The ESCRT machinery regulates retromer-dependent transcytosis of septate junction components in Drosophila
Figures
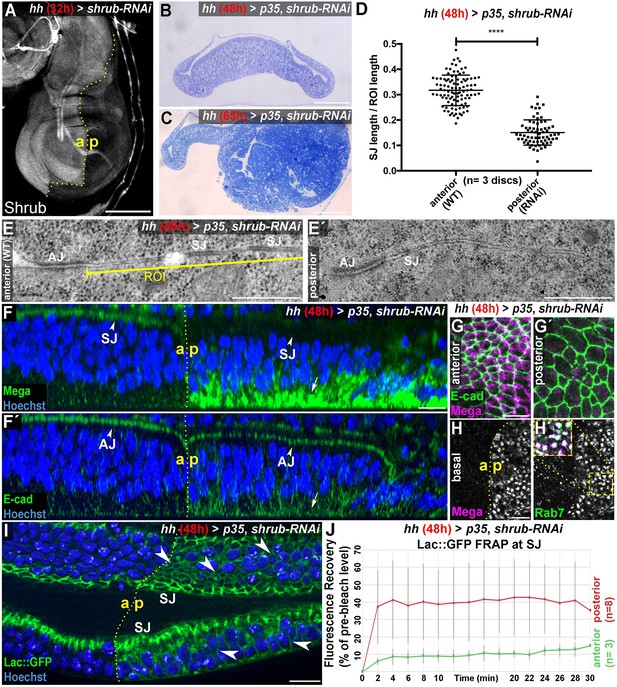
The ESCRT-III component Shrub is required for SJ integrity.
(A) 32-hr expression of shrub-RNAi under the control of hhGal4 effectively depletes Shrub levels in the posterior compartment of wing imaginal discs. (B–C) Thin sections of wing discs co-expressing shrub-RNAi and p35 for either 48 hr (B) or 65 hr (C). Note the intact epithelial monolayer at the 48 hr stage (B) vs. posterior neoplastic overgrowth after 65 hr (C). (D) Quantification of electron-dense SJ in TEM sections reveals a ~50% reduction of SJ strands within Shrub-depleted compartments (48 hr) compared to control. Each data point represents a single junctional region of interest (ROI) of 2 μm length as defined in (E) A two-tailed t-test was used for statistical analysis with the significance level **** representing a p-value<0.0001. Representative ROIs of control (E) and Shrub-depleted compartments (E') visualize the reduction in electron-dense SJ. (F) Optical section of a wing disc after 48 hr of Shrub depletion in the posterior compartment. The claudin Mega is lost from the apical SJ (compare arrowheads) and accumulates basal to the nuclei (arrow). In contrast, E-cad localization to AJ between the anterior control and the posterior compartment is largely unaffected (compare arrowheads in F') while only little accumulation is seen at the basal pole (arrow). (G) Projections of the junctional area show the close association of E-cad with Mega in anterior control compartments (G) and apparent lack of junctional Mega in the Shrub-depleted tissue (G'). In basal planes, intracellular accumulation of Mega within the shrub-RNAi compartment is evident as a punctate pattern, colocalizing with the endosomal marker Rab7 (H). Subcellular localization of the SJ component Lachesin::GFP is restricted to SJ in anterior control compartments while showing extensive spread along the entire lateral membrane in Shrub-depleted cells (arrowheads in I). (J) Fluorescence recovery after photobleaching (FRAP) analysis of Lac::GFP at the SJ level reveals increased mobility in shrub-RNAi tissue, indicating defective fence function of SJ. a: anterior/p: posterior. The red graph displays mean Lac::GFP fluorescence recovery in the posterior compartment, the green graph represents mean values for the anterior control compartment. Error bars indicate standard deviation for each individual time point. Scale bar in (A) 100 μm, (B–C) 50 μm, (E) 0.5 μm, and all other panels 10 μm.
-
Figure 1—source data 1
Quantification of SJ density in WT and Shrub-depleted (+p35) tissue.
- https://cdn.elifesciences.org/articles/61866/elife-61866-fig1-data1-v2.xlsx
-
Figure 1—source data 2
FRAP kinetics of Lac-GFP in WT and Shrub-depleted (+p35) tissue.
- https://cdn.elifesciences.org/articles/61866/elife-61866-fig1-data2-v2.xlsx
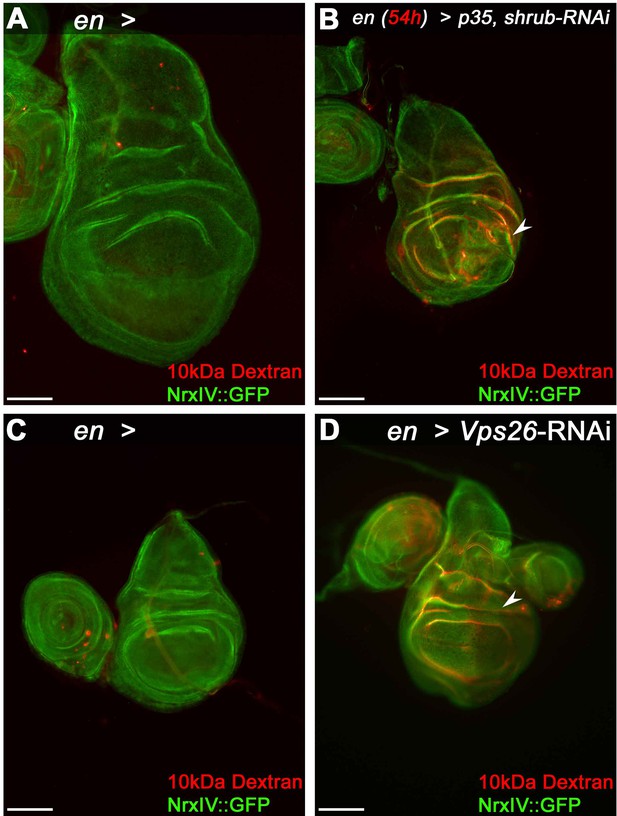
Epithelial barrier function is compromised in Shrub or Vps26-depleted wing disc tissue.
(A) Control wing disc expressing engrailed (en)-Gal4 and Nrx::IV after incubation with dsRed-labeled 10 kDa Dextran. Note the faint red staining indicative of Dextran penetration into the tissue. (B) Dextran accumulates in epithelial folds of a wing disc depleted of Shrub in the posterior compartment (arrowhead). (C) Control disc without RNAi expression and (D) after Vps26 depletion. Note the reduced NrxIV::GFP signal in the Vps26 depleted posterior compartment in (D), accompanied by Dextran infiltration of the tissue (arrowhead). Scale bars in all panels represent 100 μm.
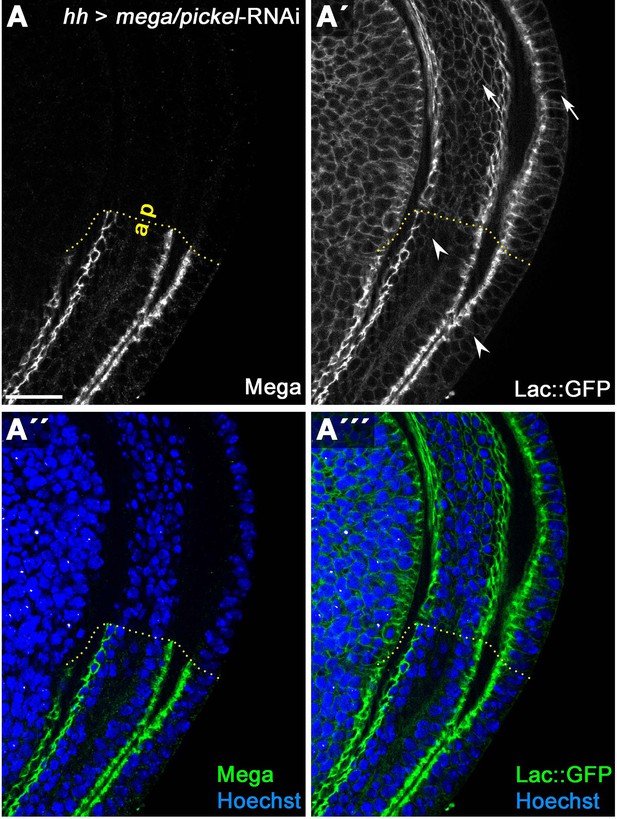
Mega depletion causes slight lateral spreading of Lachesin::GFP.
(A) HhGal4-driven posterior expression in wing discs of an RNAi construct targeting the SJ component Mega effectively depletes Mega levels as detected by antibody staining. (A') Subcellular localization of an endogenously tagged Lachesin::GFP (SJ component associated with the outer membrane leaflet) in the wildtype anterior control compartment is largely restricted to the SJ. In the Mega depleted posterior compartment, the fraction of Lac::GFP localized at the lateral membrane basal to the SJ is slightly increased (compare arrowheads/anterior with arrows/posterior). This suggests that integrity of the SJ complex is partially compromised upon loss of Mega in wing imaginal discs. Scale bar represents 20 μm.
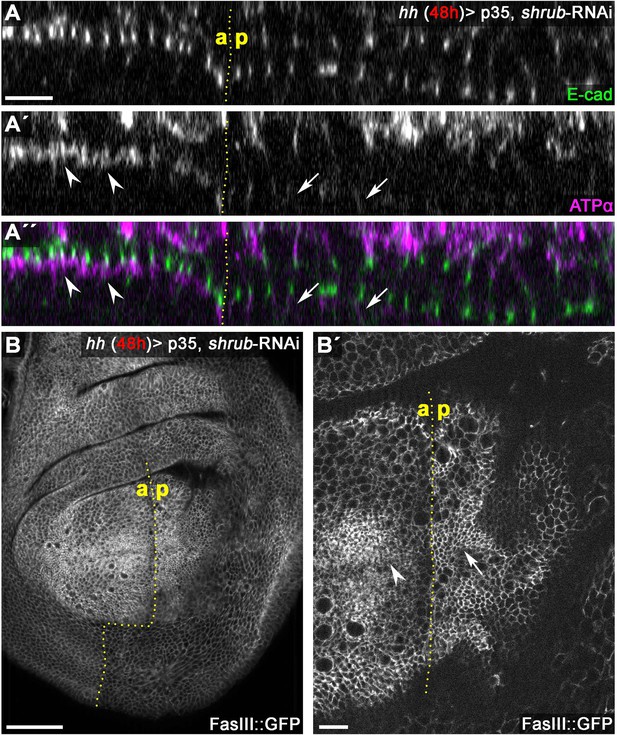
Junctional levels of the core SJ component ATPα are reduced in Shrub-depleted wing imaginal disc tissue.
(A) Optical section of the junctional region of a wing disc expressing p35 and shrub-RNAi for 48 hr under the control of hhGal4 in the posterior compartment. While E-cad levels at the AJ in the Shrub-depleted tissue appear unchanged compared to the control compartment (A), ATPα levels at the SJ are reduced in the posterior compartment (compare arrowheads/anterior to arrows/posterior in A' and A''). In contrast, apical levels of the SJ-associated TMP FasIII, visualized by expression of an endogenously GFP tagged allele (FasIII::GFP) are not reduced in the Shrub-depleted posterior compartment (compare arrowhead/anterior with arrow/posterior in B). Scale bars in all panels represent 10 μm except for (B): 50 μm.
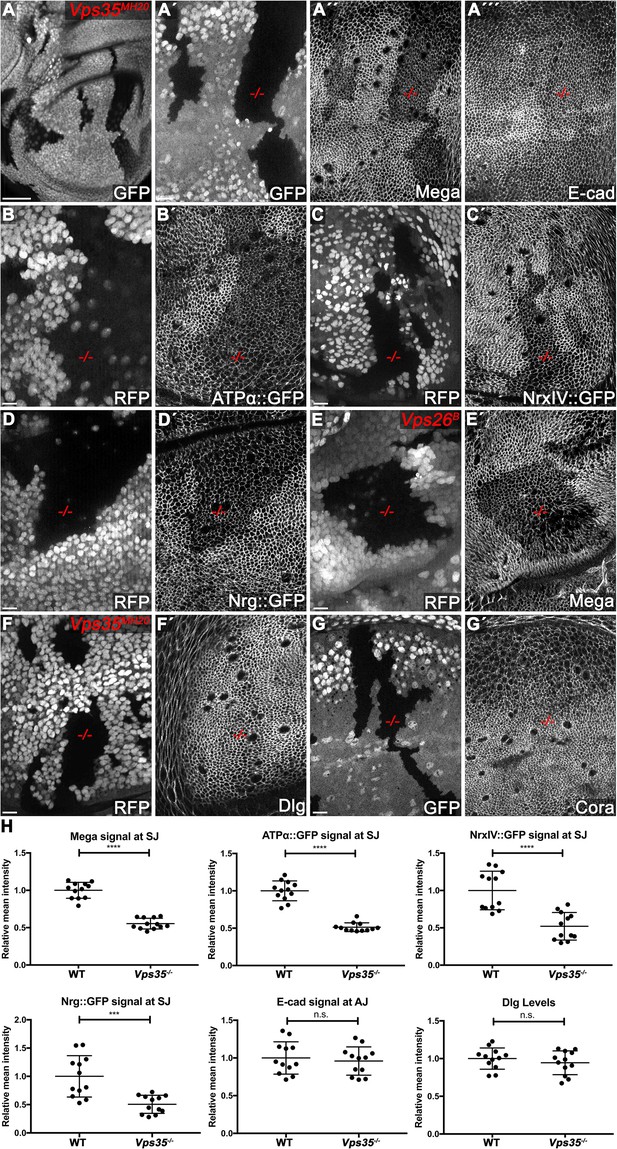
The retromer CSC regulates membrane levels of SJ core components.
(A) Null mutant clones of the retromer component Vps35 were generated in wing discs. Mutant tissue (-/-) is marked by absence of either GFP or RFP. Mega signal at the SJ is reduced in Vps35MH20 tissue (A´´), while apical E-cad levels are unaffected (A´´´). (B–D) Endogenously tagged SJ core components ATPα (B), NrxIV (C) and Nrg (D) show reduced membrane levels in Vps35MH20 tissue. (E) Vps26 clones phenocopy Vps35 clones with respect to junctional localization of Mega (compare E´ with A´´). (F–G) Junctional localizations of SJ-associated cytoplasmic proteins Dlg and Cora are unaffected in Vps35MH20 cells. (H) Quantification of junctional membrane levels of indicated proteins in wildtype and Vps35MH20 tissue. Each dot represents fluorescence intensity of a junctional region in either clonal (Vps35MH20) or surrounding wildtype tissue. n = 4 discs for each component. A two-tailed t-test was used for statistical analysis with the significance levels **** representing a p-value of <0.0001, ***<0.001, and n.s. (not significant):≥0.05. Note that the transmembrane SJ components Mega, ATPα, NrxIV, and Nrg are similarly affected, showing reduced levels at the SJ within Vps35MH20 tissue by ~50%, while E-cad and Dlg apical levels do not significantly differ between wildtype and Vps35MH20 tissue. Scale bar in (A) 50 μm, in all other panels 10 μm.
-
Figure 2—source data 1
Quantification of SJ protein apical levels in WT/Vps35 mutant clones.
- https://cdn.elifesciences.org/articles/61866/elife-61866-fig2-data1-v2.xlsx
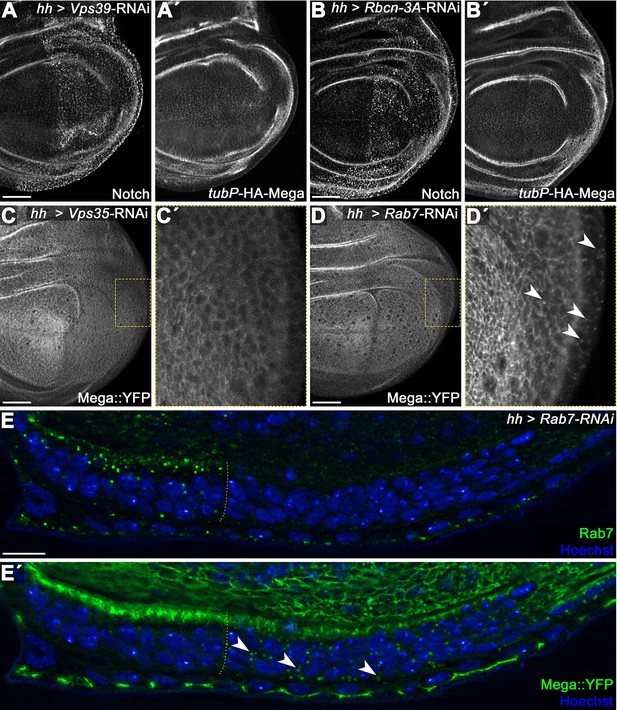
Mega is not target of lysosomal degradation under steady-state conditions.
(A) Posterior knockdown of the HOPS (homotypic fusion and vacuole protein) complex component Vps39 required for endosome-lysosome fusion. Cargo destined for lysosomal degradation such as the Notch receptor (A) accumulates in a punctate pattern upon Vps39 depletion due to failure of endosome-lysosome fusion. (A') In contrast, a tubulin-Promotor expressed HA-Mega construct (tubP-HA-Mega) that faithfully recapitulates endogenous Mega localization is unaffected by Vps39-RNAi expression. This indicates that HA-Mega is not undergoing lysosomal degradation under steady-state condition. (B) Similarly, impeding endolysosomal function by expressing an RNAi against Rabconnectin-3A (Rbcn-3A) accumulates Notch in the posterior Rbcn-3A-depleted tissue (B), while HA-Mega retains a wildtype localization throughout the disc. Rbcn-3A is a regulator of the vacuolar proton pump (V-ATPase) required for endolysosomal acidification (Yan et al., 2009). (C) Depletion of Vps35 reduces Mega::YFP levels within the tissue and little vesicular signal is seen (inset, C'). (D) In contrast, Mega::YFP levels are only mildly affected by Rab7 depletion but strong vesicular accumulation is seen (arrowheads in D'). (E) Lateral view of a wing disc expressing Rab7-RNAi in the posterior compartment. (E) Rab7 depletion is effective. (E') Mega::YFP signal at the SJ is slightly reduced in the posterior compartment and vesicular accumulation is apparent (arrowheads). Rab7 activity is required for both endosome-lysosome fusion as well as for CSC endosomal recruitment (reviewed in Guerra and Bucci, 2016) Thus, Rab7 knockdown should simultaneously impede retromer trafficking and lysosomal degradation. These data are entirely consistent with Mega being targeted for lysosomal degradation only when retromer CSC function is compromised. Scale bars in all panels represent 50 μm except for (E): 10 μm.
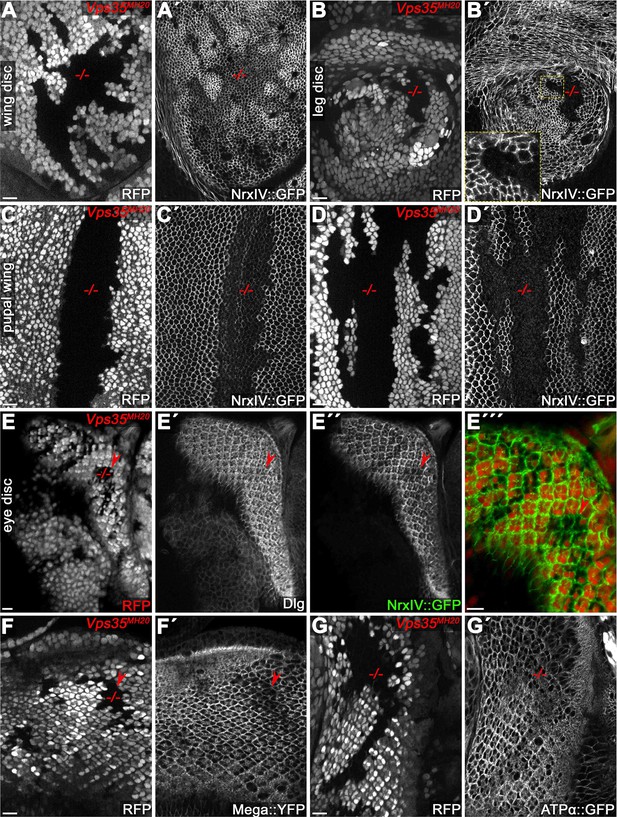
Retromer CSC-dependent transport of SJ components is conserved throughout tissues.
(A) Wing imaginal disc containing Vps35MH20 clones. Clones in all panels are marked by the absence of RFP. (A') NrxIV::GFP is reduced within the clonal area. (B) Leg imaginal disc containing Vps35MH20 clones showing stronger loss of NrxIV::GFP from the SJ. Note the complete absence of junctional NrxIV::GFP in this clone compared to modestly reduced levels in the wing disc clone (A'). (C–D) Vps35MH20 clones in pupal wings. Note the variable expressivity of the NrxIV::GFP phenotype within the clones, ranging from still detectable junctional levels (C') to complete loss of NrxIV::GFP from the junction (D'). (E–G) Vps35MH20 clones in eye imaginal discs. Similar to wing imaginal discs (see Figure 2), Dlg levels are not affected by Vps35 loss whereas NrxIV::GFP shows reduced membrane levels (compare arrowheads in E' and E"). (F–G) Similar to NrxIV::GFP, Mega::YFP (F') as well as ATPα::GFP (G') display reduced membrane levels within Vps35MH20 clones. Scale bars in all panels represent 10 μm.
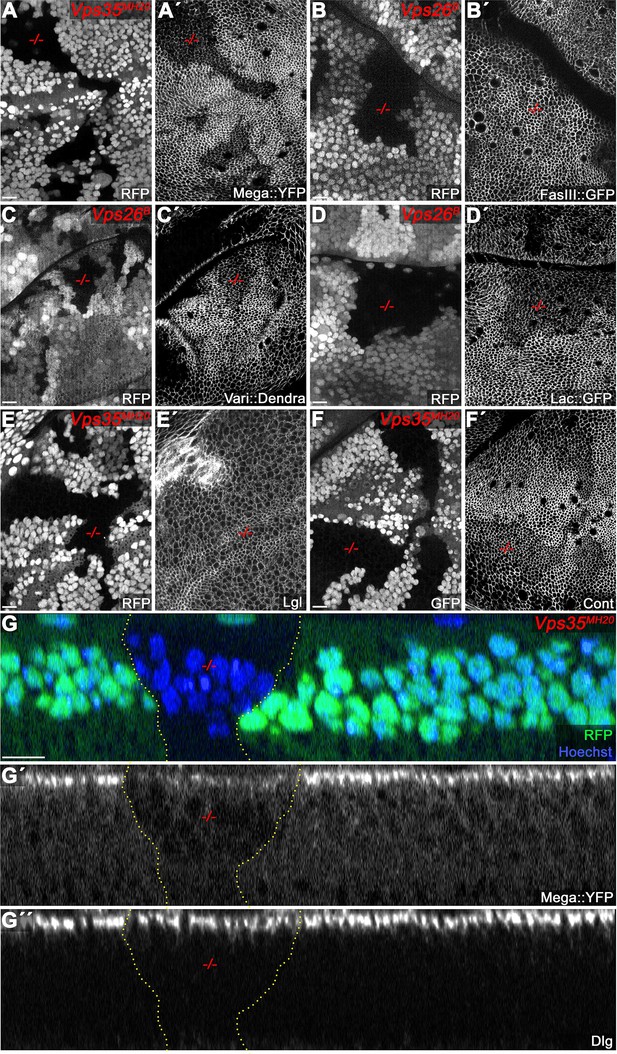
Analysis of further SJ components in retromer CSC mutant clones.
(A) Endogenously tagged Mega::YFP recapitulates reduction of Mega membrane levels within Vps35MH20 tissue detected by antibody staining (see also Figure 2A). (B) FasIII::GFP membrane levels remain wildtype within Vps26B mutant clones. (C–D) Endogenously tagged Varicose (C) and Lachesin (D) membrane levels are reduced within Vps26B mutant tissue. (E) The basolateral cell polarity protein Lethal giant larvae (Lgl) retains a wildtype localization in Vps35MH20 mutant clones. (F) Contactin (Cont) membrane levels are reduced within Vps35MH20 tissue. (G) Optical section of a wing disc containing a Vps35MH20 mutant clone. (G') Mega::YFP signal at the SJ and throughout the tissue is reduced within the Vps35MH20 clone. (G'') In contrast to Mega::YFP, apical Dlg signal at the SJ retains wildtype levels in the retromer mutant clone. Scale bars in all panels represent 10 μm.
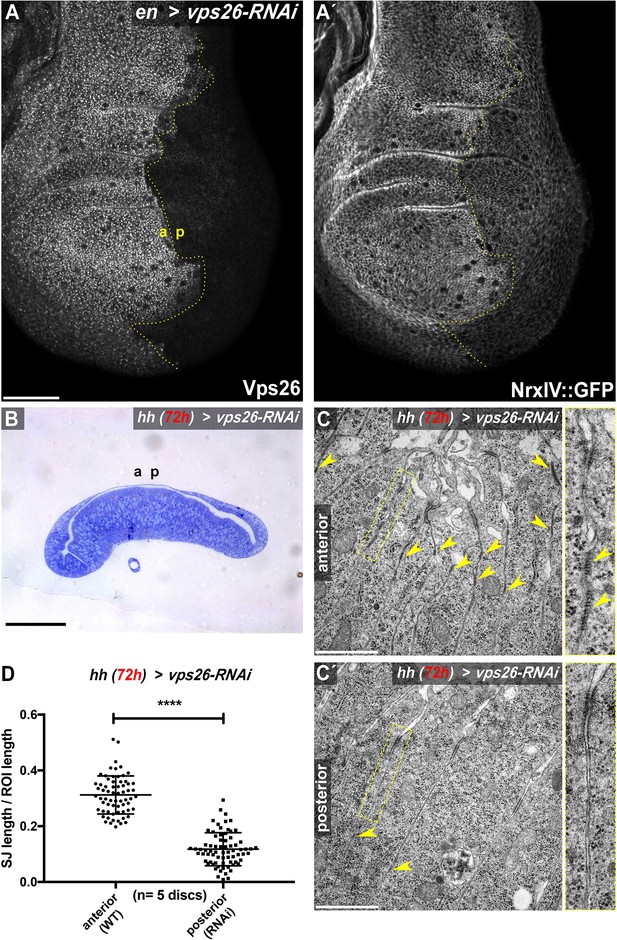
Retromer CSC function maintains SJ density.
(A) RNAi-mediated depletion of Vps26 in the posterior compartment of wing imaginal discs efficiently reduces Vps26 (A) and junctional NrxIV::GFP levels (A'). (B) Toluidin blue stained thin section of a wing imaginal disc depleted of Vps26 for 72 hr in the posterior compartment. Epithelial monolayer integrity is not affected upon depletion of this retromer component. (C) TEM analysis of junctions in control (C) and Vps26-depleted (C') wing imaginal disc tissue. Expression of Vps26-RNAi was restricted to 72 hr, since continuous hhGal4-driven expression yielded discs with high levels of apoptotic cells, hampering analysis of the junctions. Note the high density of septa in the anterior control compartment compared to the Vps26-depleted posterior compartment (arrowheads in C and C', respectively). (D) Quantification of electron-dense SJ in 2 μm ROIs (basal to the AJ) along the juxtaposed membranes. Each data point represents total SJ length within one single ROI. Note that while approximately 30% of ROI length within anterior control compartments is occupied by SJ, the SJ length/ROI length ratio is reduced to about 12% in posterior Vps26-RNAi compartments, indicating a reduction of SJ by about 60%. The significance level **** represents a p-value <0.0001. Scale bars in (A) and (B) represent 50 μm, in (C): 0.5 μm. The online version of this article includes the following figure supplements for (A-D).
-
Figure 2—figure supplement 4—source data 1
Quantification of SJ density in WT and Vps26/retromer-depleted tissue.
- https://cdn.elifesciences.org/articles/61866/elife-61866-fig2-figsupp4-data1-v2.xlsx
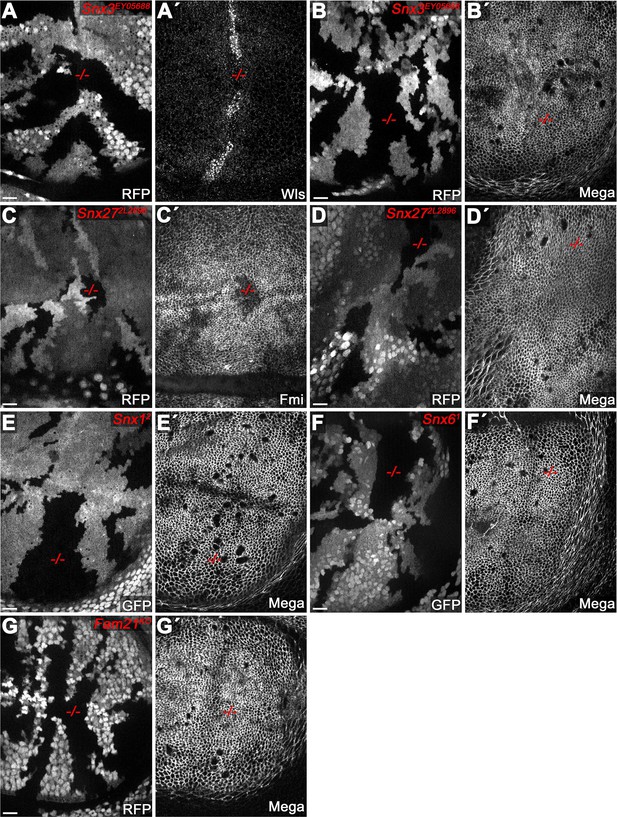
Retromer CSC-associated sorting nexins (SNX) are dispensable for SJ delivery of Mega.
(A–F) Null mutant clones of SNX family genes in wing imaginal discs, marked by absence of either RFP or GFP. (A–B) Null mutant clone of Snx3 affects levels of the control cargo Wntless (Wls) (Harterink et al., 2011). (A') while Mega retains a wildtype junctional level (B'). Mutant clones of Snx27 affect apical levels of the control cargo Flamingo (Fmi) (Strutt et al., 2019) (C') but not that of Mega (D'). Mutant clones of SNX-Bar family genes Snx1 (E) and Snx6 (F) (Zhang et al., 2011) do not affect junctional levels of Mega. (G) Mega membrane levels are wildtype in clones mutant for the WASH (WASP and SCAR Homolog complex) complex component Fam21 (Family with sequence similarity 21) (Strutt et al., 2019). Scale bars in all panels represent 10 μm.
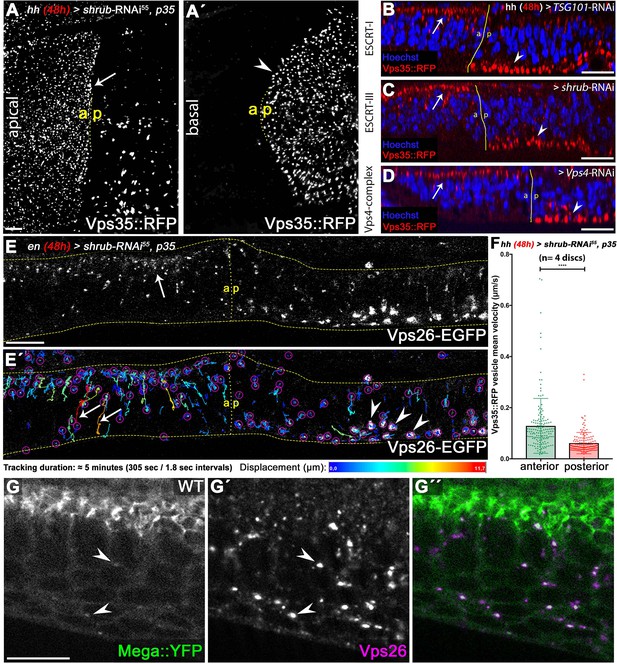
ESCRT function is required for CSC retromer apical hub localization and mobility.
(A) Endogenously tagged Vps35::RFP localizes in a vesicular pattern within the apical hub in wildtype control compartments (arrow). After 48 hr of Shrub depletion, Vps35::RFP apical hub localization is largely abolished and basal accumulation is seen (arrowhead in A'). (B–D) The shift in subcellular localization of Vps35::RFP from the apical hub (wildtype, arrows) to basal aggregates (ESCRT-depleted compartment, arrowheads) is detected upon knockdown of ESCRT components TSG101, Shrub as well as Vps4. This indicates that regulation of retromer subcellular localization is a general ESCRT function in wing discs. (E) Live imaging and vesicle tracking of a genomic Vps26-EGFP construct at the anterior/posterior boundary of a wing disc expressing shrub-RNAi for 48 hr in the posterior compartment under control of enGal4. Note the apical hub in the anterior control compartment and the large basally localized Vps26-EGFP aggregates in the posterior Shrub-depleted compartment (compare with Vps35::RFP in C). (E') Individual vesicle trajectories visualize Vps26-EGFP movement within the tissue after a 5 min imaging acquisition. Note the high mobility of several vesicles along the apicobasal axis in the anterior control compartment (arrows). In contrast to long-distance shuttling of Vps26-EGFP in the control tissue, basal Vps26-EGFP aggregates (arrowheads) in the Shrub-depleted posterior compartment are largely static and overall vesicle dynamic is reduced. (F) Quantification of Vps35::RFP mobility in wildtype (anterior) and Shrub-depleted (posterior) tissue. Each dot represents mean velocity of a single tracked vesicle. Note the reduced mean velocity of Vps35::RFP and the high ratio of low mobility vesicles within Shrub-depleted compartments. A two-tailed t-test was used for statistical analysis with the significance level **** representing a p-value <0.0001. (G) Endogenously tagged Mega::YFP colocalizes with Vps26 in vesicular structures residing in the apical as well as basal cytoplasm (arrowheads). Scale bars in all panels represent 10 μm.
-
Figure 3—source data 1
TrackMate raw data of Vps35::RFP vesicle tracking in WT and Shrub-RNAi +p35 tissue.
- https://cdn.elifesciences.org/articles/61866/elife-61866-fig3-data1-v2.xlsx
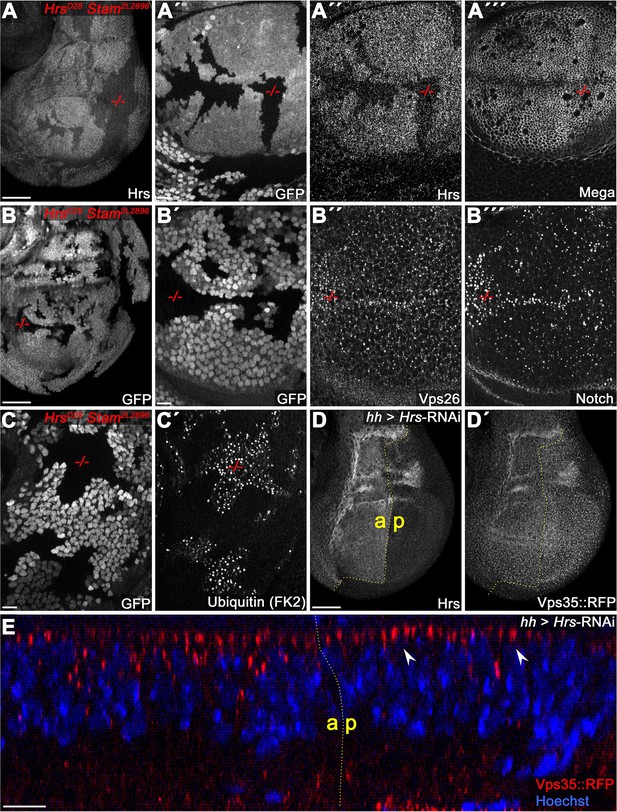
ESCRT-0 is not required for SJ delivery of Mega.
(A) Hrs, Stam double mutant clones (marked by the absence of GFP). (A") Reduction of Hrs within the clonal tissue. (A"') Mega retains a wildtype membrane level in Hrs, Stam tissue. (B) Notch accumulates in ESCRT-0 deficient clones (B"') while Vps26 vesicular localization is largely unaffected (B"). (C) Ubiquitinated cargo fails to degrade in Hrs, Stam clones. (D–E) Knockdown of Hrs in the posterior compartment does not affect Vps35::RFP levels or subcellular localization. Note the intact apical hub in an optical section (arrowheads in E). Scale bars in (A, B, D) represent 50 μm, in all other panels: 10 μm.
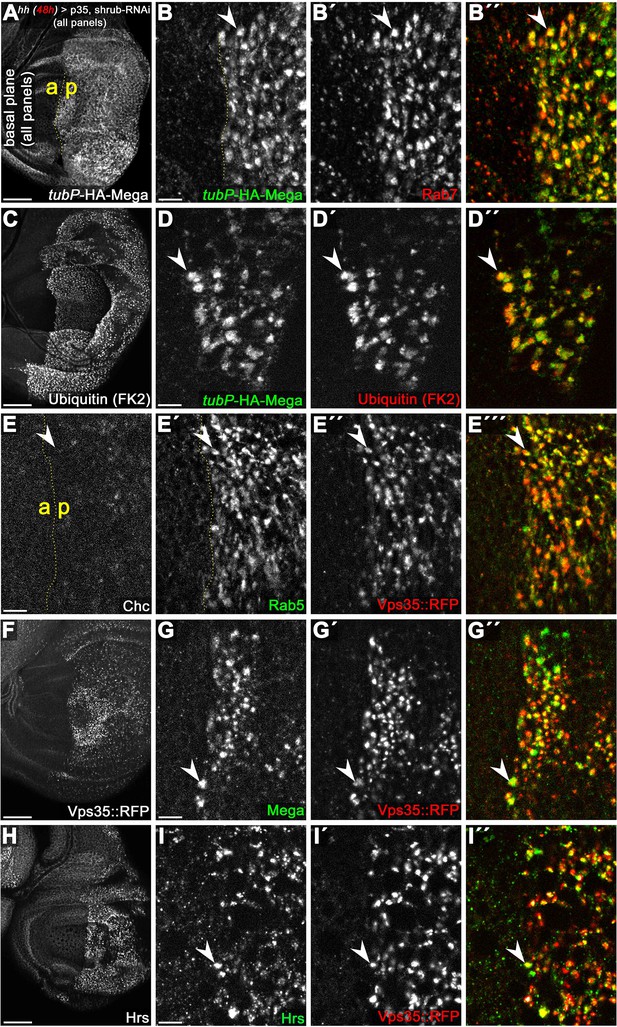
Characterization of basal ‘class E-like’ endosomal compartments induced by Shrub-RNAi.
All panels show basal planes of wing discs expressing p35, shrub-RNAi for 48 hr under control of hhGal4. (A–B) In posterior Shrub-depleted compartments, tubP-HA-Mega accumulates in basal aberrant endosomes which label positive for Rab7 (arrowheads). (C–D) tubP-HA-Mega overlaps with ubiquitin at basal aggregates (arrowheads), indicating that these contain large amounts of degradative cargo. (E) Basal aggregates are coated with Rab5 and Vps35::RFP but lack clathrin (arrowheads). (F–G) Mega and Vps35::RFP largely overlap within basal compartments. (H–I) ESCRT-0 component Hrs accumulates on basal aberrant endosomes (arrowheads). Scale bars in (A, C, F, H) represent 50 μm, in all other panels: 10 μm.
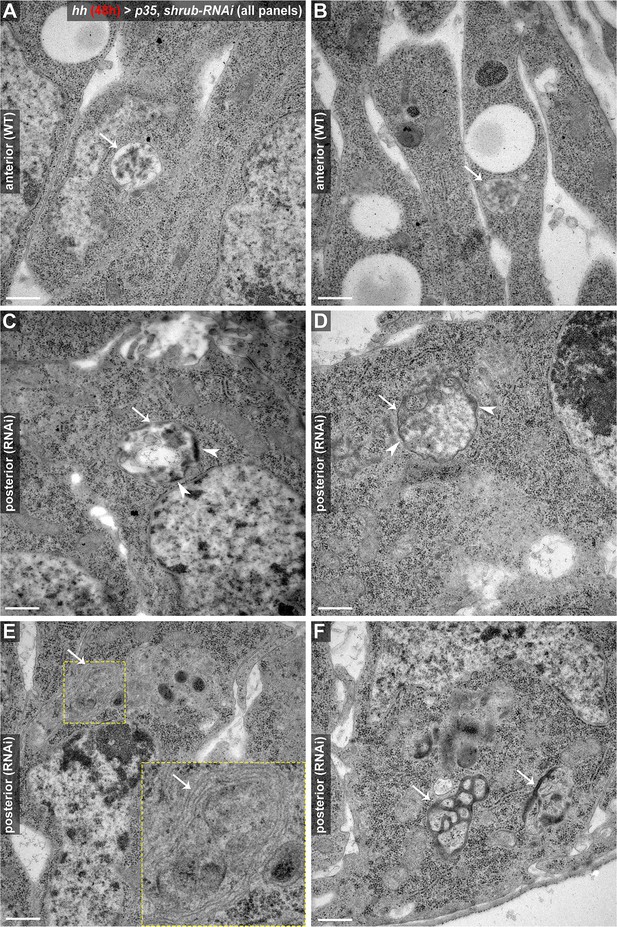
TEM analysis of aberrant endosomal compartments induced by 48 hr of Shrub depletion.
All panels show electron micrographs of wing disc cells in either wildtype anterior control compartments (A, B) or posterior (p35, shrub-RNAi) expressing compartments (C–F). (A, B) In control compartments, endosomes appear as stereotypical, spherical shaped MVBs containing electron-dense ILVs (arrows). (C–D) In contrast, MVB-like compartments induced by Shrub depletion appear enlarged and irregularly shaped (arrows). Note that the limiting membrane of the endosome is lined with patches of electron-dense material (arrowheads), which could be aggregating endosomal machineries and/or cargo proteins failing to get retrieved or internalized. (E) Multicisternal endosomal structure with a stacked membrane morphology (arrow), reminiscent of class E compartments in mammalian cells (Doyotte et al., 2005). (F) Aberrant, electron-dense endosomal compartments (arrows) in proximity to the basal membrane. These structures likely correlate with the basal endosomal aggregates characterized in Figure 3—figure supplement 2 above. Scale bars in all panels represent 0.5 μm.
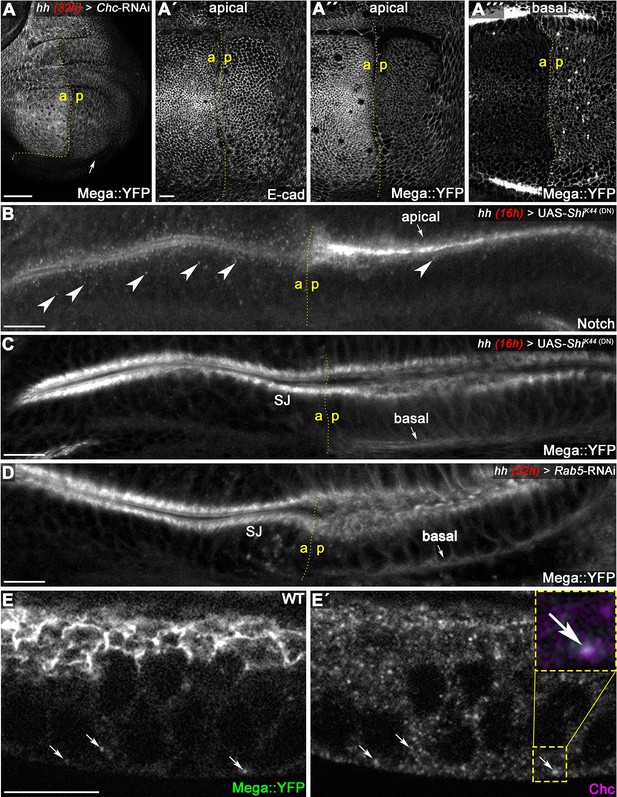
Mega is endocytosed at the basodistal membrane.
(A) Depletion of clathrin via Chc-RNAi expression for 32 hr in the posterior compartment reduces apical membrane levels of Mega::YFP. While E-cad-based AJ appear unaffected following Chc depletion (A'), the junctional Mega::YFP pool is reduced (A''). In a basal focal plane, Mega::YFP is seen accumulating in the basodistal part of the lateral membrane in the Chc-RNAi expressing posterior compartment. (B–C) Expression of a dominant negative Dynamin construct (ShiK44) leads to apical accumulation of Notch within an epithelial fold of the hinge region (arrow in B) and reduces the amount of Notch positive vesicles (arrowheads). In contrast, posterior Mega::YFP signal at the SJ is reduced and a basal pool is visible (arrow in C). (D) Depleting Rab5 in the posterior compartment yields a similar Mega::YFP phenotype with respect to junctional levels and basal accumulation (arrow in D). (E) In line with internalization from the basodistal membrane, Mega::YFP colocalizes with clathrin at vesicular structures near the basal cell pole and along the apicobasal axis (arrows in E and E'). Scale bars in all panels represent 10 μm except for (A), in which it represents 50 μm.
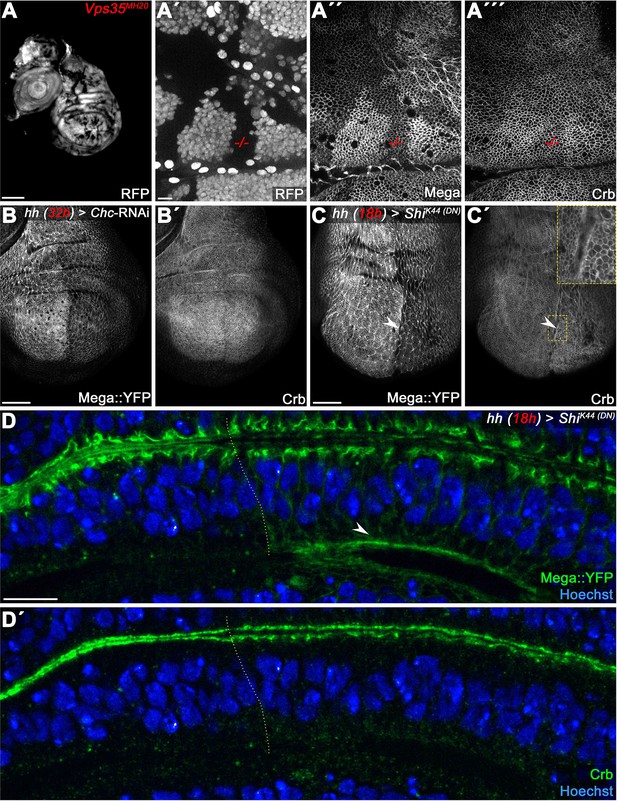
Comparison of Mega phenotypes with the retromer CSC cargo Crumbs.
(Pocha et al., 2011) (A) Vps35MH20 clones, marked by the absence of RFP (A'), differently affect membrane levels of Mega (A") and Crb (A"'). Mega appears to be more severely affected by retromer loss of function. (B) Inhibition of clathrin-dependent endocytosis by Chc-RNAi expression in the posterior compartment reduces apical Mega::YFP levels (B) while Crb levels are unaffected (B'). Similarly, expression of dominant negative Dynamin (ShiK44) reduces Mega::YFP levels (arrowhead in C), while Crb seems to accumulate slightly at the apical membrane (arrowhead in C'). This is consistent with Crb entering the endosomal system via apical internalization. (D) Optical section of a wing disc expressing ShiK44 for 18 hr in the posterior compartment reveals Mega::YFP accumulation at the basal pole of the cells (arrowhead in D). (D') In contrast, no basal accumulation of Crb is detected. These data are consistent with Mega and Crb utilizing dissimilar retromer-dependent transport pathways. Scale bar in (A) represents 100 μm, in (B, C): 50 μm, in (A', D): 10 μm.
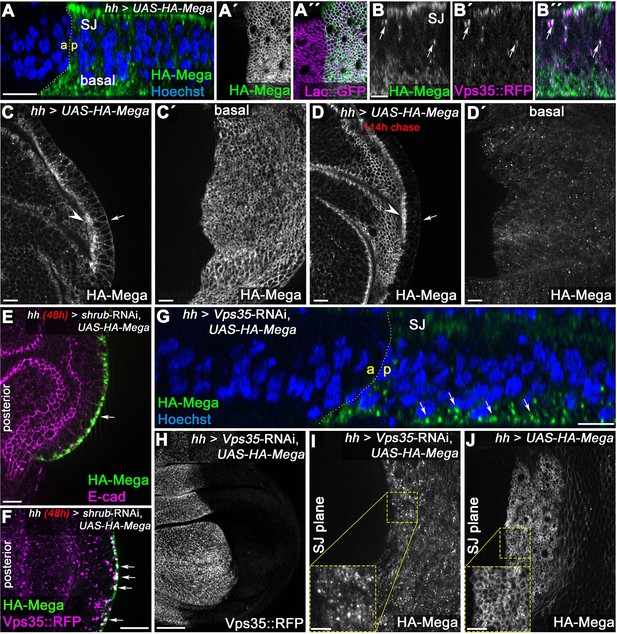
Basal to apical transcytosis of Mega requires ESCRT and retromer function.
(A) UAS-driven overexpression of HA-tagged Mega reveals a basal pool opposing the apical SJ fraction. Similarly to endogenously tagged Mega::YFP (see Figure 3G), HA-Mega colocalizes with the CSC on vesicles along the apicobasal axis (arrows in B). (C) Continuous overexpression of HA-Mega yields a strong SJ signal (arrowhead) as well as lateral and basal (arrow) staining of HA-Mega. (C') In a basal focal plane, the strong accumulation of HA-Mega in a junction like fashion at the basodistal membrane is seen. (D) Following a 14 hr chase at 18°C to halt further expression, HA-Mega signal is almost exclusively confined to the SJ (arrowhead) with very little basal staining (arrow). (D') The basodistal membrane pool of HA-Mega (compare with C') vanished after 14 hr chase, in line with a transient localization of HA-Mega at this membrane domain. (E) In Shrub-depleted tissue, overexpressed HA-Mega does not reach the SJ and is found exclusively in large basal aggregates (arrow). (F) These basal aggregates are positive for Vps35::RFP (arrows). (G) Similarly, in Vps35-depleted tissue, apical SJ signal of HA-Mega is faint and vesicular accumulation in the basal cytoplasm is seen (arrows). (H) Knockdown efficiency of the Vps35-RNAi. (I) No distinct membrane staining of HA-Mega is visible in the SJ plane of Vps35-depleted tissue, in contrast to HA-Mega junctional localization in wildtype tissue (J). Scale bar in (B) represents 5 μm, in (G): 50 μm and in all other panels: 10 μm.
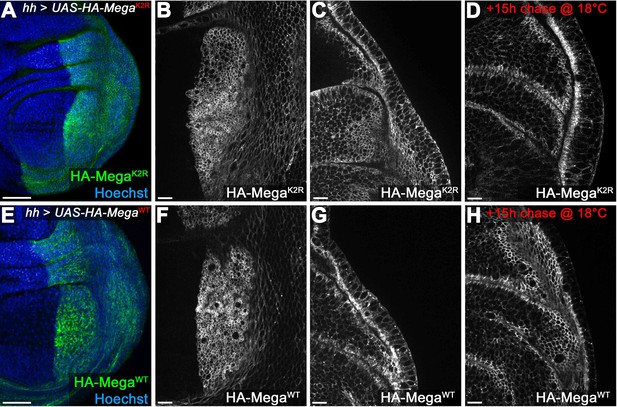
Mega transport toward the SJ is independent of intracellular lysines.
(A–D) hhGal4 driven posterior expression of an HA-tagged Mega construct in which all intracellular lysines were exchanged with arginines (UAS-HA-MegaK2R). Junctional (B) and lateral (C) plane of continuously expressed HA-MegaK2R reveals wildtype like membrane localization (compare with F, G). (D) After 15 hr chase at 18°C to allow intracellular transport of newly synthesized protein, HA-MegaK2R signal is largely confined to the SJ, much like the wildtype (HA-MegaWT) construct (H). (E–H) Expression of the wildtype control HA-MegaWT construct under identical conditions. The two constructs are largely indistinguishable with HA-MegaK2R retaining slightly more lateral and basal membrane signal after the 15 hr chase (compare D with H). The data are consistent with Mega transport to the SJ being independent of lysines (which should mediate direct interactions of cargo proteins with ESCRT components via ubiquitination). Scale bars in (A, E) represent 50 μm, in all other panels: 10 μm.
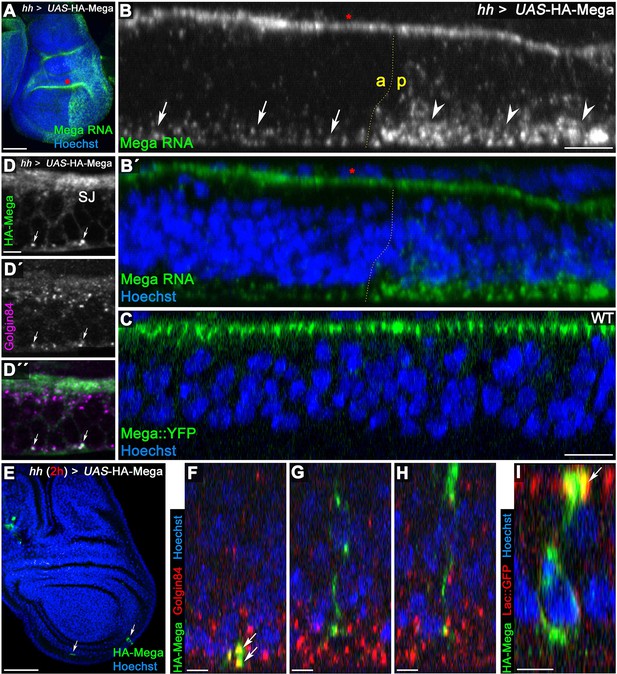
Mega is secreted from Golgi stacks within the basal cytoplasm.
(A) Fluorescence in situ hybridization (FISH) signal from a Mega RNA probe after posterior overexpression of UAS-HA-Mega. Note the strong posterior signal, indicating probe specificity. Unspecific signal between peripodial membrane and disc proper is visible in the epithelial fold of the hinge region (red asterisks in A and B). (B) Mega RNA is detected primarily in the basal cytoplasm of both the wildtype and the UAS-HA-Mega overexpressing posterior compartment (arrows and arrowheads, respectively). Note the higher abundance of fluorescence signal in the posterior compartment with a subcellular localization comparable to endogenous Mega RNA within the anterior tissue. (C) In stark contrast to basally localized mRNA, Mega protein (Mega::YFP) localizes within the SJ at the apical pole of the cells. (D) Overexpressed Mega colocalizes with the Golgi resident protein Golgin84 in vesicular structures within the basal cytoplasm (arrows), consistent with basal secretion of Mega. Note the high abundance of Golgi stacks in proximity to the SJ but lack of overlap with HA-Mega in this apical region. (E–I) A short expression of HA-Mega for 2 hr yields discs that contain single cells with detectable HA-Mega expression (arrows). (F) Some cells stain for HA-Mega exclusively in basal, punctate spots which overlap with Golgin84 (arrows). This is in line with initial appearance of HA-Mega at the basal cell pole during secretion from local Golgi stacks. (G–H) Intracellular spreading of HA-Mega along the apicobasal axis toward the SJ. (I) HA-Mega extensively colocalizes with Lac::GFP at the apical pole, indicating efficient SJ targeting (arrow). Scale bars in (A, B) represent 50 μm, in (B), (C): 10 μm and in all other panels: 5 μm.
Videos
Live imaging time series of a wildtype wing imaginal disc expressing Mega::YFP and Vps35::RFP.
Note the close association of Mega and Vps35 at dynamic vesicular structures along the apico-basal axis. Time stamp indicates passed time in minutes, starting from the initial frames of the acquisition.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Genetic reagent (D. melanogaster) | UAS-HA-Mega (II.) and (III.) | This paper | N-terminally HA-tagged WT Mega for Gal4/UAS-driven expression | |
| Genetic reagent (D. melanogaster) | tubP-HA-Mega (II.) | This paper | N-terminally HA-tagged WT Mega expressed under the control of a tubulin promotor | |
| Genetic reagent (D. melanogaster) | UAS-HA-MegaK2R | This paper | N-terminally HA-tagged Mega (intracellular lysines exchanged with arginines) for Gal4/UAS-driven expression | |
| Genetic reagent (D. melanogaster) | UAS-Rab5-RNAi | Vienna Drosophila Resource Center | V34096 | |
| Genetic reagent (D. melanogaster) | UAS-Vps35-RNAi | Bloomington Drosophila Stock Center | BL22180 | |
| Genetic reagent (D. melanogaster) | UAS-Chc-RNAi | Vienna Drosophila Resource Center | V103383 | |
| Genetic reagent (D. melanogaster) | UAS-Vps4-RNAi | Vienna Drosophila Resource Center | V101722 | |
| Genetic reagent (D. melanogaster) | UAS-TSG101-RNAi | Bloomington Drosophila Stock Center | BL35710 | |
| Genetic reagent (D. melanogaster) | UAS-shrub-RNAi | Sweeney et al., 2006 | ||
| Genetic reagent (D. melanogaster) | UAS-Vps26-RNAi | Bloomington Drosophila Stock Center | BL38937 | |
| Genetic reagent (D. melanogaster) | UAS-Vps39-RNAi | Bloomington Drosophila Stock Center | BL42605 | |
| Genetic reagent (D. melanogaster) | Rbcn-3A-RNAi | Vienna Drosophila Resource Center | V108547 | |
| Genetic reagent (D. melanogaster) | Rab7-RNAi | Vienna Drosophila Resource Center | V40337 | |
| Genetic reagent (D. melanogaster) | UAS-Hrs-RNAi | Vienna Drosophila Resource Center | V20933 | |
| Genetic reagent (D. melanogaster) | UAS-mega/pickel-RNAi | Vienna Drosophila Resource Center | V36306 | |
| Genetic reagent (D. melanogaster) | hhGal4 | Bloomington Drosophila Stock Center | BL67046 | |
| Genetic reagent (D. melanogaster) | tubGal80ts | Bloomington Drosophila Stock Center | BL7108 | |
| Genetic reagent (D. melanogaster) | UAS-p35 | Bloomington Drosophila Stock Center | BL5072 | |
| Genetic reagent (D. melanogaster) | enGal4 | Bloomington Drosophila Stock Center | BL30564 | |
| Genetic reagent (D. melanogaster) | Lac::GFP | Gift from Christian Klämbt, University of Münster, Germany | Endogenously GFP-tagged lac allele | |
| Genetic reagent (D. melanogaster) | ATPα::GFP | Bloomington Drosophila Stock Center | BL6834 | |
| Genetic reagent (D. melanogaster) | NrxIV::GFP | Edenfeld et al., 2006 | ||
| Genetic reagent (D. melanogaster) | Nrg::GFP | Bloomington Drosophila Stock Center | BL6844 | |
| Genetic reagent (D. melanogaster) | FasIII::GFP | Bloomington Drosophila Stock Center | BL59809 | |
| Genetic reagent (D. melanogaster) | Vari::Dendra | Babatz et al., 2018 | Endogenously Dendra-tagged vari allele | |
| Genetic reagent (D. melanogaster) | Mega::YFP | Gift from Reinhard Schuh, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany | Endogenously YFP-tagged mega allele | |
| Genetic reagent (D. melanogaster) | Vps35::RFP | Bloomington Drosophila Stock Center | BL66527 | |
| Genetic reagent (D. melanogaster) | Vps26-EGFP | Bloomington Drosophila Stock Center | BL67153 | |
| Genetic reagent (D. melanogaster) | UAS-ShiK44 | Bloomington Drosophila Stock Center | BL5811 | |
| Genetic reagent (D. melanogaster) | Ubx-Flp | Bloomington Drosophila Stock Center | BL42730 | |
| Genetic reagent (D. melanogaster) | hsFlp | Bloomington Drosophila Stock Center | BL8862 | |
| Genetic reagent (D. melanogaster) | hsFlp FRT19A ubi-nls-RFP | Bloomington Drosophila Stock Center | BL31418 | |
| Genetic reagent (D. melanogaster) | FRT40A 2xGFP | Bloomington Drosophila Stock Center | BL5189 | |
| Genetic reagent (D. melanogaster) | FRT42D 2xGFP | Bloomington Drosophila Stock Center | BL5625 | |
| Genetic reagent (D. melanogaster) | FRT42D ubi-nls-RFP | Bloomington Drosophila Stock Center | BL35496 | |
| Genetic reagent (D. melanogaster) | FRT82B ubi-nls-RFP | Bloomington Drosophila Stock Center | BL30555 | |
| Genetic reagent (D. melanogaster) | FRT42D Vps35MH20 | Bloomington Drosophila Stock Center | BL67202 | |
| Genetic reagent (D. melanogaster) | Vps26B FRT19A | Bloomington Drosophila Stock Center | BL57140 | |
| Genetic reagent (D. melanogaster) | HrsD28Stam2L2896 FRT40A | Tognon et al., 2014 | BL56816 | |
| Genetic reagent (D. melanogaster) | Dmon1Mut4 FRT40A | Yousefian et al., 2013 | ||
| Genetic reagent (D. melanogaster) | Snx2725 FRT19A | Strutt et al., 2019 | ||
| Genetic reagent (D. melanogaster) | FRT42 Fam21KO | Strutt et al., 2019 | ||
| Genetic reagent (D. melanogaster) | Snx1Δ2 FRT40A | Zhang et al., 2011 | ||
| Genetic reagent (D. melanogaster) | Snx61 FRT40A | Zhang et al., 2011 | ||
| Genetic reagent (D. melanogaster) | FRT82B Snx3EY05688 | Harterink et al., 2011 | ||
| Genetic reagent (D. melanogaster) | Rab7GAL4-KO | Chan et al., 2011 | ||
| Antibody | anti-ATPα (mouse monoclonal) | DSHB | a5 | IF (1:50) |
| Antibody | anti-Shrub (rabbit polyclonal) | Bäumers et al., 2019 | IF (1:100) | |
| Antibody | anti-Mega (mouse monoclonal) | Jaspers et al., 2012 | IF (1:50) | |
| antibody | anti-E-cad (rat monoclonal) | DSHB | 5D3 | IF (1:50) |
| Antibody | anti-Rab7 (mouse monoclonal) | DSHB | Rab7 | IF (1:100) |
| Antibody | anti-Dlg (mouse monoclonal) | DSHB | 4F3 | IF (1:500) |
| antibody | anti-Cora (mouse monoclonal) | DSHB | C566.9 | IF (1:100) |
| Antibody | anti-Vps26 (guinea pig polyclonal) | Wang et al., 2014 | IF (1:1500) | |
| Antibody | anti-Chc (rat polyclonal) | Wingen et al., 2009 | IF (1:50) | |
| Antibody | anti-Golgin84 (mouse monoclonal) | DSHB | 12–1 | IF (1:100) |
| Antibody | anti-Lgl (guinea pig polyclonal) | Shahab et al., 2015 | IF (1:500) | |
| Antibody | anti-Cont (guinea pig polyclonal) | Faivre-Sarrailh et al., 2004 | IF (1:2000) | |
| Antibody | anti-Notch (mouse monoclonal) | DSHB | C458.2H | IF (1:100) |
| antibody | anti-Hrs (guinea pig polyclonal) | Lloyd et al., 2002 | IF (1:500) | |
| Antibody | anti-Ubiquitin (FK2) (mouse monoclonal) | Enzo Life Sciences | BML-PW8810 | IF (1:100) |
| Antibody | anti-Crb (rat polyclonal) | Richard et al., 2006 | rat anti-Crb 2.8 | IF (1:500) |
| Antibody | anti-HA (rabbit monoclonal) | Cell Signalling Technology | C29F4 | IF (1:1500) |
| Antibody | anti-Rab5 (rabbit polyclonal) | Abcam | ab31261 | IF (1:250) |
| Antibody | anti-Fmi (mouse monoclonal) | DSHB | #74 | IF (1:10) |
| Sequence-based reagent | fw-NotI-ATG-HA-3xGly-Mega | This paper | PCR primer | See method section for sequence |
| Sequence-based reagent | rev-Mega-XhoI | This paper | PCR primer | See method section for sequence |
| Other | Hoechst 33258 | Sigma Aldrich/Merck | B2883 | IF (1:10.000), DNA/Nucleus staining |





