NusG is an intrinsic transcription termination factor that stimulates motility and coordinates gene expression with NusA
Figures
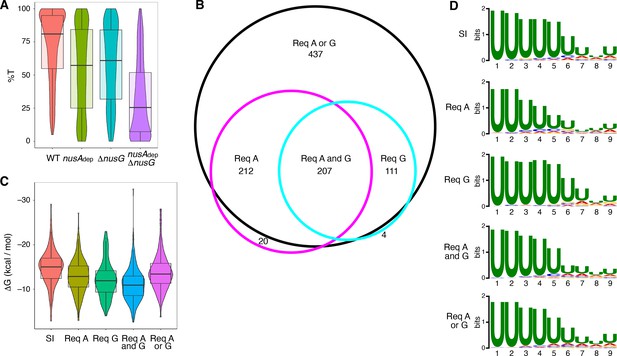
NusG is an intrinsic termination factor that works with NusA to stimulate suboptimal terminators.
(A) Violin plots overlayed with box plots showing the distribution of termination efficiency (%T) in wild-type (WT), nusAdep, ΔnusG, and nusAdep ΔnusG strains. For this and all box plots, boundaries of the box designate the interquartile range (IQR), while upper/lower whiskers extend from the 75th/25th percentile to the largest/smallest value no further than 1.5*IQR in either direction. All pairwise p-values can be obtained from Supplementary file 3. (B) Venn diagram showing the number and overlap of terminators that were classified as being dependent (Δ%T ≥ 25) on NusA (Req A) and/or NusG (Req G). The black circle contains all terminators that were classified as dependent in the nusAdep ΔnusG strain, the magenta circle contains all terminators that were classified as dependent in the nusAdep strain, and the cyan circle contains all terminators that were classified as dependent in the ΔnusG strain. Intrinsic terminator subpopulations that require NusA and/or NusG in any fashion to terminate efficiently are specified. (C) Violin plots overlayed with box plots showing the distribution of predicted hairpin strength as reported in ΔG (kcal/mol) for all identified subpopulations including the strong and independent (SI) terminators. All pairwise p-values can be obtained from Supplementary file 3. (D) Sequence logos of the U-rich tracts generated from the nine nucleotide (nt) window downstream of the predicted hairpins for all identified subpopulations including the SI terminators. All pairwise comparisons can be found in Supplementary file 3.
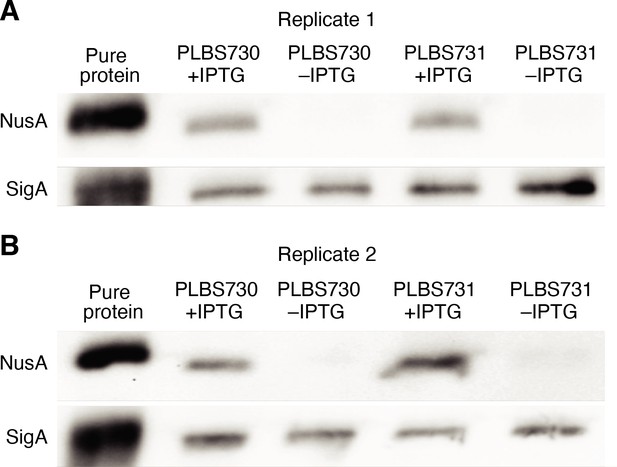
Western blot analysis of NusA depletion.
(A) Western blot for all samples used for Term-seq replicate 1. Top panel, image after probing for NusA. Bottom panel, image after probing for SigA as a loading control. Purified NusA and SigA are in the left lane. Lanes containing protein extracted from strains PLBS730 ± IPTG and PLBS731 ± IPTG are specified. (B) Identical to panel (A) except samples from replicate 2.
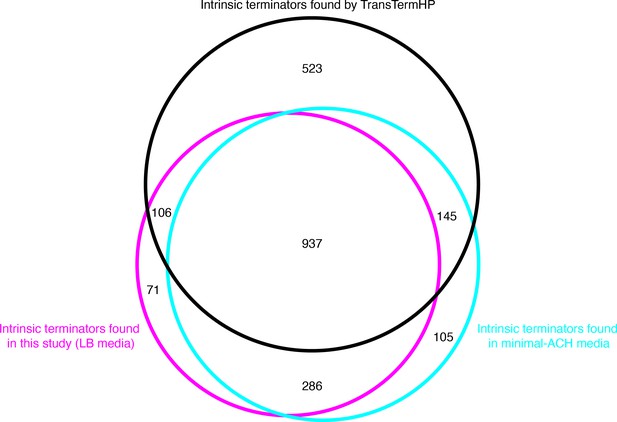
Benchmarking the intrinsic terminators identified in this study.
Venn diagram showing the number and overlap of Bacillus subtilis terminators identified within three distinct datasets. The black circle contains all terminators that were identified by the in silico tool TransTermHP, the magenta circle contains all terminators that were identified here via Term-seq conducted in wild-type (WT) B. subtilis grown in LB media, and the cyan circle contains all terminators that were identified previously via Term-seq conducted in WT B. subtilis grown in Minimal-ACH media (Mondal et al., 2016).
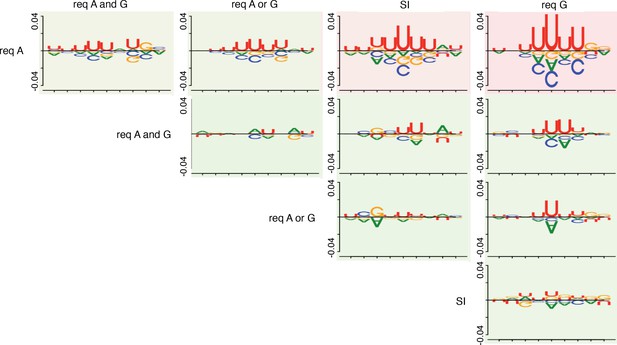
Pairwise comparative analysis of U-rich tract sequence logos.
The DiffLogo (Nettling et al., 2015) toolkit was used to determine the per-nucleotide Jensen-Shannon divergences for each U-rich tract sequence logo that was generated for the strong and independent (SI), requiring NusA (Req A), requiring NusG (Req G), Req A and G, and Req A or G terminator subpopulations in a pairwise fashion. For each pairwise comparison, the sequence above the line represents the nucleotide enrichments of each position within the U-rich tract sequence logo generated for the terminator subpopulation specified above the comparison, compared to the U-rich tract sequence logo generated for the terminator subpopulation specified to the side of the comparison. Likewise, the sequence below the line represents the nucleotide enrichments for each position within the U-rich tract sequence logo generated for the terminator subpopulation specified to the side of the comparison, compared to the U-rich tract sequence logo generated for the terminator subpopulation specified above the comparison. The y axes display the precise divergence values. More similar sequence logos rank as more green, while more divergent logos rank as more red.
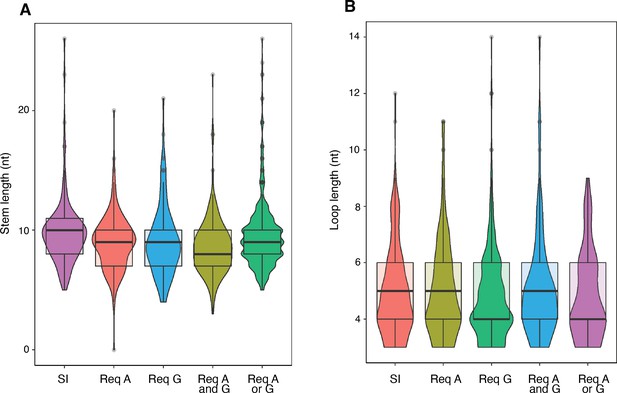
Intrinsic terminator hairpin stem length and loop length.
(A) Violin plots overlayed with box plots showing the distribution of terminator hairpin stem lengths of the strong and independent (SI), requiring NusA (Req A), requiring NusG (Req G), Req A and G, and Req A or G terminator subpopulations identified in Figure 1B. (B) Violin plots overlayed with box plots showing the distribution of terminator hairpin loop lengths of the SI, Req A, Req G, Req A and G, and Req A or G terminator subpopulations identified in Figure 1B.
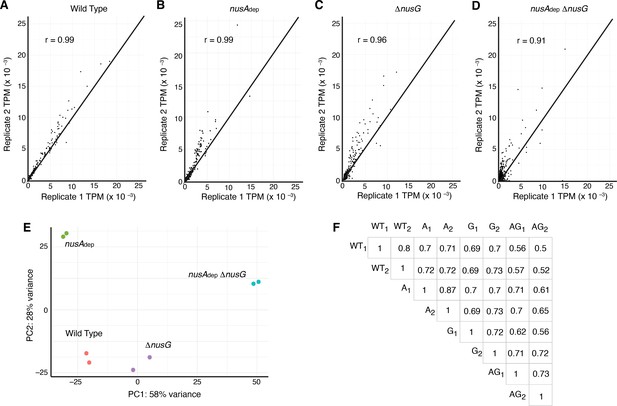
Transcriptomics data showing that Term-seq replicates are highly correlated and data from each strain is distinct.
(A) Scatter plot of all protein coding sequence transcripts per million (TPM) values calculated from the wild-type (WT) strain replicates. Line is through the diagonal (y = x) and Spearman’s correlation value specified in the plot. (B) Identical to panel (A) except for nusAdep strain replicates. (C) Identical to panel (A) except for ΔnusG strain replicates. (D) Identical to panel (A) except for nusAdep ΔnusG strain replicates. (E) Principal component analysis (PCA) plot of transcriptomics data collected from each sample. (F) A correlational matrix plot displaying the pairwise Spearman’s correlational r-values obtained when comparing all intrinsic terminator %T values between replicates.
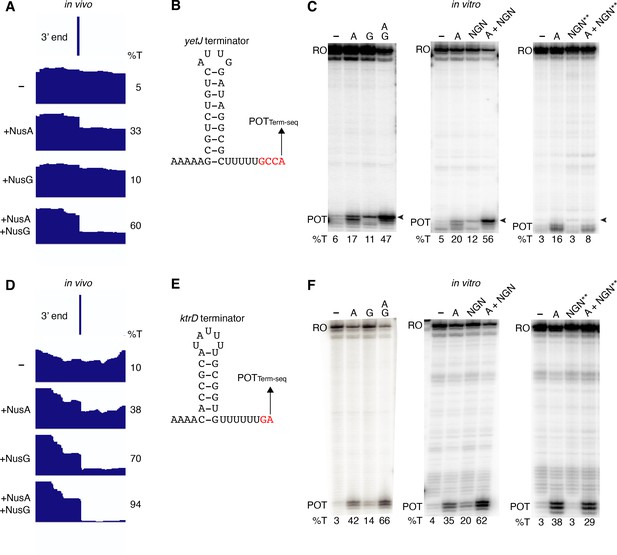
NusG stimulates intrinsic termination via its NGN domain.
(A) IGV screenshot of a genomic window centered around the yetJ terminator. Top track is the 3’ end identified by Term-seq. Bottom tracks are the RNA-seq coverage data for the nusAdep ΔnusG (-), ΔnusG (+NusA), nusAdep (+NusG), and wild-type (WT) (+NusA +NusG) strains. %T in each strain is shown on the right of each track. Transcription proceeds from left to right. (B) YetJ terminator showing the point of termination identified by Term-seq in vivo (POTTerm-seq). Disruptions in the U-rich tract are shown in red. The upstream A tract is also shown. (C) Single-round in vitro termination assay with the yetJ terminator. Experiments were performed in the absence (–) or presence of NusA (A), NusG (G), WT NGN domain, and/or mutant NGN domain as indicated (mutant NGN domain is signified as NGN**). Positions of terminated (POT) and run-off (RO) transcripts are marked. The arrowhead marks the most distal POT. %T is shown below each lane. (D–F) Identical to panels (A–C) except that it is the ktrD terminator.
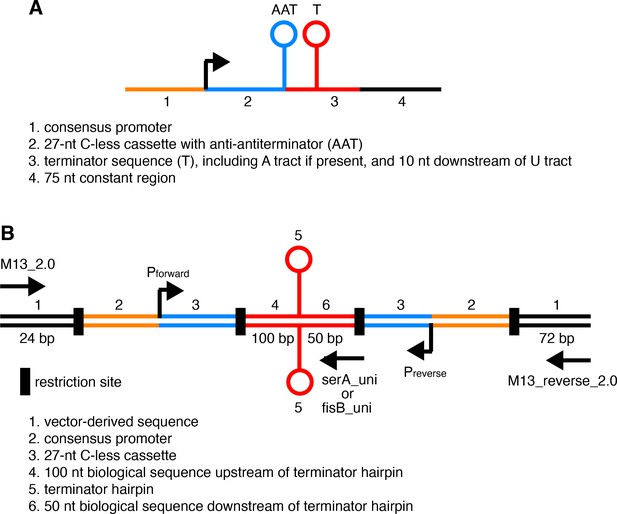
Template design for in vitro transcription.
(A) Schematic representation of templates used for all in vitro transcription experiments except for those involving convergent transcription. Template features are described below. (B) Schematic representation of templates used for in vitro transcription experiments that involve convergent transcription. Template features are described below including locations of restriction sites and primer annealing for the generation of templates.
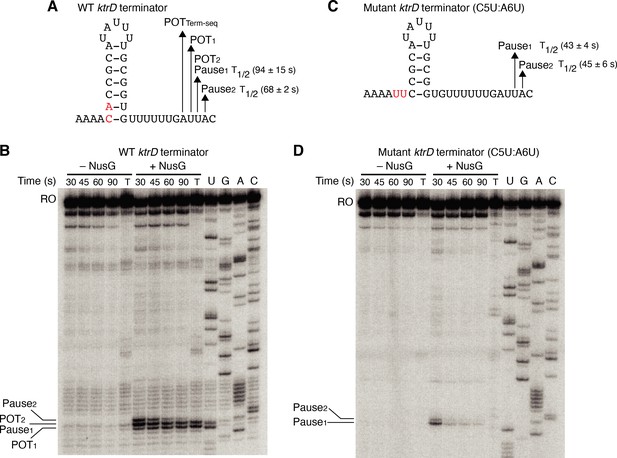
NusG stimulates termination through its role as a sequence-specific pause factor.
(A) KtrD terminator showing the points of termination (POT1 and POT2) and pause sites (Pause1 and Pause2) identified in vitro. The POT identified in vivo by Term-seq is also specified (POTTerm-seq). The upstream A tract is also shown. The average half-life (T1/2) of each pause ± standard deviation based on in vitro pausing data are specified to the right of each pause in parentheses. (B) Single-round in vitro pausing and termination assay using the wild-type (WT) ktrD terminator ± NusG. Time points of elongation are indicated above each lane. T, 30 min termination assay. RNA sequencing lanes (U, G, A, C) are labeled. Positions of NusG-dependent pause bands, termination sites, and run-off (RO) transcripts are marked. (C, D) Identical to (A–B) except that it is the C5U:A6U mutant ktrD terminator. The WT (A) and mutated (C) residues are highlighted in red.
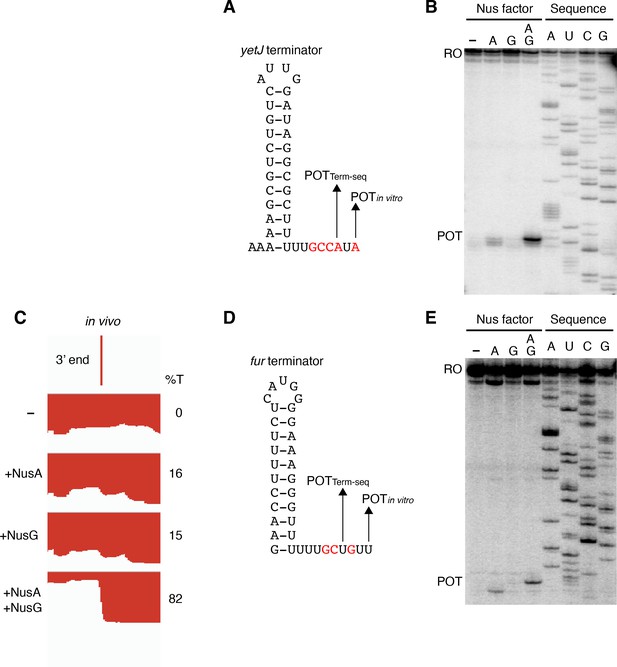
NusG stimulates terminators with particularly weak terminal base pairings.
(A) YetJ terminator showing the point of termination identified in vivo by Term-seq (POTTerm-seq) and by in vitro transcription in the +A+G condition (POTin vitro). Disruptions in the U-rich tract are shown in red. The upstream A tract is also shown. An IGV screenshot of this terminator is shown in Figure 2A. (B) Single-round in vitro termination assay with the yetJ terminator. Experiments were performed in the absence (–) or presence of NusA (A) and/or NusG (G) as indicated. Positions of terminated (POT) and run-off (RO) transcripts are marked. RNA sequencing lanes (A, U, C, G) are labeled. (C) IGV screenshot of a genomic window centered around the fur terminator. Top track is the 3’ end identified by Term-seq. Bottom tracks are the RNA-seq coverage data for the nusAdep ΔnusG (-), ΔnusG (+NusA), nusAdep (+NusG), and WT (+NusA +NusG) strains. %T in each strain is shown on the right of each track. Transcription proceeds from right to left. (D, E) Identical to panels (A, B) except that it is the fur terminator. (D) Note that the terminal three base pairs contain the A tract and one G residue.
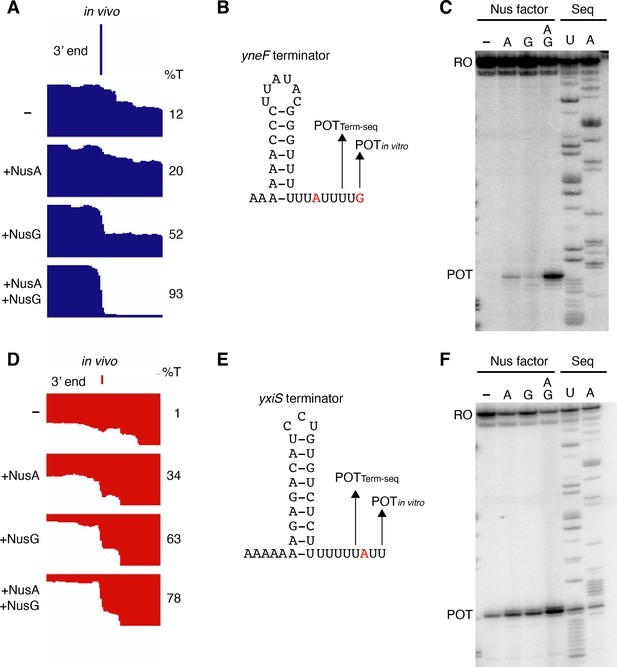
NusG stimulates termination at terminators containing A-U base pairs at the base of the hairpin.
(A) IGV screenshot of a genomic window centered around the yneF terminator. Top track is the 3’ end identified by Term-seq. Bottom tracks are the RNA-seq coverage data for the nusAdep ΔnusG (-), ΔnusG (+NusA), nusAdep (+NusG), and wild-type (WT) (+NusA +NusG) strains. %T in each strain is shown on the right of each track. Transcription proceeds from left to right. (B) YneF terminator showing the point of termination identified in vivo by Term-seq (POTTerm-seq) and by in vitro transcription in the +A+G condition (POTin vitro). Disruptions in the U-rich tract are shown in red. The upstream A tract is also shown. (C) Single-round in vitro termination assay with the yneF terminator. Experiments were performed in the absence (–) or presence of NusA (A) and/or NusG (G) as indicated. Positions of terminated (POT) and run-off (RO) transcripts are marked. RNA sequencing lanes (U, A) are labeled. (D–F) Identical to panels (A–C) except that it is for the yxiS terminator. (D) Transcription is from right to left.
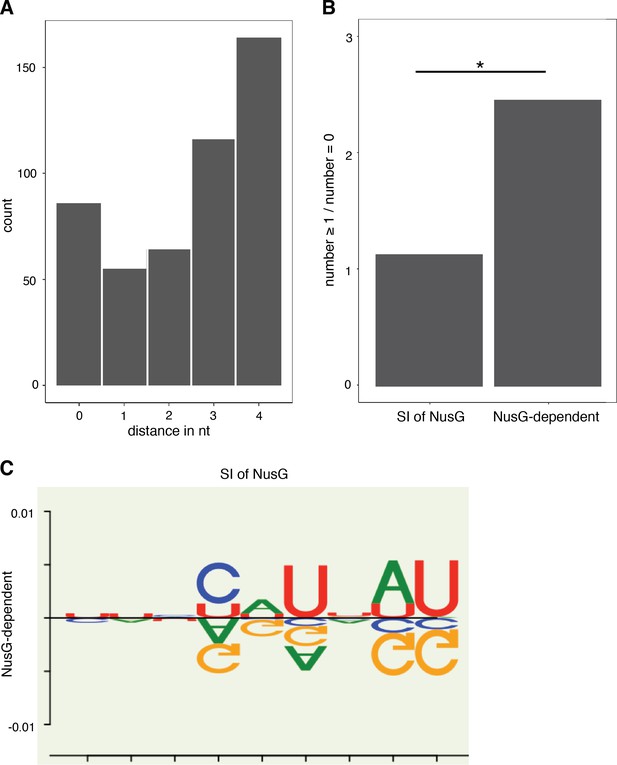
RNET-seq analysis of intrinsic termination.
(A) Histogram showing the number of intrinsic terminators at which an RNET-seq 3’ end was identified 0, 1, 2, 3, or 4 nucleotide (nt) downstream from the 3’ end identified by Term-seq. (B) Histogram showing the number of intrinsic terminators with ≥1 consecutive terminal A-U/G-U base pair (bp) divided by the number of intrinsic terminators with 0 consecutive terminal A-U/G-U bp for intrinsic terminators classified as strong and independent of NusG (SI of NusG) and terminators classified as NusG-dependent. Statistical p-value obtained from Fisher’s exact test displayed above the horizontal bar. (C) The DiffLogo (Nettling et al., 2015) toolkit was used to determine the per-nucleotide Jensen-Shannon divergences for the revised U-rich tract sequence logos that were generated for the SI of NusG and NusG-dependent terminator subpopulations. The sequence above the line represents the nucleotide enrichments of each position within the U-rich tract sequence logo generated for the SI of NusG terminator subpopulation, compared to the U-rich tract sequence logo generated for the NusG-dependent terminator subpopulation. Likewise, the sequence below the line represents the nucleotide enrichments for each position within the U-rich tract sequence logo generated for the NusG-dependent terminator subpopulation, compared to the U-rich tract sequence logo generated for SI of NusG terminator subpopulation. The y axes display the precise divergence values.
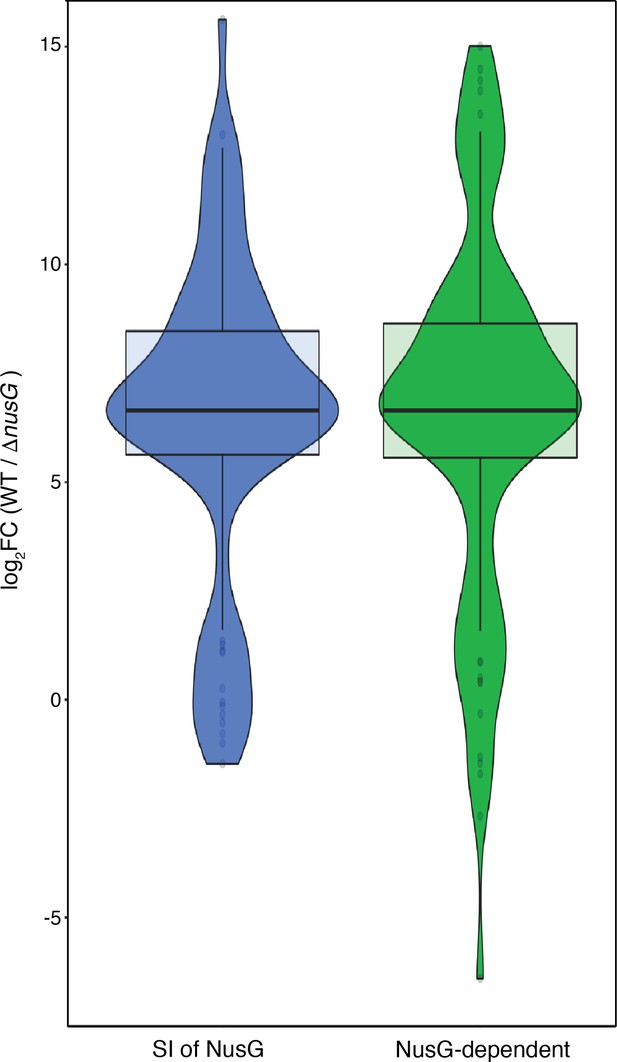
RNET-seq analysis of NusG-dependent pausing at intrinsic terminators.
Violin plots overlayed with a box plot showing the NusG dependency of RNET-seq 3’ ends found at intrinsic terminators as calculated by the log2 transformed ratio of the normalized 3’ end abundance in the wild-type (WT) strain to the normalized 3’ end abundance in the ΔnusG strain.
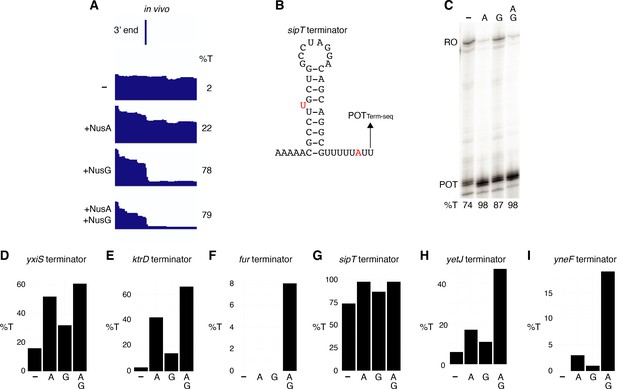
NusA is a more potent termination factor in vitro than NusG.
(A) IGV screenshot of a genomic window centered around the sipT terminator. Top track is the 3’ end identified by Term-seq. Bottom tracks are the RNA-seq coverage data for the nusAdep ΔnusG (-), ΔnusG (+NusA), nusAdep (+NusG), and wild-type (WT) (+NusA +NusG) strains. %T in each strain is shown on the right of each track. Transcription proceeds from left to right. (B) SipT terminator showing the point of termination identified in vivo by Term-seq (POTTerm-seq). Disruptions in the hairpin and U-rich tract are shown in red. The upstream A tract is also shown. (C) Single-round in vitro termination assay with the sipT terminator. Experiments were performed in the absence (–) or presence of NusA (A) and/or NusG (G) as indicated. Positions of terminated (POT) and run-off (RO) transcripts are marked. %T is shown below each lane. (D–I) In vitro termination efficiencies for the specified terminators in the absence (–) or presence of NusA (A) and/or NusG (G) as indicated.
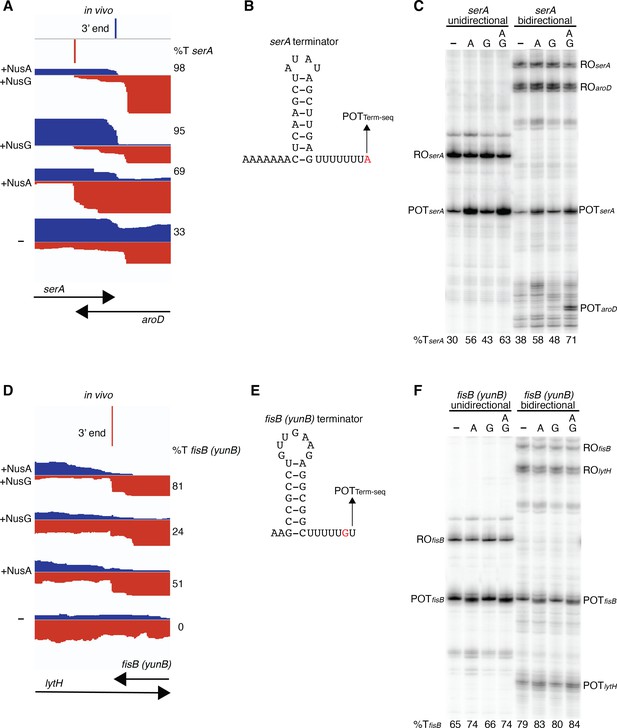
Convergent transcription does not modify the impact of NusG in vitro.
(A) IGV screenshot of a genomic window showing the intersection of serA and aroD. Top track is the 3’ end of serA identified by Term-seq. Bottom tracks are the RNA-seq coverage data for the wild-type (WT) (+NusA +NusG), nusAdep (+NusG), ΔnusG (+NusA), and nusAdep ΔnusG (–) strains. %T in each strain is shown on the right of each track. Transcription proceeds from left to right for serA (blue) and right to left for aroD (red). (B) SerA terminator showing the point of termination identified in vivo by Term-seq (POTTerm-seq). The upstream A tract is also shown. (C) In vitro transcription of the serA terminator (serA unidirectional) or both the serA and aroD terminators (serA bidirectional). Experiments were performed in the absence (–) or presence of NusA (A) and/or NusG (G) as indicated. Positions of terminated (POT) and run-off (RO) products are marked. %T is shown below each lane. (D–F) Identical to panels (A–C) except that it is the fisB (yunB) terminator at the fisB-lytH intersection.
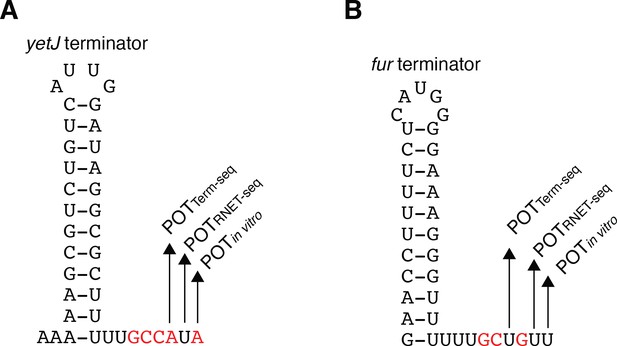
Comparison of Term-seq 3’ ends, RNET-seq 3’ ends, and in vitro 3’ ends.
(A) Model of the yetJ terminator showing the point of termination identified in vivo by Term-seq (POTTerm-seq), in vivo by RNET-seq (POTRNET-seq), and by in vitro transcription in the +A+G condition (POTin vitro). (B) Identical to panel A except that it is for the fur terminator.
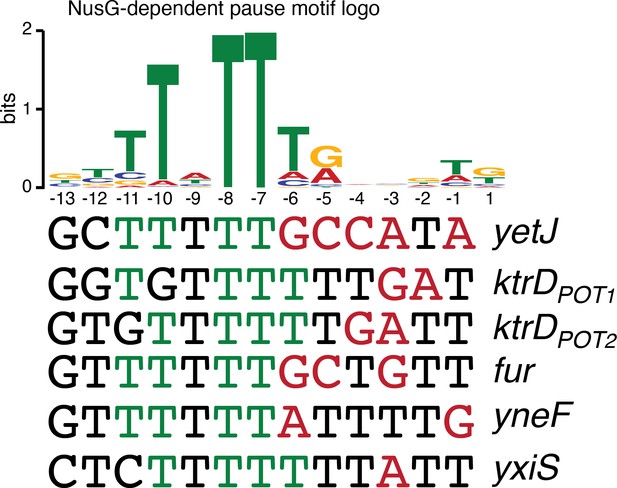
NusG-dependent pause motif.
The NusG-dependent pause motif logo of the non-template DNA (ntDNA) strand is pictured on the top. ntDNA strand sequence upstream of the 3’ end identified by in vitro transcription for NusG-dependent terminators identified in vivo. Green nucleotide (nt) are T residues that fit the NusG-dependent pause motif logo (TTNTTT). Red nt are non-U residues within the U-rich tract, which extends from positions −9 to −1.
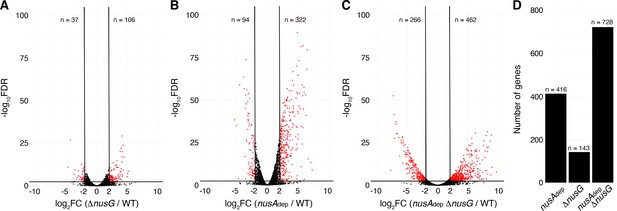
NusG coordinates global gene expression with NusA.
(A) Volcano plot derived from differential expression analysis comparing steady-state gene expression levels in the wild-type (WT) and ΔnusG strains. Cutoffs are log2 fold change (log2FC) of 2 and a −log10 of the false discovery rate (−log10FDR) of 2.3. Number (n) of genes downregulated and upregulated are specified. (B) Identical to panel (A) except comparing the WT and nusAdep strains. (C) Identical to panel (A) except comparing the WT and nusAAdep ΔnusG strains. (D) Total number of differentially expressed genes in the nusAdep, ΔnusG, and nusAdep ΔnusG strains; 2604 transcripts in which the transcripts per million (TPM) >10 in the WT strain were used in this analysis (Supplementary file 6).
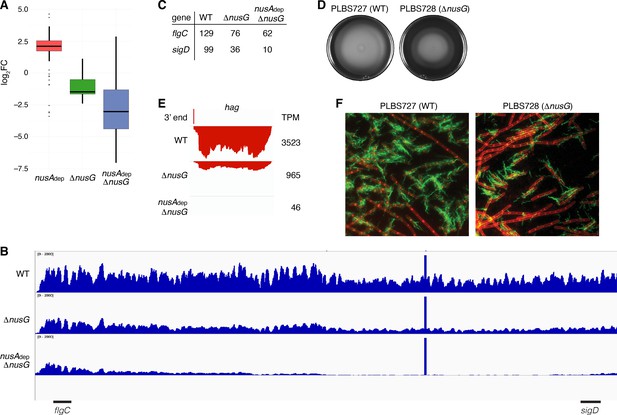
NusG is a motility factor in Bacillus subtilis.
(A) Box plot showing the effect of NusA and/or NusG for all transcripts within the motility regulon; log2 fold change (log2FC) (mutant:WT [wild type]). (B) IGV screenshot of the fla/che operon. Each track is the RNA-seq coverage data for the WT, ΔnusG, and nusAdep ΔnusG strains. Locations of the flgC and sigD genes are specified below the screenshot. (C) Transcripts per million (TPM) values calculated for the flgC and sigD genes in the WT, ΔnusG, and nusAdep ΔnusG strains. (D) Swimming motility assay for PLBS727 (WT) and PLBS728 (ΔnusG). (E) IGV screenshot of the hag transcript. Top track is the 3’ end identified by Term-seq. Bottom tracks are the RNA-seq coverage data for the WT, ΔnusG, and nusAdep ΔnusG strains. Transcription proceeds from right to left. TPM values were calculated for the hag transcript in each strain and are specified to the right of each track. (F) Fluorescence microscopy performed on PLBS727 (WT) and PLBS728 (ΔnusG) strains. Membrane is stained with FM4-64 (false colored red) and flagella are stained with Alexa Fluor 488 C5 maleimide (false colored green).
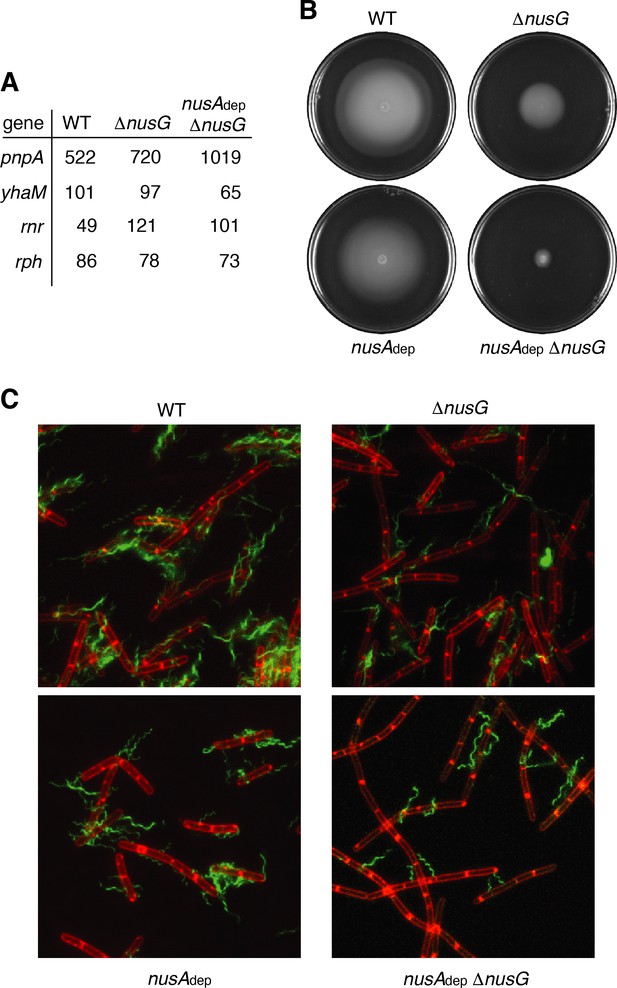
NusG is a motility factor in Bacillus subtilis.
(A) Transcripts per million (TPM) values calculated for pnpA, yhaM, rnr, and rph, four genes that encode 3’–5’ exoribonuclease, in wild-type (WT), ΔnusG, and nusAdep ΔnusG strains. (B) Swimming motility assay for WT, nusAdep, ΔnusG, and nusAdep ΔnusG strains. (C) Florescence microscopy performed on WT, nusAdep, ΔnusG, and nusAdep ΔnusG strains. Membrane is stained with FM4-64 (false colored red) and flagella are stained with Alexa Fluor 488 C5 maleimide (false colored green).
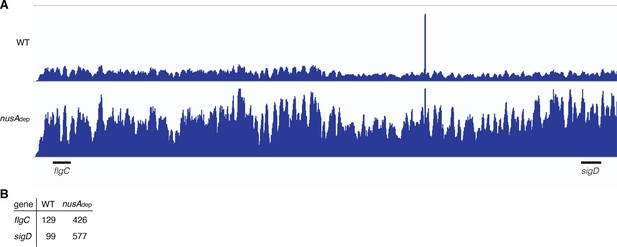
NusA may serve as a transcription destabilization factor.
(A) IGV screenshot of the fla/che operon. Each track is the RNA-seq coverage data for the wild-type (WT) and nusAdep strains. Locations of the flgC and sigD genes are specified below the screenshot. (B) Transcripts per million (TPM) values calculated for the flgC and sigD genes in the WT and nusAdep strains.
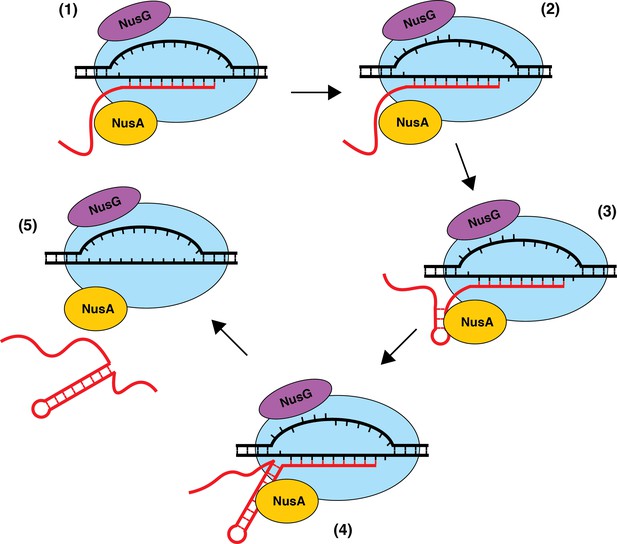
Model of NusA-dependent and NusG-dependent intrinsic termination.
(1) During transcription elongation, NusA binds to the β flap domain of RNA polymerase (RNAP) near the RNA exit channel, whereas NusG binds to the β' clamp helices in close proximity to the non-template DNA (ntDNA) strand within the transcription bubble. NusA stimulates pausing during transcription of a suboptimal U-rich tract containing distal U tract interruptions. (2) NusG shifts RNAP to the post-translocated state and increases the pause half-life by making sequence-specific contacts with the TTNTTT motif within the ntDNA strand of the paused transcription bubble. The T residues in this motif are shown as being flipped out toward the NGN domain of bound NusG, but evidence for base flipping has not been obtained. (3) NusA assists with hairpin formation within the RNA exit channel. (4) NusG-dependent pausing provides time for weak A-U and/or G-U base pairs to form at the base of the terminator hairpin such that the hairpin to 3’ end distance is reduced to 7–9 nt. (5) The combination of the weak RNA-DNA hybrid, the close proximity of the hairpin to the RNA 3’ end, and the long-lived pause contributes to transcript release.
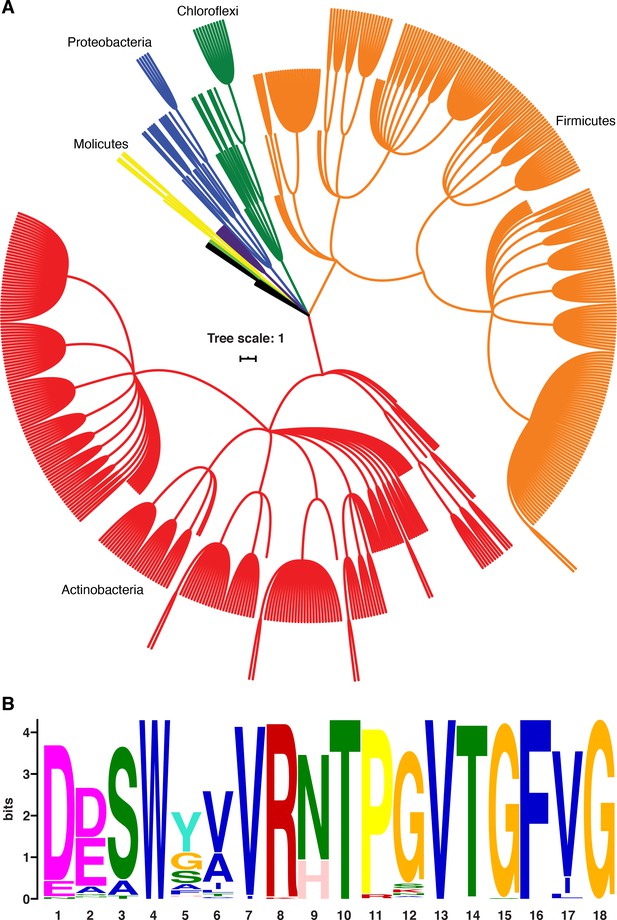
NusG homologs exhibit the capacity to contact the ntDNA strand across the bacterial domain.
(A) Phylogenetic tree constructed from the 16S rRNA sequences of all bacteria found to encode a NusG homolog that contains the Bacillus subtilis-like dipeptide residues (NT and HT). Each different phylum of bacteria that is represented with three or more species is highlighted a different color. Actinobacteria branches are red, Firmicute branches are orange, Chloroflexi branches are dark green, Proteobacteria branches are blue, Synergistaceae branches are purple, Mollicute branches are yellow, and Bacteroidete branches are light green. Phyla with fewer than three representative species are in black. (B) Sequence logo constructed from the portion of NusG that interacts with the ntDNA strand in B. subtilis for all NusG homologs present in (A). Critical dipeptide NT/HT is located at positions 9 and 10.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background (Bacillus subtilis) | MH5636 | Qi and Hulett, 1998 | N/A | See Supplementary file 8 for derivatives |
| Strain, strain background (Bacillus subtilis) | PLBS338 | Yakhnin et al., 2004 | N/A | See Supplementary file 8 for derivatives |
| Strain, strain background (Bacillus subtilis) | 3610 | LaVallie and Stahl, 1989 | N/A | See Supplementary file 8 for derivatives |
| Antibody | Rabbit polyclonal anti-NusA | Peter Lewis | N/A | WB (1:5000) |
| Antibody | Rabbit polyclonal anti-SigA | Masaya Fujita | N/A | WB (1:5000) |
| Antibody | Goat polyclonal peroxidase labeled anti-rabbit | GenScript | Cat# A00098 | WB (1:1000) |
| Recombinant DNA reagent | pTZ19R | Thermo Fisher | Cat# SD0141 | See Supplementary file 9 for derivatives |
| Recombinant DNA reagent | pNC018 | David Rudner | N/A | See Supplementary file 9 for derivatives |
| Recombinant DNA reagent | pNE4 | Blair et al., 2008 | N/A | See Supplementary file 9 for derivatives |
| Sequence-based reagent | Primers | This study | N/A | See Supplementary file 10 |
| Software, algorithm | Python | N/A | v.3.83 | https://www.python.org/ |
| Software, algorithm | R | N/A | v.4.03 | https://www.r-project.org/ |
| Software, algorithm | ImageQuant | N/A | v.5.2 | N/A |
| Software, algorithm | TransTermHP | Kingsford et al., 2007 | v.2.09.1 | http://transterm.ccb.jhu.edu/ |
| Software, algorithm | DiffLogo | Nettling et al., 2015 | v.3.12 | https://bioconductor.org/packages/release/bioc/html/DiffLogo.html |
| Software, algorithm | DESeq2 | Love et al., 2014 | v.1.26.0 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| Software, algorithm | Kallisto | Bray et al., 2016 | v.0.46.2 | https://pachterlab.github.io/kallisto/ |
| Software, algorithm | Trimmomatic | Bolger et al., 2014 | v.0.38 | http://www.usadellab.org/cms/?page=trimmomatic |
| Software, algorithm | Cutadapt | Marcel, 2011 | v.1.16 | https://cutadapt.readthedocs.io/en/stable/ |
| Software, algorithm | Bedtools | Quinlan and Hall, 2010 | v.2.26.0 | https://bedtools.readthedocs.io/en/latest/ |
| Software, algorithm | Samtools | Li et al., 2009 | v.0.1.19–44428 cd | http://www.htslib.org/ |
| Software, algorithm | bwa-mem | Li, 2013 | v.0.7.12-r1034 | http://bio-bwa.sourceforge.net/bwa.shtml |
| Software, algorithm | BLASTp | Altschul et al., 1990 | v.2.11 webtool | https://blast.ncbi.nlm.nih.gov/Blast.cgi |
| Software, algorithm | iTOL | Letunic and Bork, 2016 | v.5 webtool | https://itol.embl.de/ |
| Software, algorithm | NCBI taxonomic tree webtool | Sayers et al., 2009 | webtool | https://www.ncbi.nlm.nih.gov/guide/howto/gen-com-tree/ |
| Software, algorithm | MEME | Bailey and Elkan, 1994 | v.4.12.0 | https://meme-suite.org/meme/tools/meme |
| Software, algorithm | BaseSpace | Illumina | N/A | https://basespace.illumina.com/dashboard |
| Software, algorithm | BioPython | Cock et al., 2009 | v.1.77 | https://biopython.org/ |
| Software, algorithm | numpy | Harris et al., 2020 | v.1.19.0 | https://numpy.org/ |
| Software, algorithm | scipy | Virtanen et al., 2020 | v.1.5.1 | https://www.scipy.org/ |
| Software, algorithm | RNAStructure | Reuter and Mathews, 2010 | v.6.0.1 | https://rna.urmc.rochester.edu/RNAstructureWeb/Servers/Predict1/Predict1.html |
| Software, algorithm | ggplot2 | Wickham, 2016 | v.3.2.1 | https://ggplot2.tidyverse.org/ |
| Software, algorithm | IGV | Robinson et al., 2011 | v.2.4.14 | http://software.broadinstitute.org/software/igv/ |
| Software, algorithm | Term-seq peak calling pipeline | ‘Term-seq’ Github repository | N/A | https://github.com/zfmandell/Term-seq |
| Other | CIP | NEB | (Discontinued) | Term-seq |
| Other | Ribo-zero | Illumina | (Discontinued) | Term-seq |
| Other | T4 RNA Ligase 1 | NEB | Cat# M0204S | Term-seq |
| Other | α-32P UTP | PerkinElmer | Cat# BLU007H25OUC | Urea-PAGE |
| Other | Agarose | Dot Scientific | Cat # DSA20090-50 | Agarose gels |
| Other | Polyacrylamide | Fisher Sci | Cat# HBGR337500 | Urea-PAGE |
| Other | FM-64 | Invitrogen | Cat# T13320 | Fluorescence Microscopy |
| Other | Maleimide dye | Invitrogen | Cat# A10254 | Fluorescence Microscopy |
| Other | RNeasy columns | Qiagen | Cat# 74106 | Term-seq |
| Other | Lysozyme | Sigma-Aldrich | Cat# L6876 | Term-seq |
| Other | RNA sequencing reagents | Illumina | Cat# 20020594 | Term-seq |
| Other | Urea | EMD | Cat# 666122–2.5 kg | Urea-PAGE |
| Other | PVDF membrane | Thermo | Cat# 88585 | Western blot |
| Other | ECL substrate | Thermo Fisher | Cat# 32209 | Western blot |
| Chemical compound, drug | IPTG | Dot Scientific | Cat# DSI56000-25 | Cell Culture – NusA production |
| Chemical compound, drug | Chloramphenicol | Sigma-Aldrich | Cat# C0378-25 g | Cell Culture |
| Peptide, recombinant protein | NusA | This study | N/A | 1 µM |
| Peptide, recombinant protein | NusG | This study | N/A | 1 µM |
| Peptide, recombinant protein | NGN only NusG | This study | N/A | 1 µM |
| Peptide, recombinant protein | Mutant NGN only NusG | This study | N/A | 1 µM |
| Peptide, recombinant protein | RNAP core | This study | N/A | 0.19 µM |
| Peptide, recombinant protein | SigA | This study | N/A | 0.38 µM |
Additional files
-
Supplementary file 1
3’ ends identified in each strain.
Column A (3’ end), genomic coordinate of 3’ end identified by Term-seq. Column B (strand), strand information of identified 3’ end. Column C (Cv), the coverage variation (Cv) calculated at the genomic coordinate of the identified 3’ end. Column D (3’ end and upstream 50 nucleotide [nt]), 3’ end position and 50 nt upstream of 3’ end (51 nt total) of the ntDNA strand. Column E (ΔG-51nt (kcal/mol)), the free energy of RNA folding obtained from the 50 nt upstream of the 3’ end and the 3’ end. Column F (%T), the termination efficiency calculated at the identified 3’ end. Column G (relative position), the transcriptomic context of the 3’ end.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp1-v2.xlsx
-
Supplementary file 2
All intrinsic terminators identified by Term-seq in the wild-type (WT) strain.
Column A (point of termination [POT]), genomic coordinate of the identified intrinsic terminator. Column B (strand), strand information of the identified intrinsic terminator. Column C (relative position), distance of identified intrinsic terminator from nearest upstream coding sequence and identity of coding sequence. Column D (%T [WT]), termination efficiency of identified intrinsic terminator in the WT strain. Column E (%T [mutant]), termination efficiency of identified intrinsic terminator in the mutant strain. Column F (Δ%T), change in termination efficiency due to loss of factor(s). Column G (upstream sequence), nucleotide composition of the 9 nt upstream of the predicted terminator hairpin. Column H (predicted hairpin sequence), nucleotide composition of the predicted terminator hairpin. Column I (downstream sequence), nucleotide composition of the 9 nt downstream of the predicted terminator hairpin. Column J (ΔG-hairpin [kcal/mol]), the free energy of RNA folding the predicted terminator hairpin.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp2-v2.xlsx
-
Supplementary file 3
Pairwise p-values.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp3-v2.xlsx
-
Supplementary file 4
All intrinsic terminators identified by RNET-seq in the wild-type (WT) strain.
Column A (Term-seq point of termination [POT]), genomic coordinate of the intrinsic terminator identified by Term-seq. Column B (strand), strand information of the identified intrinsic terminator. Column C (dist), the distance in nucleotide (nt) of the RNET-seq 3’ end from the Term-seq 3’ end. Column D (relative position), distance of the intrinsic terminator identified by RNET-seq from nearest upstream coding sequence and identity of coding sequence. Column E (revised upstream sequence), nucleotide composition of the 9 nt upstream of the predicted terminator hairpin revised using the RNET-seq 3’ end. Column F (revised predicted hairpin sequence), nucleotide composition of the predicted terminator hairpin revised using the RNET-seq 3’ end. Column G (revised downstream sequence), nucleotide composition of the 9 nt downstream of the predicted terminator hairpin revised using the RNET-seq 3’ end. Column H (classification), whether the terminators were classified as SI of NusG or NusG-dependent. Column I (log2 fold change [log2FC]), the log2 transformed ratio of the pause strength in the WT strain compared to the ΔnusG strain.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp4-v2.xlsx
-
Supplementary file 5
Results of differential expression analysis.
Column A (transcript), transcript, naming system = ILL (transcriptome was built using Illumina sequencing data), official gene symbol, Locus, location of coding sequence within transcript. Column B (baseMean), normalized counts for transcript in column A, averaged across all samples. Column C (log2 fold change), the log2 transformation of the fold change in expression of the transcript in column A. Column D (lfcSE), standard error of the log2(fold change) calculated in column C. Column E (stat), Wald statistic for log2(fold change) calculated in column C. Column F (p-value), Wald p-value for log2(fold change) calculated in column C. Column G (padj), Benjamni-Hochberg p-adjusted value for log2(fold change) calculated in column C.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp5-v2.xlsx
-
Supplementary file 6
Transcriptome-wide expression data collected for all strains as reported in transcripts per million (TPM) and raw counts.
Column A (transcript), transcript, naming system = ILL (transcriptome was built using Illumina sequencing data), official gene symbol, Locus, location of coding sequence within transcript. Column B (WT Rep. 1), data collected from wild-type (WT) replicate 1. Column C (WT Rep. 2), data collected from WT replicate 2. Column D (WT Merged), data collected from merged WT replicates. Column E (nusAdep Rep. 1), data collected from nusAdep replicate 1. Column F (nusAdep Rep. 2), data collected from nusAdep replicate 2. Column G (nusAdep Merged), data collected from merged nusAdep replicates. Column H (ΔnusG Rep. 1), data collected from ΔnusG replicate 1. Column I (ΔnusG Rep. 2), data collected from ΔnusG replicate 2. Column J (ΔnusG Merged), data collected from merged ΔnusG replicates. Column K (nusAdep ΔnusG Rep. 1), data collected from nusAdep ΔnusG replicate 1. Column L (nusAdep ΔnusG Rep. 2), data collected from nusAdep ΔnusG replicate 2. Column M (nusAdep ΔnusG Merged), data collected from merged nusAdep ΔnusG replicates.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp6-v2.xlsx
-
Supplementary file 7
Expression data for the motility regulon.
Column A (transcript), transcript, naming system = ILL (transcriptome was built using Illumina sequencing data), official gene symbol, Locus, location of coding sequence within transcript. Column B (wild-type), normalized TPM values collected from merged wild-type replicates for the transcript specified in column A. Column C (nusAdep), normalized transcripts per million (TPM) values collected from merged nusAdep replicates for the transcript specified in column A. Column D (ΔnusG), normalized TPM values collected from merged ΔnusG replicates for the transcript specified in column A. Column E (nusAdep ΔnusG), normalized TPM values collected from merged nusAdep ΔnusG replicates for the transcript specified in column A.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp7-v2.xlsx
-
Supplementary file 8
Bacillus subtilis strains used in this study.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp8-v2.xlsx
-
Supplementary file 9
Plasmids used in this study.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp9-v2.xlsx
-
Supplementary file 10
Primers used in this study.
- https://cdn.elifesciences.org/articles/61880/elife-61880-supp10-v2.xlsx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61880/elife-61880-transrepform-v2.pdf





