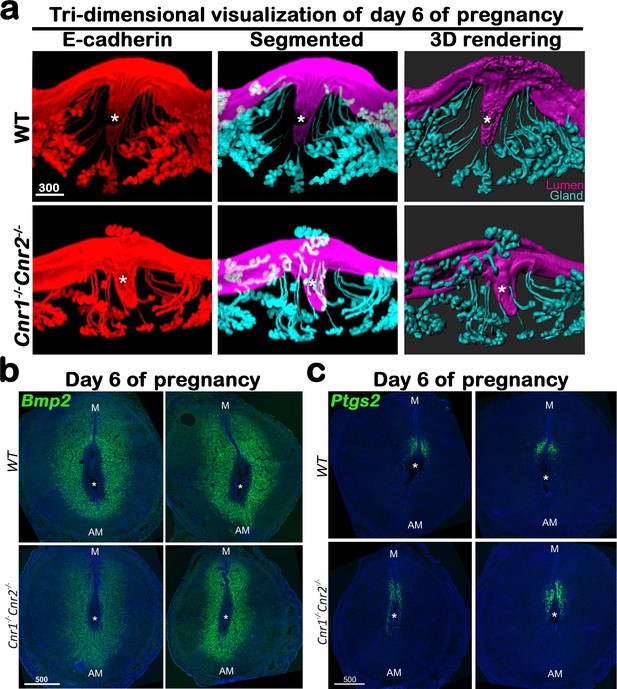
Cnr1-/-Cnr2-/- females have suboptimal decidualization.
(a) 3D visualization of day 6 implantation sites in WT and Cnr1-/-Cnr2-/- females. Images of E-cadherin immunostaining, segmented, and 3D rendered images of day 6 implantation sites in each genotype show compromised decidual responses in Cnr1-/-Cnr2-/- females. Scale bars, 300 μm. (b and c) In situ hybridization of Bmp2 and Ptgs2 in uteri on day 6 of pregnancy. M, mesometrial; AM, anti-mesometrial; Asterisks, positions of embryos; Scale bars, 500 μm. All images are representative of three independent experiments.
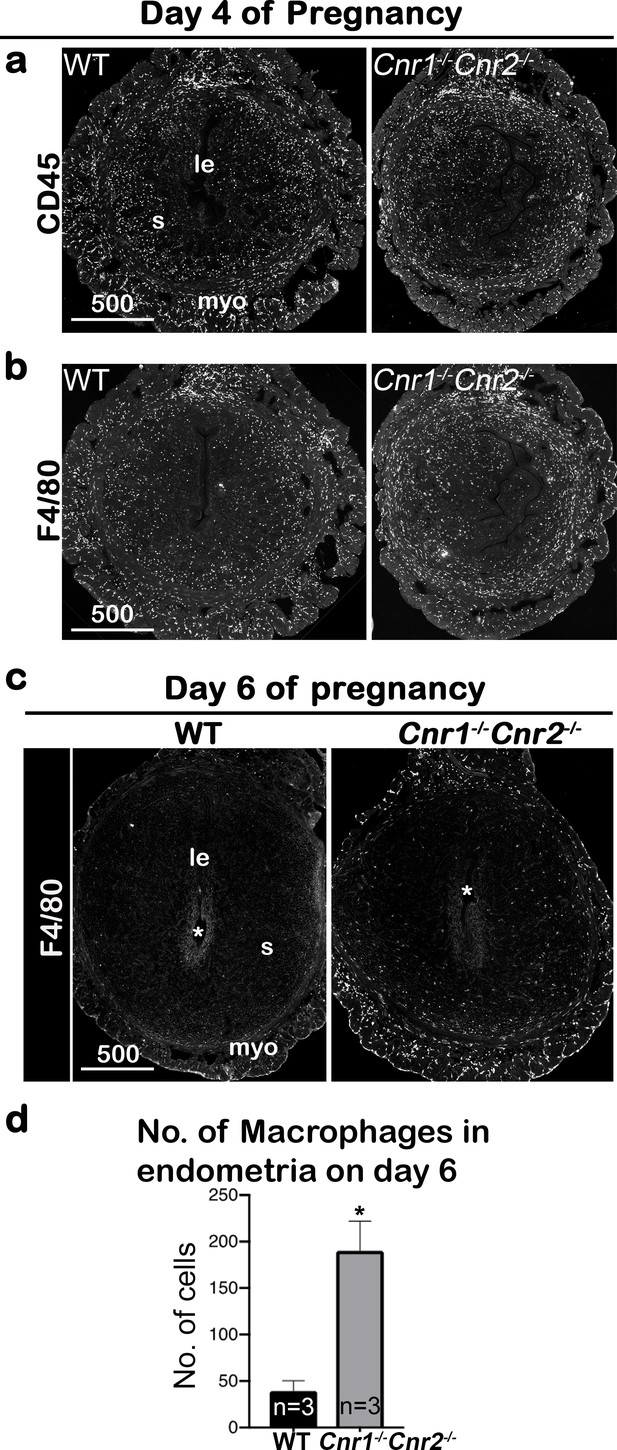
Macrophages are retained around implantation chambers of compromised deciduae in Cnr1-/-Cnr2-/- females.
(a) Uterine leukocytes in pre-implantation are highlighted by CD45, which is expressed on all leukocytes. (b) Immunofluorescent staining of F4/80 on day 4 of pregnancy to mark macrophages. (c) Immunofluorescent staining of F4/80 on day 6 of pregnancy. (d) The numbers of macrophages within the endometrial domains are quantified using three sections obtained from three different animals in each genotype. le, luminal epithelium; s, stroma; myo, myometrium; Asterisks, positions of embryos; Scale bars, 500 μm; *p<0.05, Student’s t-tests. All images are representative of three independent experiments.
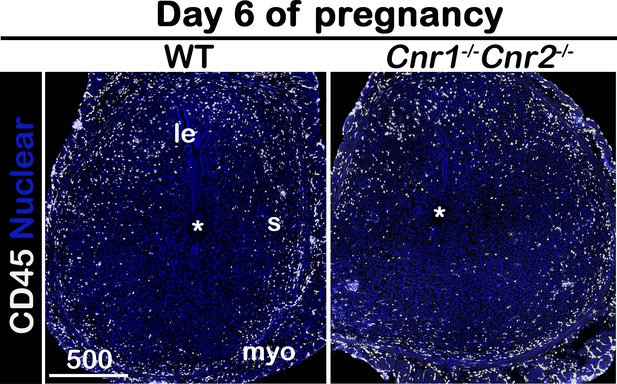
Immunofluorescence of CD45 on day 6 of pregnancy.
le, luminal epithelium; s, stroma; myo, myometrium; Asterisks, positions of embryos; Scale bars, 500 μm. All images are representative of three independent experiments.
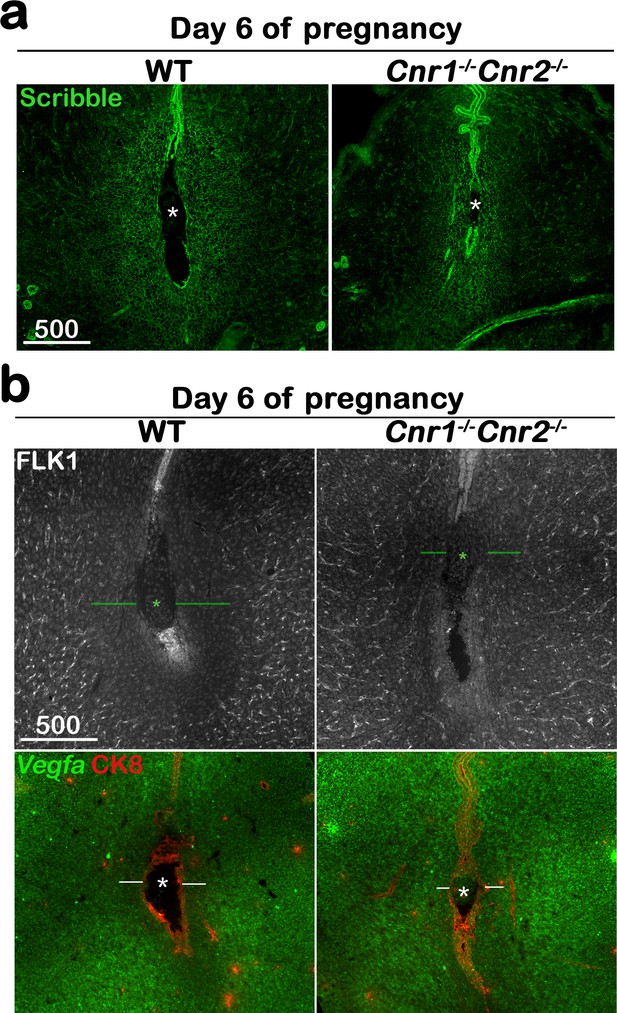
The formation of the primary decidual zone is compromised in Cnr1-/-Cnr2-/- implantation sites.
(a) The primary decidual zone is highlighted by Scribble on day 6 of pregnancy. (b) Immunofluorescence staining of FLK1 and in situ hybridization of Vegfa in day 6 pregnant uteri. Blood vessels and higher Vegfa signals are observed in the PDZ area (indicated by green or white lines) of Cnr1-/-Cnr2-/- implantation sites. CK8 staining outlines epithelial cells. Asterisks, positions of embryos; Scale bars, 500 μm. All images are representative of three independent experiments.
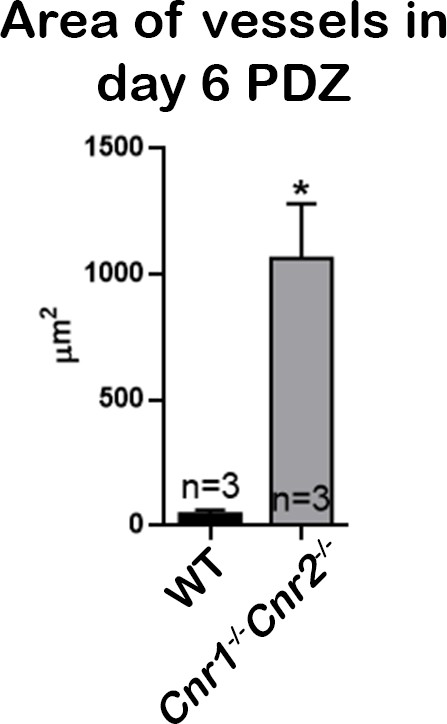
Quantitation of blood vessel area in PDZs on day 6 of pregnancy.
Areas of blood vessels in PDZs of three sections from each genotype were quantified using ImageJ. Numbers on bars are sample sizes; *p<0.05, Student’s t-tests.
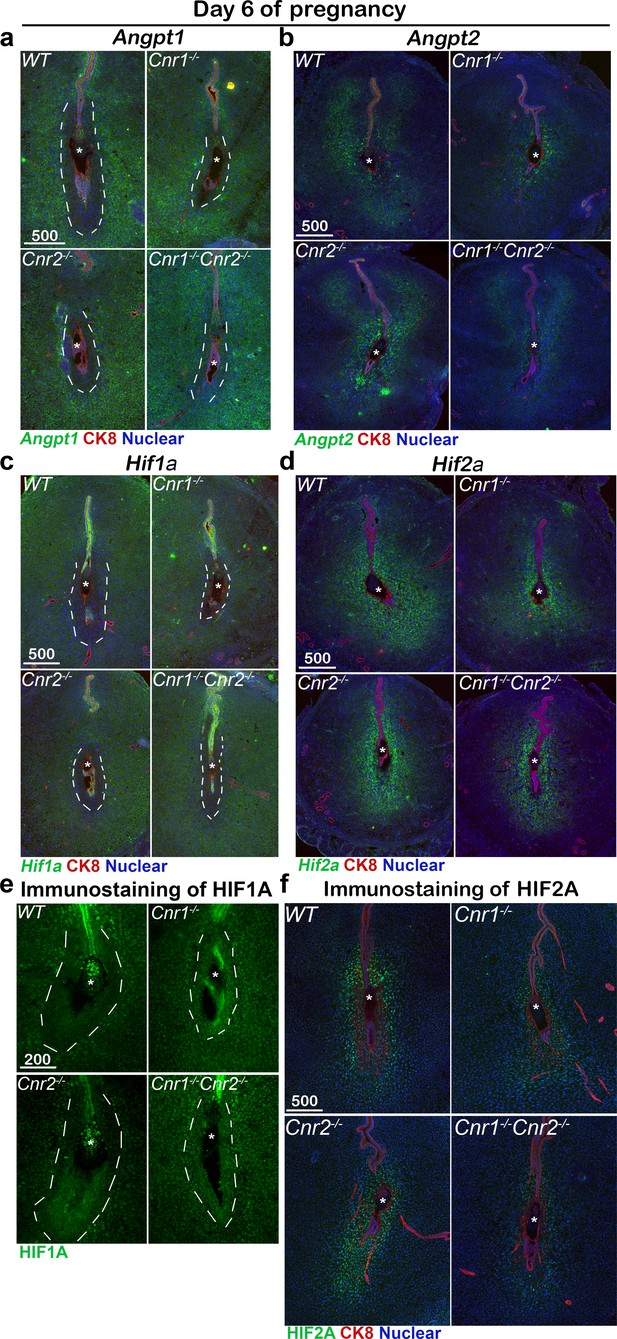
Angiogenic factors are misregulated in Cnr1-/-Cnr2-/- and Cnr1-/- PDZ.
(a–d) In situ hybridization of Angpt1, Angpt2, Hif1a and Hif2a on day 6 of pregnancy. CK8 staining outlines uterine epithelial cells. (e and f) Immunostaining of HIF1A and HIF2A on day 6 of pregnancy. Dotted lines outline the PDZs. Asterisks, positions of embryos; Scale bars, 500 μm. All images are representative of three independent experiments.
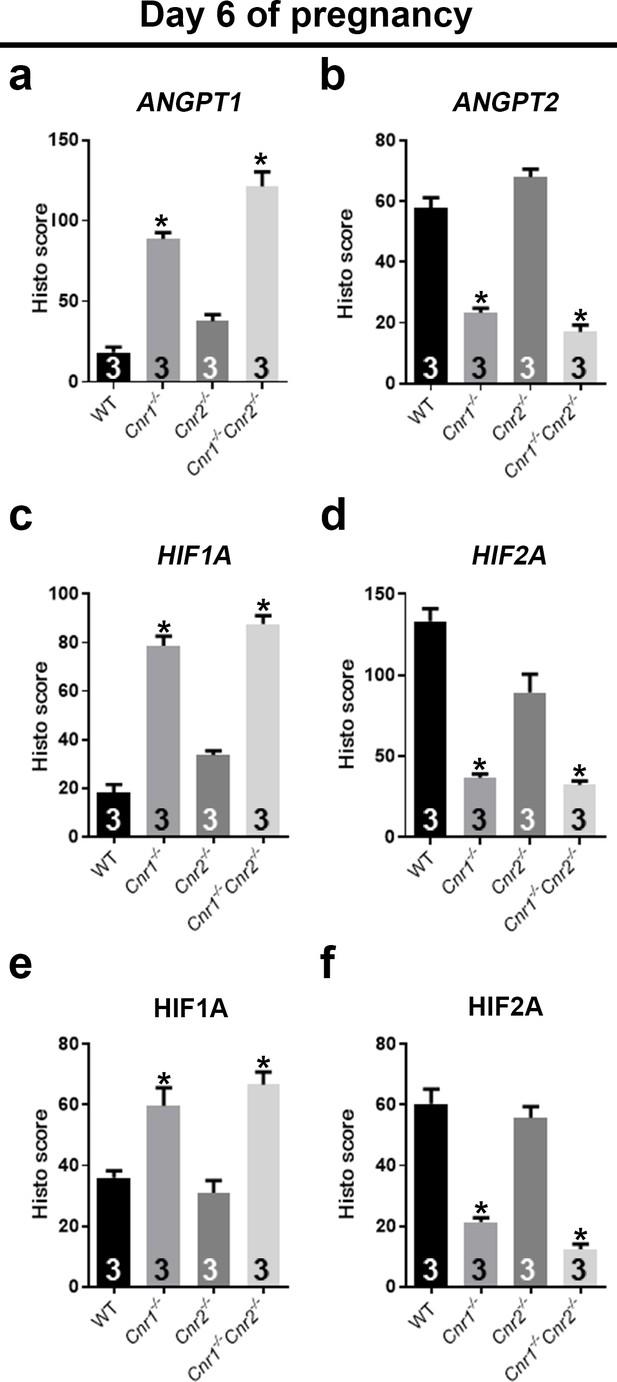
Quantification of signals of angiogenic factors on day 6 of pregnancy.
Quantification of in situ hybridization signals (a–d) and immunostaining signals (e and f) is plotted. H scores of signals in PDZs as outlined by dotted white lines are calculated for ANGPT1 in panel a and HIF1A in panels c and e. H scores for ANGPT2 signals are plotted in panel b. H scores for HIF2A signals are plotted in panels d and f. Values are mean ± SEM; Numbers on bars are sample sizes; *p<0.05, Unpaired t-tests with Welch’s correction.
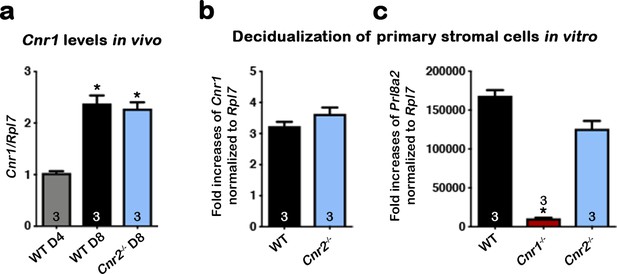
CB1 expression in mouse uterine stromal cells increases during decidualization.
(a) Levels of Cnr1 are higher at the peak of decidual response on day 8 of pregnancy as compared to day 4 of pregnancy. Samples were from three different females in each genotyping group. (b) Cnr1 is induced in primary culture of WT and Cnr2-/- stromal cells undergoing decidualization in vitro. (c) Decidualization is severely reduced in primary Cnr1-/- stromal cells after decidual stimulation in vitro. Prl8a2 expression served as an indicator of decidual responses. Primary cells were collected from three different females in each genotyping groups. Values are mean ± SEM; Numbers on bars represent sample sizes; *p<0.05, Unpaired t-tests with Welch’s correction.
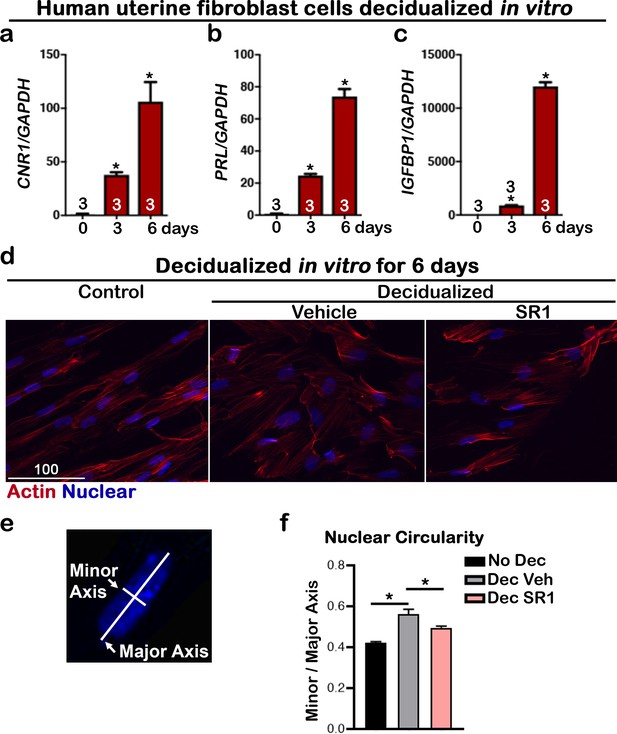
CNR1 levels are upregulated in human uterine fibroblasts during in vitro decidualization.
(a) Levels of CNR1 gradually increase in Huf cells undergoing decidualization. (b and c) Decidual responses are marked by significant increases in PROLACTIN and IGFBP1 levels in Huf cells. Sample sizes on bars are the number of culture wells. (d) Morphology of Huf cells changes from spindle shapes to more round shape; SR1 treatment compromises the morphological change in Huf cells. Cells are outlined with actin staining. Scale bar, 100 μm. All images are representative of three culture wells. (e) A scheme depicts the measurement of minor and major axes for calculating nuclear circularity. (f) The nuclear circularity of Huf cells before and after decidualization. SR1 treatment compromises the decidual process. Three fields were randomly chosen in three different culture wells. Values are mean ± SEM; Numbers on bars are sample sizes; *p<0.05, Unpaired t-tests with Welch’s correction.
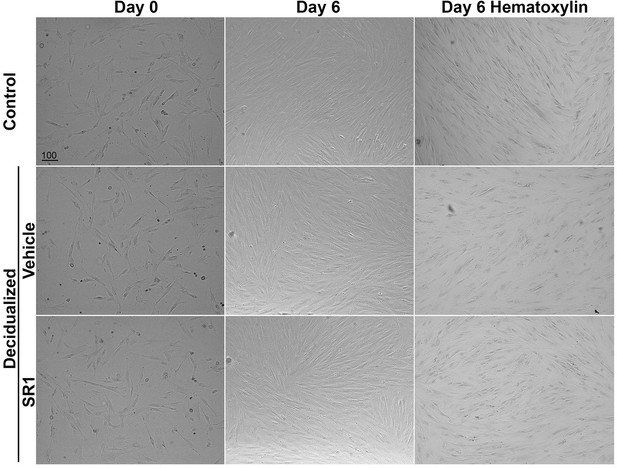
Morphology of Huf cells before and after decidualization.
Phase-contrast pictures of Huf cells treated with SR1 or vehicle before (day 0) and after (day 6) decidualization were shown. After decidualization, cells were stained with hematoxylin, and pictures were taken after mounting. All images are representative of three independent experiments. Scale bars, 100 μm.
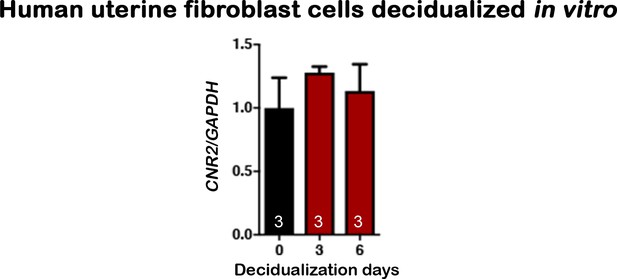
CNR2 shows no significant change in human uterine fibroblasts during in vitro decidualization.
Numbers on bars are sample sizes; Values are mean ± SEM.
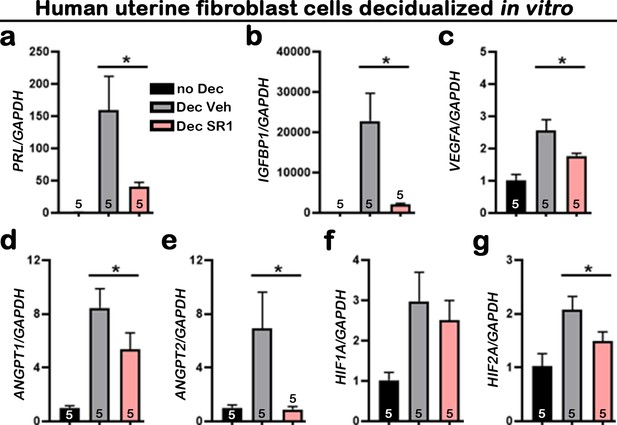
Suppression of CB1 impairs normal Huf cell decidualization and expression of angiogenic factors.
(a–g) qPCR of PRL, IGFBP1, VEGFA, ANGPT1, ANGPT2, HIF1A, and HIF2A using RNA collected 3 d after decidualization. Values are mean ± SEM; Numbers on bars are numbers of culture wells; *p<0.05, Unpaired t-tests with Welch’s correction.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-E-cadherin (Rabbit monoclonal) | Cell Signaling Technology | Cat# 3195S, RRID:AB_2291471 | Whole mount IF(1:100) |
| Antibody | Alexa Fluor 594 AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson Immuno Research | Cat# 711-585-152, RRID:AB_2340621 | Whole mount IF(1:300) |
| Antibody | Anti-Cytokeratin 8, TROMA-1 (Rat monoclonal) | Developmental Studies Hybridoma Bank | Cat# TROMA-I, RRID:AB_531826 | IF (1:100) |
| Antibody | Anti-CD45, (Rat monoclonal) | Biolegend | Cat# 103101, RRID:AB_312966 | IF (1:200) |
| Antibody | Anti-HIF1A (Rabbit monoclonal) | Cell Signaling Technology | Cat# 36169T, RRID:AB_2799095 | IF (1:200) |
| Antibody | Anti-HIF2A (Rabbit polyclonal) | Novus | Cat# NB100-122SS, RRID:AB_10002593 | IF (1:500) |
| Antibody | Anti-F4/80 (Rat monoclonal) | Biolegend | Cat# MCA497R, RRID:AB_323279 | IF (1:200) |
| Antibody | Anti-Scrib (Rabbit polyclonal) | Santa Cruz | Cat# sc-28737, RRID:AB_2184807 | IF (1:500) |
| Antibody | Alexa Fluor 594 AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson Immuno Research | Cat#712-585-150, RRID:AB_2340688 | IF (1:400) |
| Antibody | Alexa Fluor 488 AffiniPure Donkey Anti-Rabbit IgG (H+L) | Jackson Immuno Research | Cat# 711-545-152, RRID:AB_2313584 | IF (1:400) |
| Antibody | Cy2 AffiniPure Donkey Anti-Rat IgG (H+L) | Jackson Immuno Research | Cat# 712-225-153, RRID:AB_2340674 | IF (1:400) |
| Biological sample (M. musculus) | Primary WT,Cnr1-/- and Cnr2-/- mouse uterine stromal cells | The current laboratory | Primary mouse uterine stromal cells freshly isolated from mouse uteri | |
| Biological sample (Homo-sapiens) | Human uterine fibroblasts | Cincinnati Children’s Hospital | A gift from Handwerger laboratory. Aliquots maintained as frozen cells | |
| Sequence-based reagent | Mouse Prl8a2_F | This paper | PCR primers | ACGTGATGAGGAGGTTCT |
| Sequence-based reagent | Mouse Prl8a2_R | This paper | PCR primers | AATCTTGCCCAGTTATGC |
| Sequence-based reagent | Mouse Cnr1_F | This paper | PCR primers | CATTGGGACTATCTTTGCGG |
| Sequence-based reagent | Mouse Cnr1_R | This paper | PCR primers | GGTTCTGGAGAACCTGCTGG |
| Sequence-based reagent | Mouse Rpl7_F | This paper | PCR primers | GCAGATGTACCGCACTGAGATTC |
| Sequence-based reagent | Mouse Rpl7_R | This paper | PCR primers | ACCTTTGGGCTTACTCCATTGATA |
| Sequence-based reagent | Human CNR1_F | This paper | PCR primers | gctgcctaaatccactctgc |
| Sequence-based reagent | Human CNR1_R | This paper | PCR primers | tggacatgaaatggcagaaa |
| Sequence-based reagent | Human CNR2_F | This paper | PCR primers | GATTGGCAGCGTGACTATGA |
| Sequence-based reagent | Human CNR2_R | This paper | PCR primers | GATTCCGGAAAAGAGGAAGG |
| Sequence-based reagent | Human IGFBP1_F | This paper | PCR primers | ccaaactgcaacaagaatg |
| Sequence-based reagent | Human IGFBP1_R | This paper | PCR primers | gtagacgcaccagcagag |
| Sequence-based reagent | Human PRL_F | This paper | PCR primers | aagctgtagagattgaggagcaaa |
| Sequence-based reagent | Human PRL_R | This paper | PCR primers | tcaggatgaacctggctgacta |
| Sequence-based reagent | Human VEGFA_F | This paper | PCR primers | aaggaggagggcagaatcat |
| Sequence-based reagent | Human VEGFA_R | This paper | PCR primers | cacacaggatggcttgaaga |
| Sequence-based reagent | Human ANGPT1_F | This paper | PCR primers | ggacagcaggaaaacagagc |
| Sequence-based reagent | Human ANGPT1_R | This paper | PCR primers | cacaagcatcaaaccaccat |
| Sequence-based reagent | Human ANGPT2_F | This paper | PCR primers | ataagcagcatcagccaacc |
| Sequence-based reagent | Human ANGPT2_R | This paper | PCR primers | aagttggaaggaccacatgc |
| Sequence-based reagent | Human HIF1A_F | This paper | PCR primers | tcatccaagaagccctaacg |
| Sequence-based reagent | Human HIF1A_R | This paper | PCR primers | cgctttctctgagcattctg |
| Sequence-based reagent | Human HIF2A_F | This paper | PCR primers | gaacagcaagagcaggttcc |
| Sequence-based reagent | Human HIF2A_R | This paper | PCR primers | ggcagcaggtaggactcaaa |
| Sequence-based reagent | Human GAPDH_F | This paper | PCR primers | gaaggtgaaggtcggagt |
| Sequence-based reagent | Human GAPDH_R | This paper | PCR primers | gatggcaacaatatccactt |
| Chemical compound, drug | SR141716 | Cayman Chemical | Cat# 9000484 | 2 µM in culture |
| Other | RPMI 1640 Medium | Corning | Cat# 15040-cv | |
| Other | Fetal bovine serum | Fisher | Cat# FB12999102 |





