XLF acts as a flexible connector during non-homologous end joining
Figures
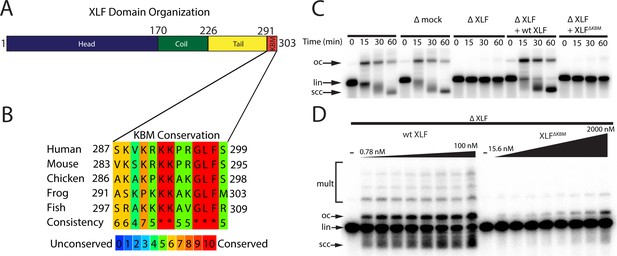
The Ku binding motif (KBM) of XLF is critical for end joining in Xenopus egg extract.
(A) A schematic of the domain organization of XLF. The residue number at the boundary of each region is shown. (B) A protein sequence alignment of the KBM from human (Homo sapiens UniProt ID Q9H9Q4), mouse (Mus musculus UniProt ID Q3KNJ2), chicken (Gallus gallus UniProt ID F1NVP8), frog (Xenopus laevis see note below), and fish (Danio rerio UniProt ID Q6NV18). The Xenopus laevis XLF sequence and translation start site was determined previously by immunoprecipitation from extract and subsequent trypsin digestion and analysis by mass spectrometry (Graham et al., 2016). The alignment was performed using PRALINE multiple sequence alignment and its default settings (Simossis and Heringa, 2005). A colored scale shows the degree of conservation. (C) Ensemble time course end joining assay in either mock-depleted (immunodepletion with non-specific rabbit IgG) or XLF-depleted Xenopus egg extract. Recombinant wild-type (wt) XLF and XLFΔKBM were added back at 75 nM final concentration. DNA species: scc, supercoiled closed circular; lin, linear; oc, open circle. (D) Ensemble end point titration end joining assay in Xenopus egg extract. XLF-depleted extract was supplemented with recombinant protein, either wt XLF or XLFΔKBM, at varying concentrations. The reactions were stopped after 20 min. DNA species: scc, supercoiled closed circular; lin, linear; oc, open circle; mult, multimer.
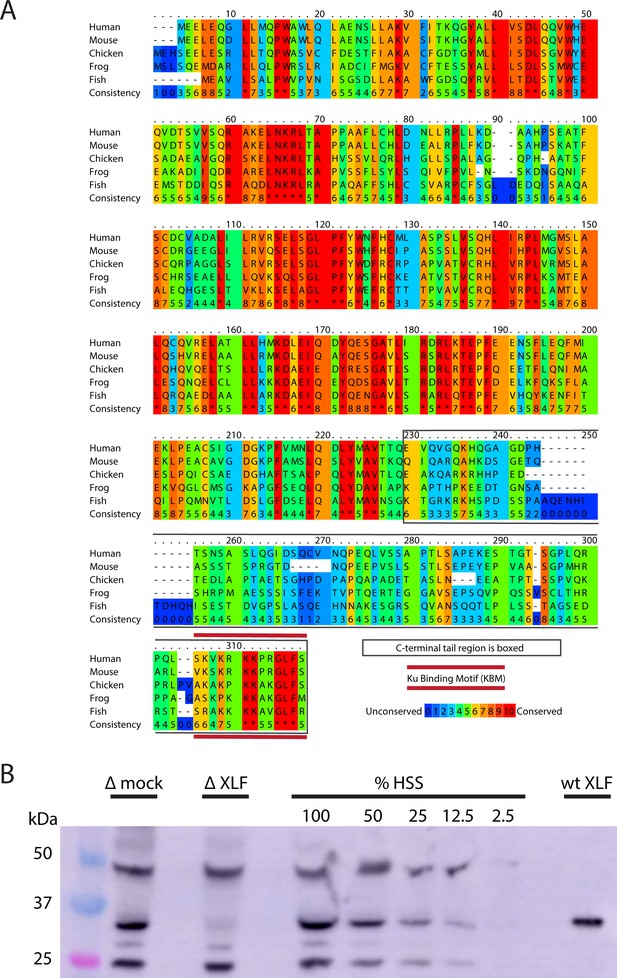
Xenopus laevis XLF is evolutionarily conserved and can be depleted from egg extract.
(A) A protein sequence alignment of XLF from human (Homo sapiens UniProt ID Q9H9Q4), mice (Mus musculus UniProt ID Q3KNJ2), chicken (Gallus gallus UniProt ID F1NVP8), frog (Xenopus laevis see note), and fish (Danio rerio UniProt ID Q6NV18). The Xenopus laevis sequence and translation start site was determined previously by immunoprecipitation from extract and subsequent trypsin digestion and analysis by mass spectrometry (Graham et al., 2016). The alignment was performed using PRALINE multiple sequence alignment and its default settings (Simossis and Heringa, 2005). A colored scale shows the degree of conservation. The C-terminal tail is boxed in gray, and the KBM is highlighted by a red bar above and below the sequences. (B) Western blotting for XLF showing its presence in extract and its relative absence in XLF-depleted extract. Unidentified cross-reactive proteins from egg extract are also visible, but the band corresponding to XLF is the only one where we see significant loss of signal in the XLF-depleted extract. Purified recombinant wt XLF (87 nM here) runs at the same molecular weight as the endogenous XLF from extract.
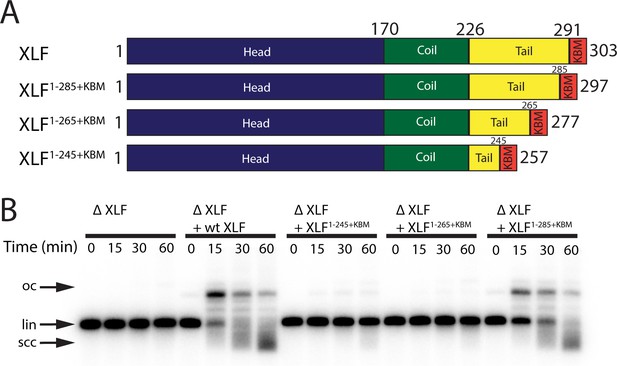
The tail region of XLF is essential for DNA end joining.
(A) Schematics of the domain organization of wt XLF and truncation mutants (XLF1-285+KBM, XLF1-265+KBM, and XLF1-245+KBM). The residue number at the boundary of each region of the protein is indicated. (B) Ensemble time course end joining assay in XLF-depleted Xenopus egg extract. Recombinant wt XLF, XLF1-285+KBM, XLF1-265+KBM, and XLF1-245+KBM were added back at 75 nM final concentration to their respective reaction samples. DNA species: scc, supercoiled closed circular; lin, linear; oc, open circle.
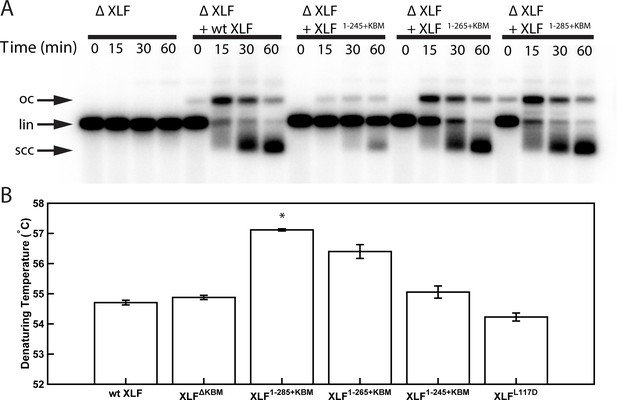
Characterization of XLF truncation mutants.
(A) Ensemble time course end joining assay in XLF-depleted Xenopus egg extract. Recombinant wt XLF, XLF1-285+KBM, XLF1-265+KBM, and XLF1-245+KBM were added back to their respective reaction samples at 500 nM final concentration. DNA species: scc, supercoiled closed circular; lin, linear; oc, open circle. (B) Denaturing temperatures of XLF constructs as measured by differential scanning fluorimetry. The average of two replicates is shown with error bars representing the minimum and maximum. Only XLF1-285+KBM had a denaturing temperature statistically different from wt XLF (p=0.0066), while the denaturing temperature of the other constructs (XLFΔKBMp=0.2451, XLF1-265+KBMp=0.615, XLF1-245+KBMp=0.3091, XLFL117Dp=0.1172) were not statistically different from that of wt XLF as determined by a two-tailed, unpaired t test with unequal variance and the Bonferroni correction. The range of denaturing temperatures was less than 3°C, suggesting no substantial changes in protein folding or stability.
-
Figure 2—figure supplement 1—source data 1
Source data for graph shown in Figure 2—figure supplement 1B.
- https://cdn.elifesciences.org/articles/61920/elife-61920-fig2-figsupp1-data1-v2.xls
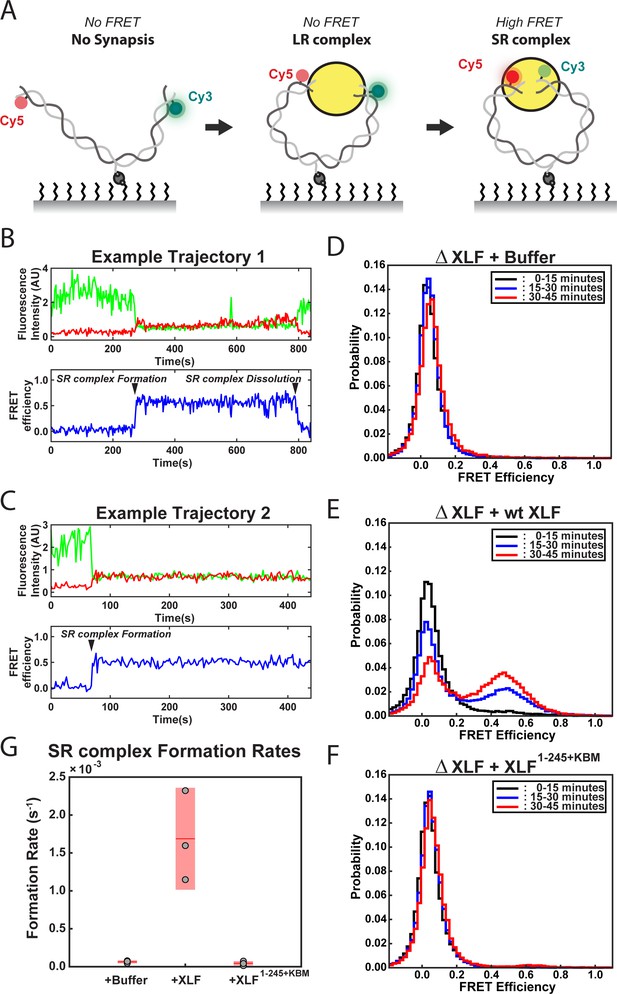
DNA end synapsis requires the tail of XLF.
(A) Schematic of the FRET-labeled DNA substrate immobilized in the flow cell via biotin-streptavidin interactions. In the absence of egg extract the DNA substrate exhibits no FRET. When the LR complex forms, the DNA ends are not positioned close enough together for energy transfer from Cy3 to Cy5 even though the ends are co-localized within the NHEJ synaptic complex (Graham et al., 2016). Upon formation of the SR complex, FRET between the fluorophores on opposing DNA ends can be observed. (B,C) Example trajectories that contain SR complex formation events. Donor and acceptor fluorescence intensity are shown in green and red, respectively. The corresponding FRET efficiency from each trace is shown in blue in a separate trajectory below. (D, E, F) Normalized FRET histograms for each experimental condition accumulated over a 15-min time window. Data from each 15 min field of view is represented by a separate curve. (G) Plot of SR complex formation rates. For each condition, individual replicates are plotted as gray circles, and the mean is represented as dark red horizontal line. The 95% confidence interval is represented for each condition as a light red rectangle centered on the mean. This plot was generated using the notBoxPlot MATLAB function (Campbell, 2020).
-
Figure 3—source data 1
Source data for histograms shown in Figure 3D–F.
- https://cdn.elifesciences.org/articles/61920/elife-61920-fig3-data1-v2.xlsx
-
Figure 3—source data 2
Source data for plot shown in Figure 3G.
- https://cdn.elifesciences.org/articles/61920/elife-61920-fig3-data2-v2.xls
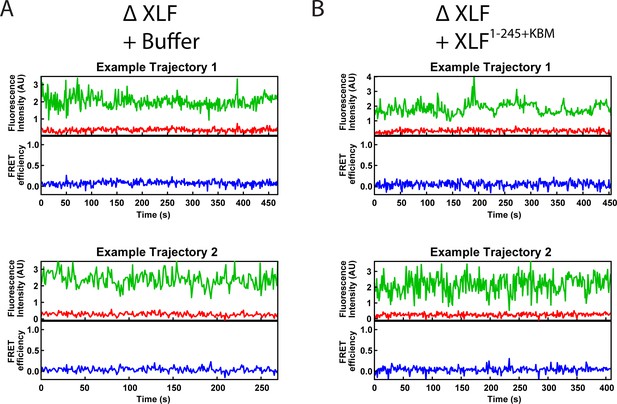
Example smFRET trajectories.
(A, B) Example trajectories from the smFRET SR Complex formation assay for the ΔXLF + Buffer and the ΔXLF + XLF1-245+KBM conditions. Donor and acceptor fluorescence intensity are shown in green and red, respectively. The corresponding FRET efficiency from each trace is shown in blue in a separate trajectory below.
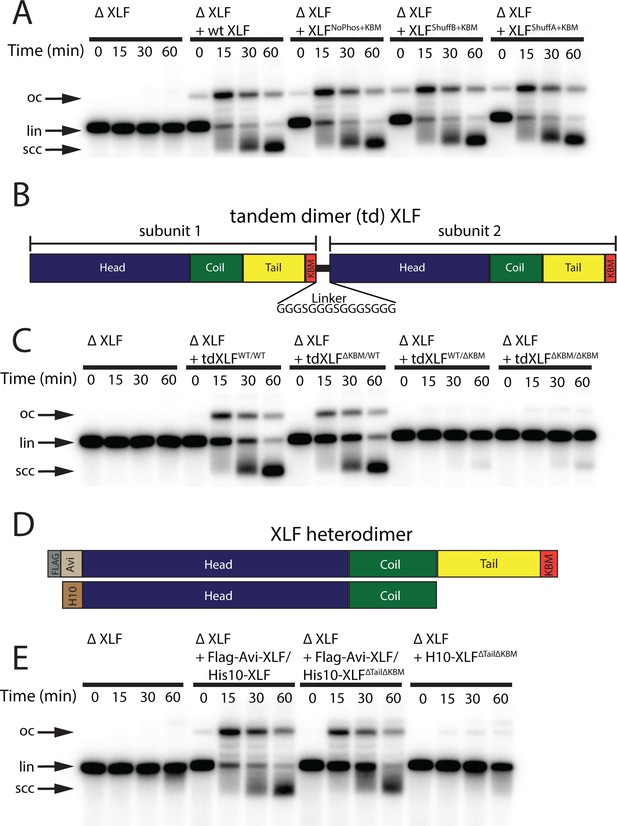
Requirements of the XLF tail in end joining.
(A) Ensemble time course end joining assay in XLF-depleted Xenopus egg extract. Recombinant wt XLF, XLFNoPhos+KBM, XLFShuffB+KBM, and XLFShuffA+KBM were added back at 75 nM to their respective reaction samples. DNA species: scc, supercoiled closed circular; lin, linear; oc, open circle. (B) Schematic of the tandem dimer (td) XLF construct. (C) Ensemble time course end joining assay in XLF-depleted Xenopus egg extract. Recombinant tdXLFWT/WT, tdXLFΔKBM/WT, tdXLFWT/ΔKBM, and tdXLFΔKBM/ΔKBM were added back at 50 nM final concentration to their respective reaction samples. DNA species: scc, supercoiled closed circular; lin, linear; oc, open circle. (D) Schematics of the individual subunits coexpressed to form the XLF heterodimer, Flag-Avi-XLF/His10-XLFΔtailΔKBM. (E) Ensemble time course end joining assay in XLF-depleted Xenopus egg extract. Recombinant heterodimers (Flag-Avi-XLF/His10-XLF and Flag-Avi-XLF/His10-XLFΔtailΔKBM) and His10-XLFΔtailΔKBM were added back at 75 nM final concentration to their respective reaction samples. DNA species: scc, supercoiled closed circular; lin, linear; oc, open circle.
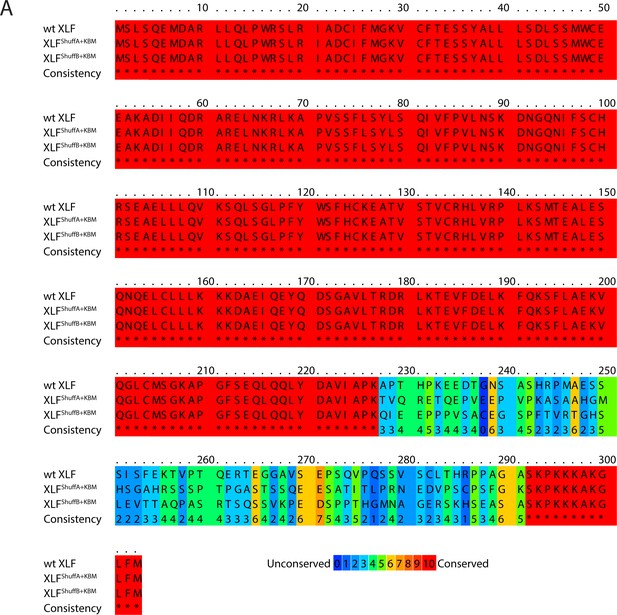
Sequence alignment of XLF shuffled tail mutants.
(A) Protein sequence alignment of wt XLF, XLFShuffA+KBM, and XLFShuffB+KBM. The alignment was performed using PRALINE multiple sequence alignment (Simossis and Heringa, 2005). Gap penalties were altered to prevent gaps in the alignment.
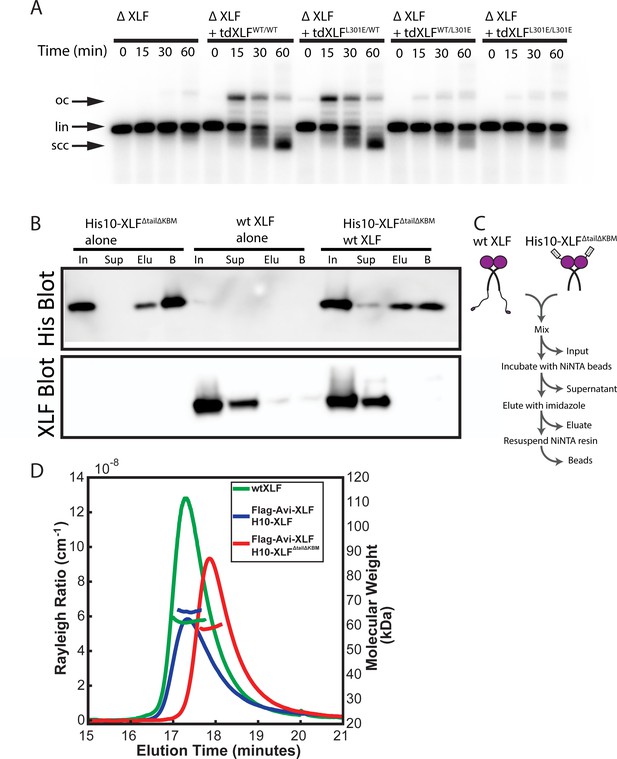
Characterization of the tdXLF and XLF heterodimer constructs.
(A) Ensemble time course end joining assay in XLF-depleted Xenopus egg extract. Recombinant tdXLFWT/WT, tdXLFL301E/WT, tdXLFWT/L301E, and tdXLFL301E/L301E were added back at 50 nM final concentration to their respective reaction samples. DNA species: scc, supercoiled closed circular; lin, linear; oc, open circle. (B) XLF heterodimer subunit exchange assay. His10-XLFΔtailΔKBM and wt XLF each alone or in combination were incubated at room temperature (input: I) before being incubated with NiNTA resin that was subsequently, spun down (supernatant: Sup), washed, eluted with imidazole (eluate: Elu), and resuspended (beads: B). Exchange between dimer subunits in the His10-XLFΔTailΔKBM + wt XLF sample would result in detecting wt XLF in the eluate and/or bead sample(s). The XLF antibody used here and throughout this study is specific to the XLF C-terminus (see Materials and methods), leading to no signal on the XLF blot for H10-XLFΔtailΔKBM. (C) Cartoon schematic of experimental workflow of the heterodimer exchange assay. (D) Analysis of XLF constructs by size-exclusion chromatography and multi-angle light scattering (SEC-MALS). Each XLF construct (wt XLF (green), Flag-Avi-XLF/H10-XLF heterodimer (blue), and Flag-Avi-XLF/H10-XLFΔtailΔKBM heterodimer (red)) eluted as a single peak (left y-axis). The corresponding molecular mass (right y-axis, shown in colors to match each sample peak) of the species present within each peak measured to within 10% of the expected mass of a dimer for each XLF sample. The XLF tandem dimer, tdXLF, has previously been shown to exist as a dimer via SEC-MALS (Graham et al., 2018).
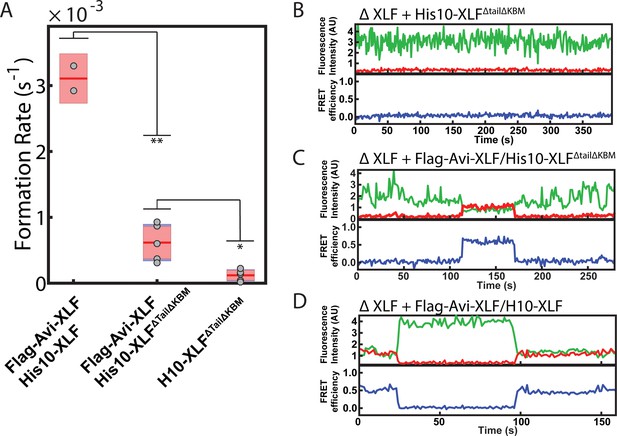
SR complex formation by XLF heterodimer constructs.
(A) Plot of SR complex formation rates. For each condition, individual replicates are plotted as grey circles, and the mean is represented as dark red horizontal line. The 95% confidence interval is represented for each condition as a light red rectangle centered on the mean. This plot was generated using the notBoxPlot MATLAB function (Campbell, 2020). A two-tailed, unpaired t test with unequal variance was performed to determine that the rate of SR complex formation by Flag-Avi-XLF/His10-XLFΔtailΔKBM is significantly slower than Flag-Avi-XLF/His10-XLF (**p=0.0092) but significantly faster than dimers of His10-XLFΔtailΔKBM (*p=0.0138). (B, C, D) Example trajectories from the smFRET SR Complex formation assay for the ΔXLF + H10-XLFΔTailΔKBM, ΔXLF + Flag-Avi-XLF/H10-XLFΔTailΔKBM, and ΔXLF + Flag-Avi-XLF/H10 XLF conditions, respectively, are shown. Donor and acceptor fluorescence intensity are shown in green and red, respectively. The corresponding FRET efficiency from each trace is shown in blue in a separate trajectory below.
-
Figure 4—figure supplement 3—source data 1
Source data for plot shown in Figure 4—figure supplement 3A.
- https://cdn.elifesciences.org/articles/61920/elife-61920-fig4-figsupp3-data1-v2.xls
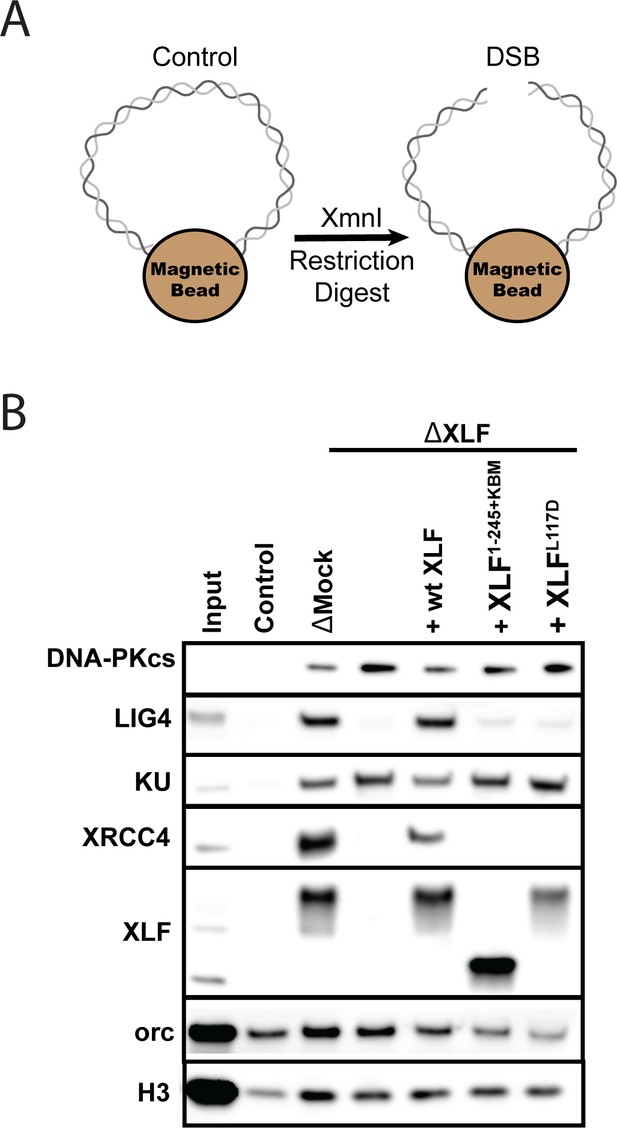
The XLF tail is required to stabilize XRCC4-Lig4.
(A) Cartoon schematic of the DNA pulldown assay. Both ends of a linear DNA substrate are conjugated to magnetic beads, and either cut with XmnI to generate a DSB with blunt ends or left uncut as a control. (B) Immunoblots of NHEJ core factors (DNA-PKcs, Lig4, Ku, XRCC4, and XLF) and the loading controls, Orc and H3, bound to DNA-beads after a 15-min incubation in egg extract. Samples run in parallel were the input (extract diluted 1:40) and control (uncut DNA substrate pulldown) as well as pulldowns with the DSB substrate in either mock-depleted extract or XLF-depleted extract with recombinant wt XLF, XLF1-245+KBM, or XLFL117D added back at 20 nM. The lower band observed in the XLF input sample is an unidentified cross-reactive species (see Figure 1—figure supplement 1B) that is not pulled down under the control or the DSB condition.
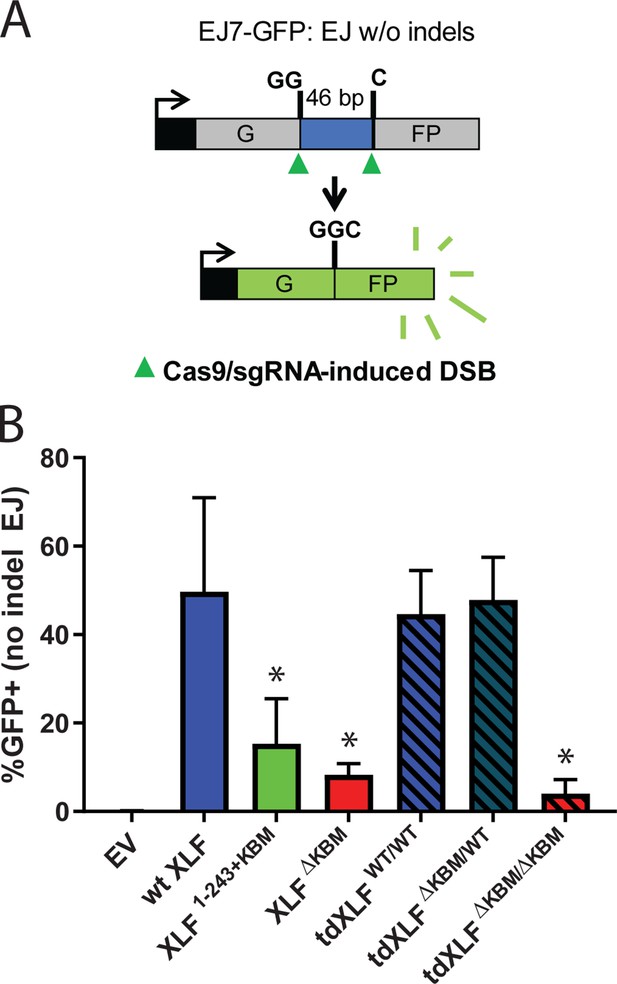
A single XLF tail is required for end joining in cells.
(A) Schematic of the cellular GFP NHEJ reporter (EJ7-GFP). A 46 bp insertion is located within a region of the GFP gene that is critical for fluorescence. Cas9 and guide RNAs are expressed so that DSBs are induced on either end of this insertion. Fluorescence is restored only if the blunt ends of the GFP gene are repaired via error-free end joining. (B) Xlf-/- mESCs were transfected with an empty vector or the same vector containing a human XLF construct. The GFP frequencies were normalized against parallel transfections of a GFP+ expression vector. The normalized mean %GFP+ and corresponding standard deviation for each condition are shown. N=6 for each condition, and an unpaired T-Test with the Holm-Sidak correction was used to determine significance. An * represents datasets that are significantly different (p≤0.015) from the wt XLF results (XLF1-243+KBMp=0.015, XLFΔKBMp=0.003, tdXLFWT/WTp=0.085, tdXLFΔKBM/WTp=0.085, tdXLFΔKBM/ΔKBMp=0.002).
-
Figure 6—source data 1
Source data for graph shown in Figure 6B.
- https://cdn.elifesciences.org/articles/61920/elife-61920-fig6-data1-v2.xlsx
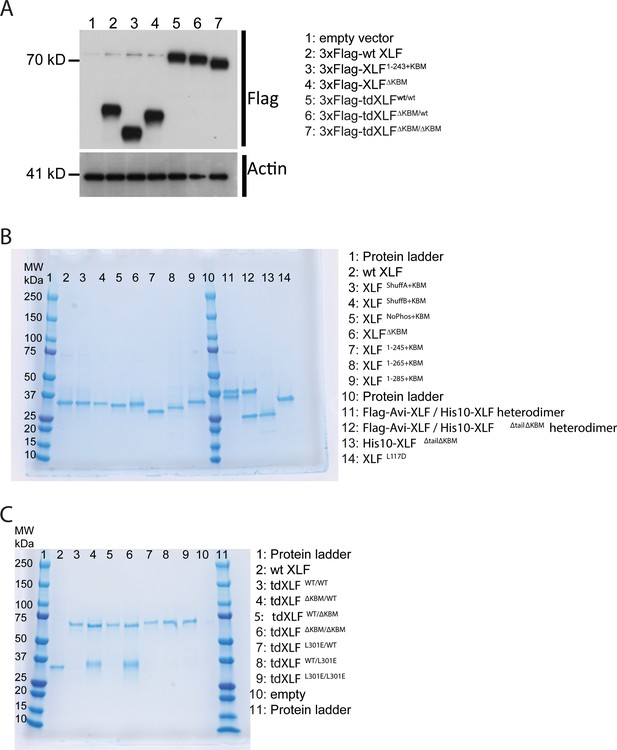
XLF constructs.
(A) Expression of XLF constructs in mESCs after transfection with the indicated constructs within the pCAGGS-BSKX vector. Samples of extract from each transfection were blotted with both anti-Flag and anti-Actin antibodies. (B) XLF mutants and heterodimer constructs. For each construct, 1500 ng was loaded on a 4–15% Mini-PROTEAN TGX Precast Protein Gel (BioRad). For each heterodimer, 3 µg construct was loaded so that individual bands should be of the same intensity as the other XLF constructs. (C) Tandem dimer XLF constructs. 750 ng of wtXLF and each tandem dimer construct were loaded and run on a 4–15% Mini-PROTEAN TGX Precast Protein Gel (BioRad). tdXLF constructs where the KBM of subunit 1 was replaced with additional linker sequence (lane four tdXLFΔKBM/WT and lane six tdXLFΔKBM/ΔKBM ) purified with a contaminants that run at ~ 35 kDa on these PAGE gels. These contaminant bands were sent for mass spectrometry (Taplin Mass Spectrometry Core Facility, Harvard Medical School). Although peptides corresponding to Xenopus XLF were detected in both cases, the majority of peptides identified were from endogenous E. coli proteins, with the bacterial lipid synthesis protein, lpxD, being responsible for 72% and 41% of peptides identified from the contaminant bands within the tdXLFΔKBM/WT and tdXLFΔKBM/ΔKBM samples, respectively.
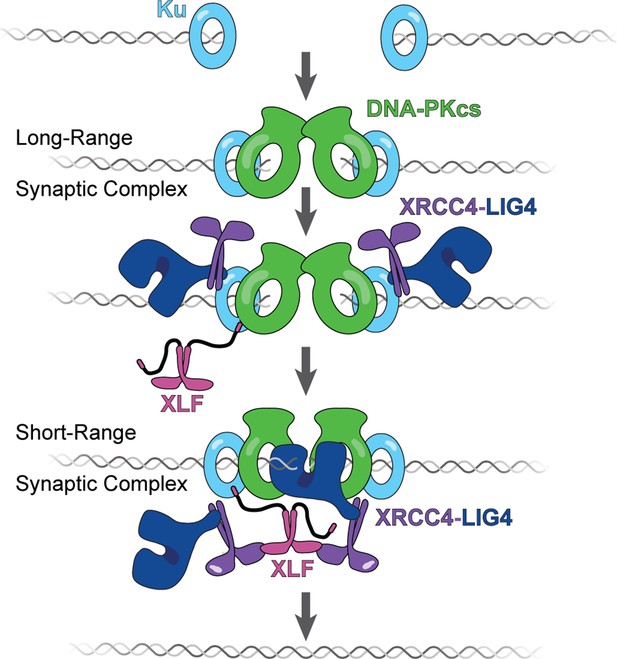
The tail enables XLF to stabilize XRCC4-Lig4 in the short-range (SR) complex.
Cartoon model representing the evolution of the NHEJ synaptic complex. Ku initially binds DNA ends and DNA-PKcs is recruited shortly after to mediate formation of the long-range (LR) complex. The XLF KBM tethers it to Ku while the other domains of XLF can diffuse locally to find and bind XRCC4. This mediates the formation of the XRCC4-XLF-XRCC4 bridge that leads to SR complex formation and likely puts Lig4 in position to engage the DNA ends.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Antibody | Anti-XLF (rabbit polyclonal) | New England Peptide, Graham et al., 2016 DOI: 10.1016/j.molcel.2016.02.010 | Loparo Lab NEP 2993 | See Materials and methods, Immunodepletion. (1:500) |
| Recombinant DNA reagent | His10-SUMO-xl XLF | Graham et al., 2016 DOI: 10.1016/j.molcel.2016.02.010 | Loparo Lab pTG296 | See Materials and methods, Plasmid Construction, XLF and XLF Truncation Mutants |
| Recombinant DNA reagent | His10-SUMO-xl XLFΔKBM | This work | Loparo Lab pSC49 | See Materials and methods, Plasmid Construction, XLF and XLF Truncation Mutants |
| Recombinant DNA reagent | His10-SUMO-xl XLF1-245+KBM | This work | Loparo Lab pSC26 | See Materials and methods, Plasmid Construction, XLF and XLF Truncation Mutants |
| Recombinant DNA reagent | His10-SUMO-xl XLF1-265+KBM | This work | Loparo Lab pSC32 | See Materials and methods, Plasmid Construction, XLF and XLF Truncation Mutants |
| Recombinant DNA reagent | His10-SUMO-xl XLF1-285+KBM | This work | Loparo Lab pSC25 | See Materials and methods, Plasmid Construction, XLF and XLF Truncation Mutants |
| Recombinant DNA reagent | His10-SUMO-xl XLFNoPhos+KBM | This work | Loparo Lab pSC48 | See Materials and methods, Plasmid Construction, XLF NoPhos and Shuffled Tail Mutants |
| Recombinant DNA reagent | His10-SUMO-xl XLFShuffB+KBM | This work | Loparo Lab pSC41 | See Materials and methods, Plasmid Construction, XLF NoPhos and Shuffled Tail Mutants |
| Recombinant DNA reagent | His10-SUMO-xl XLFShuffA+KBM | This work | Loparo Lab pSC40 | See Materials and methods, Plasmid Construction, XLF NoPhos and Shuffled Tail Mutants |
| Recombinant DNA reagent | His10-SUMO-xl XLFWT/WT | Graham et al., 2018 DOI:10.1038/s41594-018-0120-y | Loparo Lab pTG454 | See Materials and methods, Plasmid Construction, tdXLF constructs |
| Recombinant DNA reagent | His10-SUMO-xl tdXLFΔKBM/WT | This work | Loparo Lab pSC64 | See Materials and methods, Plasmid Construction, tdXLF constructs |
| Recombinant DNA reagent | His10-SUMO-xl tdXLFWT/ΔKBM | This work | Loparo Lab pSC63 | See Materials and methods, Plasmid Construction, tdXLF constructs |
| Recombinant DNA reagent | His10-SUMO-xl tdXLFΔKBM/ΔKBM | This work | Loparo Lab pSC76 | See Materials and methods, Plasmid Construction, tdXLF constructs |
| Recombinant DNA reagent | His10-SUMO-xl tdXLFL301E/WT | This work | Loparo Lab pSC65 | See Materials and methods, Plasmid Construction, tdXLF constructs |
| Recombinant DNA reagent | His10-SUMO-xl tdXLFWT/L301E | This work | Loparo Lab pSC66 | See Materials and methods, Plasmid Construction, tdXLF constructs |
| Recombinant DNA reagent | His10-SUMO-xl tdXLFL301E/L301E | This work | Loparo Lab pSC68 | See Materials and methods, Plasmid Construction, tdXLF constructs |
| Recombinant DNA reagent | His10-xl XLF: Flag-Avi-xl XLF | Graham et al., 2018 DOI:10.1038/s41594-018-0120-y | Loparo Lab pTG448 | See Materials and methods, Plasmid Construction, XLF heterodimers |
| Recombinant DNA reagent | His10-xl XLFΔTailΔKBM: Flag-Avi-xl XLF | This work | Loparo Lab pSC52 | See Materials and methods, Plasmid Construction, XLF heterodimers |
| Recombinant DNA reagent | His10-xl XLFΔTailΔKBM | This work | Loparo Lab pSC79 | See Materials and methods, Plasmid Construction, XLF heterodimers |
| Recombinant DNA reagent | His10-SUMO-xl XLFL117D | Graham et al., 2018 DOI:10.1038/s41594-018-0120-y | Loparo Lab pTG339 | See Materials and methods, Plasmid Construction, XLF and XLF Truncation Mutants |
| Recombinant DNA reagent | pCAGGS-BSKX (Empty Vector) | Bhargava et al., 2018 DOI: 10.1038/s41467-018-04867-5 | Stark Lab JS74 | |
| Recombinant DNA reagent | pCAGGS-BSKX 3xFlag-hXLF | This work | Loparo Lab pSC70 | See Materials and methods, Plasmid Construction, Human XLF and tdXLF constructs |
| Recombinant DNA reagent | pCAGGS-BSKX 3xFlag-hXLF1-243-KBM | This work | Loparo Lab pSC77 | See Materials andmethods, Plasmid Construction, Human XLF and tdXLF constructs |
| Recombinant DNA reagent | pCAGGS-BSKX 3xFlag-hXLFΔKBM | This work | Loparo Lab pSC78 | See Materials and methods, Plasmid Construction, Human XLF and tdXLF constructs |
| Recombinant DNA reagent | pCAGGS-BSKX 3xFlag-tdhXLFWT/WT | This work | Loparo Lab pSC71 | See Materials and methods, Plasmid Construction, Human XLF and tdXLF constructs |
| Recombinant DNA reagent | pCAGGS-BSKX 3xFlag-tdhXLFΔKBM/WT | This work | Loparo Lab pSC72 | See Materials and methods, Plasmid Construction, Human XLF and tdXLF constructs |
| Recombinant DNA reagent | pCAGGS-BSKX 3xFlag-tdhXLFΔKBM/ΔKBM | This work | Loparo Lab pSC73 | See Materials and methods, Plasmid Construction, Human XLF and tdXLF constructs |
| Software, algorithm | Source code 1 | Graham et al., 2016; DOI: 10.1016/j.molcel.2016.02.010 Graham et al., 2018; DOI: 10.1038/s41594-018-0120-y Graham et al., 2017; DOI: 10.1016/bs.mie.2017.03.020 | Loparo Lab circFRET_roving4_drift_thumbs | See Materials and methods, Single-Molecule FRET SR Complex Formation Assay |
| Software, algorithm | Source code 2 | Graham et al., 2016; DOI: 10.1016/j.molcel.2016.02.010 Graham et al., 2018; DOI: 10.1038/s41594-018-0120-y Graham et al., 2017; DOI: 10.1016/bs.mie.2017.03.020 | Loparo Lab simple_browselocalback_thumbnails | See Materials and methods, Single-Molecule FRET SR Complex Formation Assay |
Additional files
-
Source code 1
Peak Finder Script.
- https://cdn.elifesciences.org/articles/61920/elife-61920-code1-v2.m.zip
-
Source code 2
Trajectory Analysis Script.
- https://cdn.elifesciences.org/articles/61920/elife-61920-code2-v2.m.zip
-
Supplementary file 1
Single-molecule FRET data table.
Table containing replicate number, number of molecules tracked, number of SR complex formation events detected, average SR complex formation rate, and the standard error of the mean for the rate for each experimental condition.
- https://cdn.elifesciences.org/articles/61920/elife-61920-supp1-v2.docx
-
Supplementary file 2
Dataset replicate and statistics table.
Table containing figure panel reference, type of data, replicate number, data shown, and reference to details of any statistical analysis for each data-based figure panel.
- https://cdn.elifesciences.org/articles/61920/elife-61920-supp2-v2.docx
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/61920/elife-61920-transrepform-v2.pdf





