The essential role of Dnmt1 in gametogenesis in the large milkweed bug Oncopeltus fasciatus
Figures
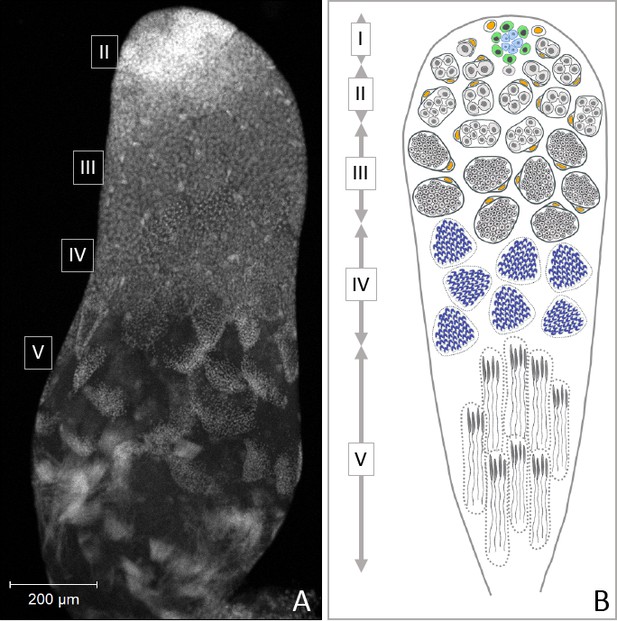
Progression of spermatogenesis in O. fasciatus.
In O. fasciatus spermatogenesis progresses from the tip of the testis tubule. (A) DAPI-stained testis tubule of stock male. (B) Diagram of the stages of spermatogenesis. At the apical tip (region I) of each of the seven testis tubules there is a ‘rosette’ of niche cells (blue) surrounded by the germline stem cells (B, green; Schmidt et al., 2002). The rosette is not typically visible in confocal images of testis tubules and markers for the rosette have not been identified yet. As spermatogonia (B, light gray) arise from division of the germline stem cells, they are enclosed by cyst cells (B, yellow). In region II, spermatogonia undergo mitotic transit amplification divisions to form spermatocysts containing 64 spermatogonia (Economopoulos and Gordon, 1971; Ewen-Campen et al., 2013). Spermatocytes (B, dark gray) in region III divide meiotically. Oncopeltus fasciatus undergoes inverted meiosis (Viera et al., 2009). Primary spermatocytes undergo the first meiotic division to produce diploid secondary spermatocytes. The meiotic division of the secondary spermatocytes produces the haploid spermatids (B, dark blue) in region IV that then differentiate into spermatozoa in the region V at the terminal end of the testis tubule.
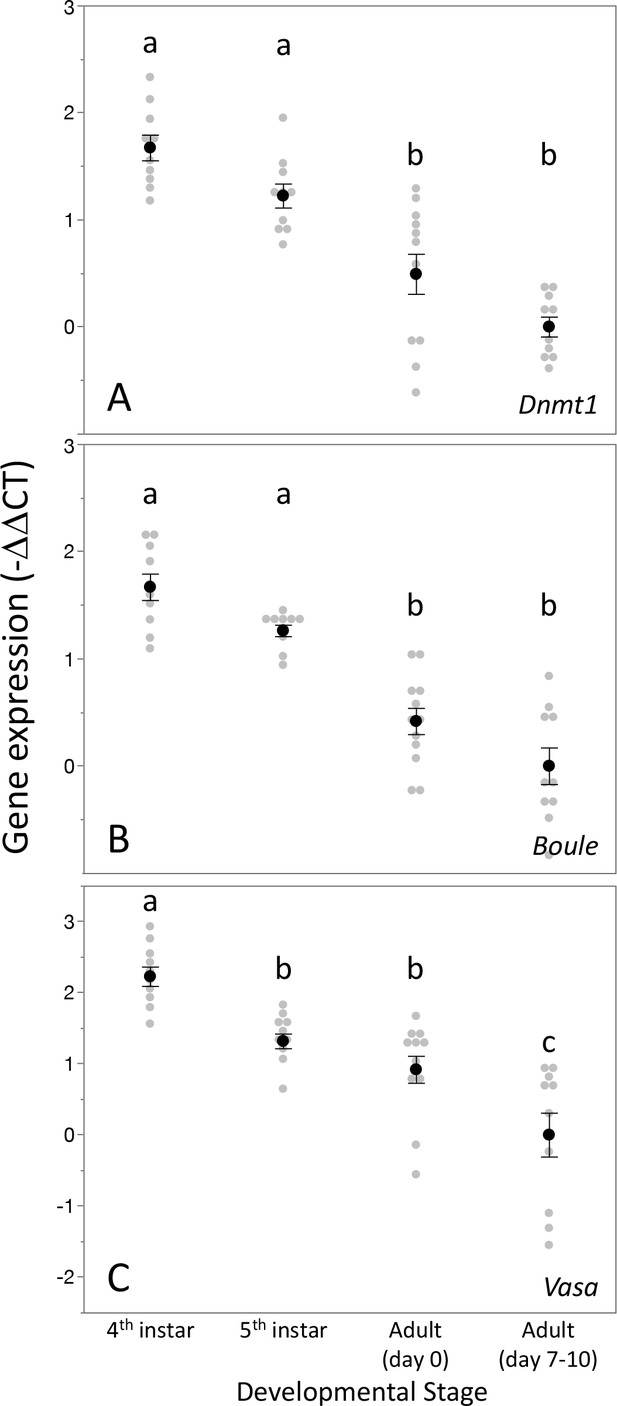
Expression pattern of Dnmt1 within the developing testes mirrored that the two gametogenesis genes Boule and Vasa.
Dnmt1 expression (A) was highest in the larval stages where spermatogenesis is initiated compared to the testes of newly emerged (day 0) and virgin sexually mature (days 7–10) adults. This is similar to the expression patterns of Boule (B), in which expression was higher in the fourth and fifth larval stages during which the transition to meiosis occurs compared to newly emerged and virgin sexually mature adults. Expression of Vasa (C) demonstrated a similar pattern, with the highest expression in the fourth instar stage and lowest in sexually mature virgin males. Black dots and bars represent mean and SE. Gray dots represent data points for each individual tested. Lowercase letters designate significant differences (p<0.05) among pairwise comparisons using post hoc Tukey–Kramer HSD test.
-
Figure 2—source data 1
Expression of Dnmt1 in testes across developmental stages.
- https://cdn.elifesciences.org/articles/62202/elife-62202-fig2-data1-v2.xlsx
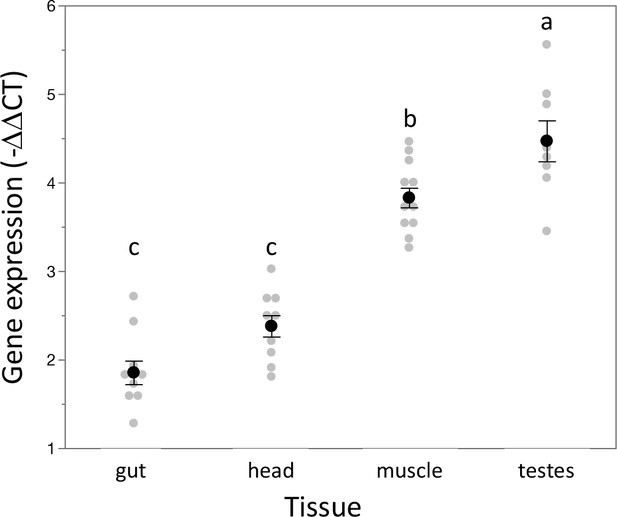
Expression of Dnmt1 is highest in reproductive tissue.
As has been seen for females (Amukamara et al., 2020), expression of Dnmt1 varied across the type of tissue in males. As in females, expression was lowest in the gut and head compared to the other somatic tissue tested, muscle. Expression is highest in the gonad (testes). Lowercase letters designate significant differences (p<0.02) among pairwise comparisons using post hoc Tukey–Kramer HSD test.
-
Figure 2—figure supplement 1—source data 1
Expression of Dnmt1 across tissues in males.
- https://cdn.elifesciences.org/articles/62202/elife-62202-fig2-figsupp1-data1-v2.xlsx
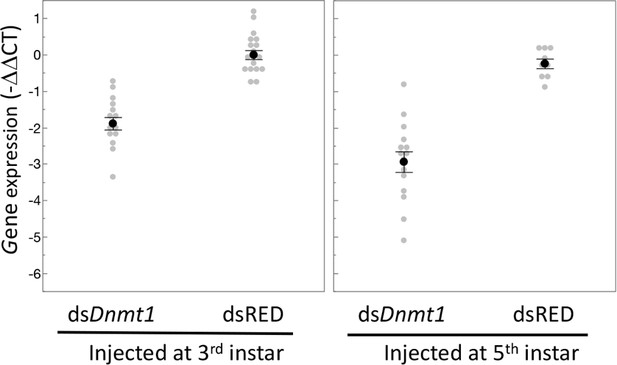
Expression of Dnmt1 was reduced in the testes of adult males treated with ds-Dnmt1 at both stages of development.
Relative gene expression is standardized to expression levels in control treatments. Black dots and bars represent mean and SE. Gray dots represent data points for each individual tested.
-
Figure 3—source data 1
Dnmt1 expression is knocked down following ds-Dnmt1 treatment.
- https://cdn.elifesciences.org/articles/62202/elife-62202-fig3-data1-v2.xlsx
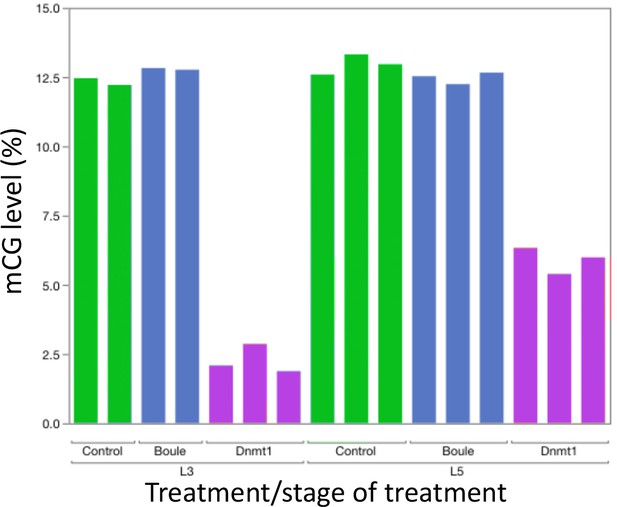
Knockdown of Dnmt1 (magenta bars), but not Boule (blue bars), reduced DNA methylation compared to controls (green bars).
Each bar represents a single individual. Across all individuals, the percent methyl CpG is reduced in the DNA isolated from the adult testis of males treated with ds-Dnmt1. Earlier injection reduces the percent methylation to a greater extent, consistent with more rounds of cell division within the testes between injection and dissection.
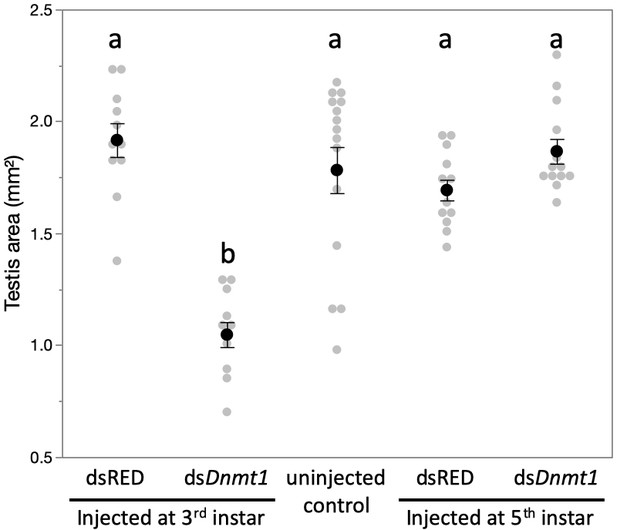
Downregulation of Dnmt1 during the third instar stage of development significantly reduced the size of the testis in sexually mature males.
There was no effect of downregulation of Dnmt1 on testis size when treatment occurred during the fifth instar stage of development after meiosis has been initiated. Black dots and bars represent mean and SE. Gray dots represent data points for each individual tested.
-
Figure 5—source data 1
Testis area from adult males treated with ds-Dnmt1 at L3 or L5 stage of development.
- https://cdn.elifesciences.org/articles/62202/elife-62202-fig5-data1-v2.xlsx
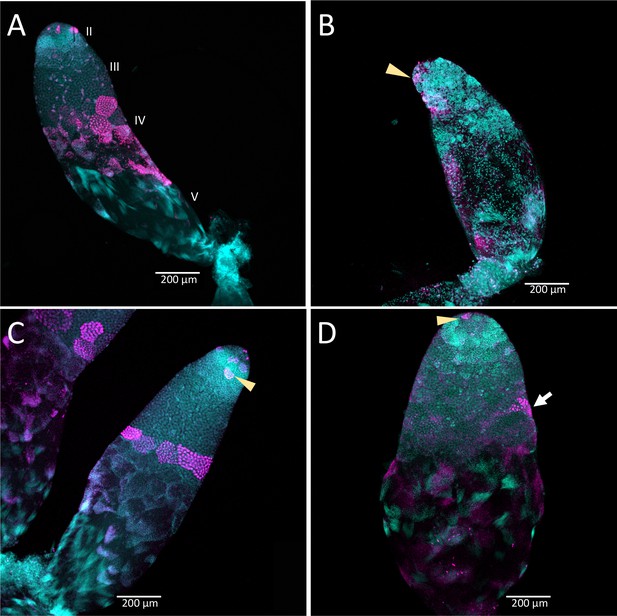
Timing of treatment during development determined the effect on testis structure of adults that develop from Dnmt1 (Panels B and D) knockdown males.
In control testis tubules from males injected with ds-RED (A and C), spermatogonia divided mitotically in region II to form spermatocysts as is observed in untreated males. Mitotic spermatogonia are labeled with anti-phosphohistone three antibodies in region II of the testis tubule (C, arrowhead). Meiotic divisions also occurred as in untreated males in ds-RED injected males. In our ds-RED control samples, a band of synchronously dividing spermatocysts, identified by positive anti-phosphohistone H3 antibody staining, was present at the interface between primary and secondary spermatocytes in testis region III (A and C). In testes from males with Dnmt1 knockdown in the 3rd instar (B), the anterior tip of the testis tubule, region II, showed evidence of mitotic activity (arrowhead). However, there were fewer spermatocytes present and the spermatocysts in this region were disorganized, and there was little evidence of the band of positive anti-phosphohistone H3 stained meiotic spermatocytes in region III. The testis tubule structure from males treated with ds-Dnmt1 following meiosis at the fifth instar stage of development (D) was more similar to the controls than those treated at the third instar stage of development. In ds-Dnmt1 males treated at the fifth instar stage, there were positive anti-phosphohistone H3 stained spermatogonia in region II (arrowhead) and spermatocytes in region III (arrow). There were differences between males treated as fifth instars with ds-RED males and ds-Dnmt1, however, including spermatocysts containing cells with highly condensed nuclei that were not present in the controls and fewer spermatocysts containing spermatids or spermatozoa. All images taken at 10× magnification.
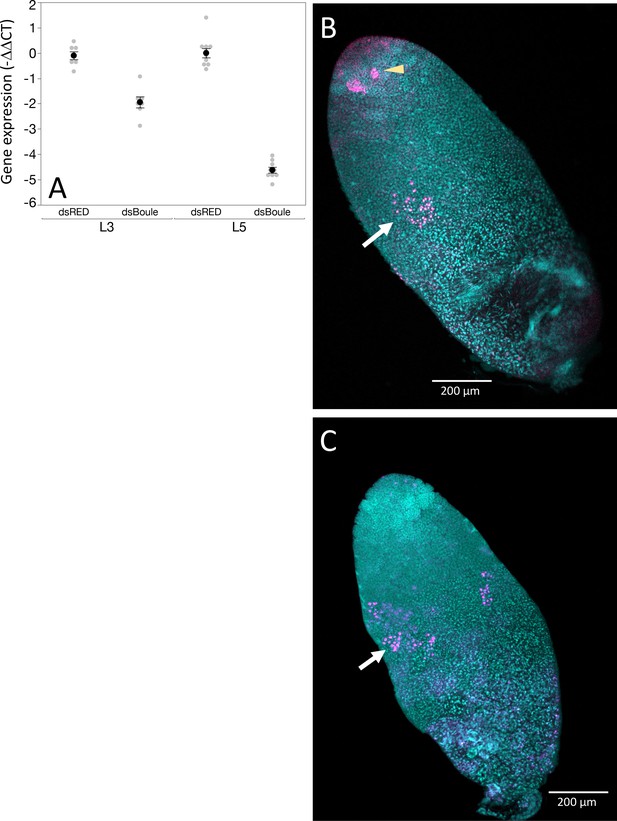
Boule knockdown with RNAi showed the expected phenotype in males.
(A) Ds-Boule RNA treatment reduced Boule expression compared to ds-RED injected males. Black dots and bars represent mean and SE. Gray dots represent data points for each individual tested. (B) While the anterior end of the testis tubule in Boule knockdown males treated at the third instar stage of development look relatively normal, with spermatogonia dividing normally (B, arrowhead), RNAi resulted in a failure to produce spermatozoa and an abnormal phenotype in region IV. While there was some evidence of anti-phosphohistone three staining in spermatocytes (B, arrow), it was reduced and did not mark a transition between spermatocytes and spermatids as is seen in control males. (C) A similar phenotype is seen in males treated with ds-Boule RNA at the fifth instar stage of development. 10× magnification.
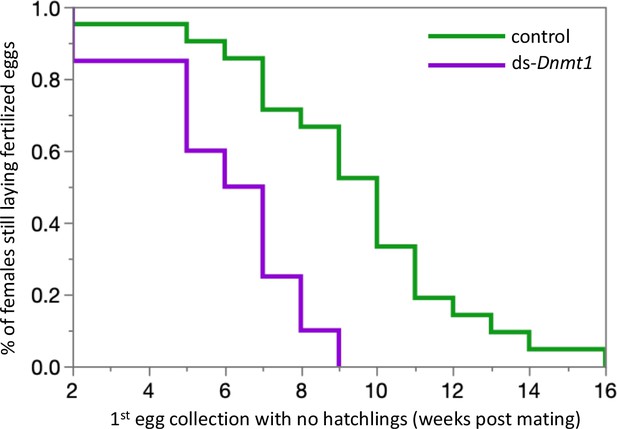
Females mated to control males lay fertilized eggs longer than females mated to Dnmt1 knockdown males.
Eggs were collected twice per week and eggs allowed to develop to hatching. Eggs that did not hatch showed no sign of development, indicating that they had not been fertilized. Both treatments demonstrate a decrease in proportion of eggs that hatch over time, but the ds-Dnmt1 treatment group shows a faster decrease.
-
Figure 7—source data 1
Fecundity data for the mates of Dnmt1 and control males.
- https://cdn.elifesciences.org/articles/62202/elife-62202-fig7-data1-v2.xlsx
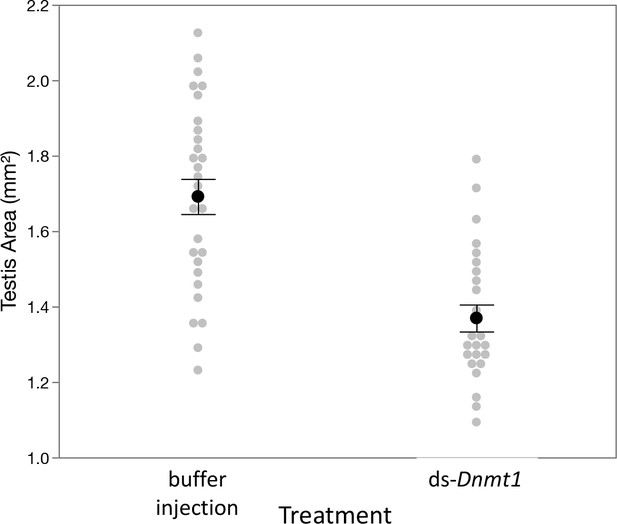
Downregulation of Dnmt1 in sexually mature males resulted in a loss of testis area following multiple matings.
Control males allowed to mate with multiple females across 3 weeks had larger testes than males treated with ds-Dnmt1 at sexual maturity. Black dots and bars represent mean and SE. Gray dots represent data points for each individual tested.
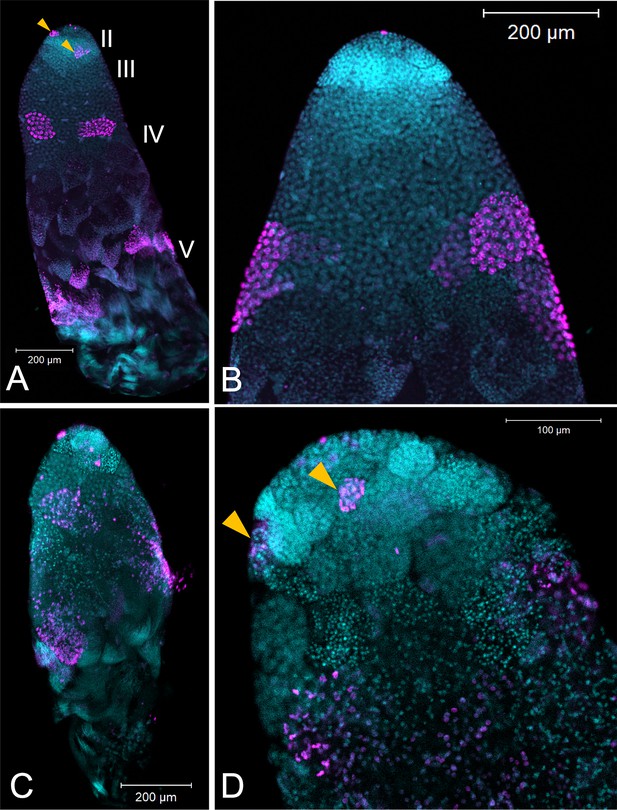
Testis structure breaks down in Dnmt1 knockdown males treated as adults after having sperm replenishment induced by mating activity.
The regions of spermatogenesis were apparent in mated males following 3 weeks of mating activity in control testis tubules (A and B) and the evidence of both mitotic division in spermatogonia in region II (B, arrowhead) and the band of meiotic divisions in region III was clear from anti-phosphohistone H3 staining. In Dnmt1 knockdown males at low magnification (C), the anterior tip, region II, of the testis tubule looked relatively normal. However, region III containing both the primary and secondary spermatocytes was disorganized. Spermatocyst structure was broken down, and the nuclei of the primary and secondary spermatocytes had lost their characteristic structure (Ewen-Campen et al., 2013). Finally, there were fewer mature spermatids in the posterior end of the testis tubule. At higher magnification (D), it was apparent that nuclear structure in the anterior tip was also affected by the knockdown, both in the spermatogonia and spermatocytes. Spermatogonia nuclei in the Dnmt1 knockdown testis tubules (D) were more condensed than in the control testis tubules (B), although they still seemed to be organized into spermatocysts. Spermatocyte nuclei, however, were fewer in number than in controls, did not have their characteristic shape (Ewen-Campen et al., 2013; Economopoulos and Gordon, 1971), and were not organized in spermatocytes. A and C:10× magnification, B and D: 20× magnification.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Strain, strain background | Oncopeltus fasciatus | Carolina Biologicals | Item # 143810 NCBI Taxon:7535 | Cultured in Moore lab for 5 years |
| Antibody | Anti-phospho-Histone H3 (Ser10) (Rabbit polyclonal) | Millipore | Cat # 06–570 RRID:AB_310177 | IF (1:1000) |
| Commercial assay or kit | Qiagen RNA easy kit with Qiazol | Qiagen | Cat # 74104 | |
| Commercial assay or kit | LightCycler 480 SYBR Green I Master | Roche | Product No. 04707516001 | |
| Commercial assay or kit | MEGAscript T7 Transcription kit | ThermoFisher Scientific | AMB13345 | |
| Sequence-based reagent | ds-Dnmt1 RNAi primer set for transcription template | Amukamara et al., 2020 | PCR primers | Sense: TGATGCTCGGCCTCAAAACAAGAT Anti-sense: ACTCCAGGAGGTGGAACAGTAGTCT |
| Sequence-based reagent | ds-Boule RNAi primer set for transcription template | Amukamara et al., 2020 | PCR primers | Sense: AGCCTCACCACCAGTATTCG Anti-sense: AGGGTGCCTAGGATTGGACT |
| Sequence-based reagent | qRT-PCR primer set: Dnmt1 | Amukamara et al., 2020 | PCR primers | Sense: GCTTGGACAAAGGCTACTACT Anti-sense: CTTCGTGGTCCCTTATCCTTATC |
| Sequence-based reagent | qRT-PCR primer set: Boule | Amukamara et al., 2020 | PCR primers | Sense: TATTCGTACCACCCTCTTCC Anti-sense: GACAATGGCTGGGTCATAAG |
| Sequence-based reagent | qRT-PCR primer set: Vasa | Amukamara et al., 2020 | PCR primers | Sense: CTGTTGCTCCTCAGGTTATT Anti-sense: CATTAAGCCTTCCAGGAGTAG |
| Sequence-based reagent | qRT-PCR primer set: actin | Amukamara et al., 2020 | PCR primers | Sense: CTGTCTCCCGAAAGAGAATATG Anti-sense: TCTGTATGGATTGGAGGATCTA |
| Sequence-based reagent | qRT-PCR primer set: GAPDH | Amukamara et al., 2020 | PCR primers | Sense: ACGGTTTCAAGGAGAAGTTAG Anti-sense: AGCTGATGGTGCAGTTATG |
| Software, algorithm | JMP Pro | SAS Institute | Version 14.1 RRID:SCR_014242 | |
| Other | DAPI stain | Invitrogen | D1306 | 0.5 µg/mL |





