Biological constraints on GWAS SNPs at suggestive significance thresholds reveal additional BMI loci
Figures
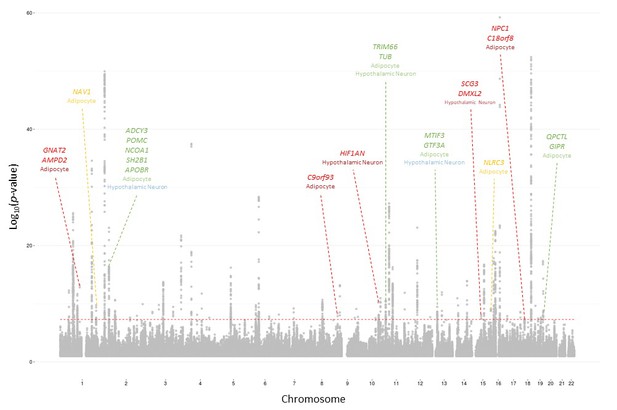
2015 BMI Manhattan plot depicting loci identified with 2010 salvaged SNPs 2015 BMI loci identifiable with 2010 salvaged SNPs.
Cell type where locus was identified indicated below locus name. Color indicates the p-value threshold where the locus became implicated (Locke et al., 2015). Color key: Green – 5×10−8≤p<5×10−7, blue – 5×10−7≤p<5×10−6, orange – 5×10−6≤p<5×10−5, red – 5×10−5≤p<5×10−4.
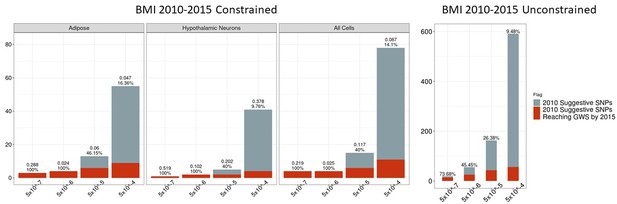
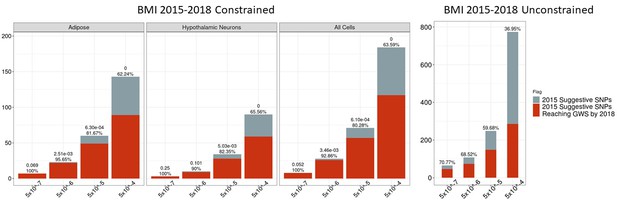
Independent 2010 BMI SNPs identified via variant-to-gene mapping that go on to reach genome-wide significance by 2015, as well as the set of unconstrained 2010 suggestive SNPs that achieve genome-wide significance by 2015.
Positive predictive value is depicted as a percentage for each bar. Above these percentages, the p-value, as identified through Fisher’s exact test, is posted. These p-values depict the probability that the proportions of salvaged SNPs using variant-to-gene mapping differ from simply salvaging all suggestive SNPs within the same suggestive bin.
-
Figure 2—source data 1
Number of 2010 loci identified by constrained method and the number that achieved GWS by 2015 in each cell type.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig2-data1-v2.csv
-
Figure 2—source data 2
Number of 2010 loci identified with no constraint and the number that achieved GWS by 2015.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig2-data2-v2.csv
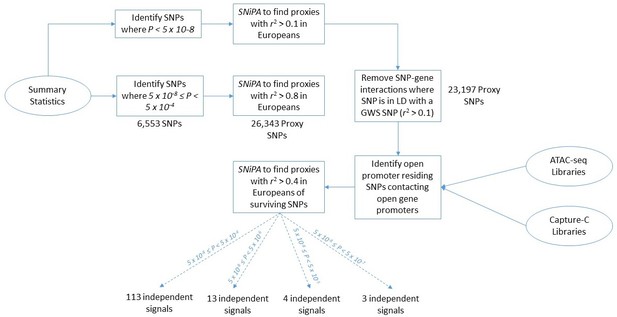
Flowchart of the pipeline describing each computational step.
BMI 2010–2015 data is utilized here as an example to report the number of SNPs and loci that occur at each step of the analysis.
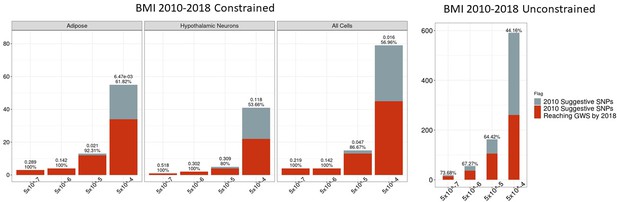
Independent 2010.
BMI SNPs salvaged via variant-to-gene mapping that go on to reach genome-wide significance by 2018, as well as the set of unconstrained 2010 suggestive SNPs that achieve genome-wide significance by 2018. Positive predictive value is depicted for each bar. Above these percentages, the p-value, as identified through Fisher’s exact test, is posted. These p-values depict the probability that the proportions of salvaged SNPs using variant-to-gene mapping differ from simply salvaging all suggestive SNPs within the same suggestive bin.
-
Figure 4—source data 1
Number of 2010 loci identified by constrained method and the number that achieved GWS by 2018 in each cell type.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig4-data1-v2.csv
-
Figure 4—source data 2
Number of 2010 loci identified with no constraint and the number that achieved GWS by 2018.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig4-data2-v2.csv
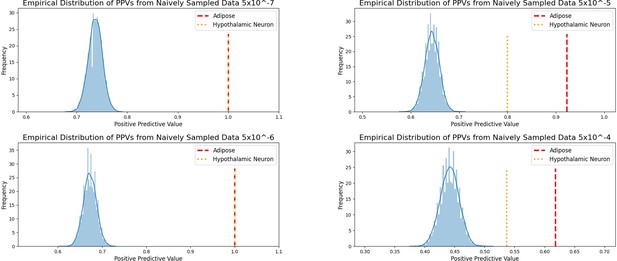
Empirical distribution of positive predictive values of suggestive 2010 BMI SNPs achieving GWS by 2018.
Independent 2015.
BMI SNPs salvaged via variant-to-gene mapping that go on to reach genome-wide significance by 2018, as well as the set of unconstrained 2015 suggestive SNPs that achieve genome-wide significance by 2018. Positive predictive value is depicted for each bar. The posterior probability that loci identified by our chromatin-based constraint more often achieve GWS than loci with no constraint is posted above these percentages.
-
Figure 5—source data 1
Number of 2015 loci identified by constrained method and the number that achieved GWS by 2018 in each cell type.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig5-data1-v2.csv
-
Figure 5—source data 2
Number of 2015 loci identified with no constraint and the number that achieved GWS by 2018.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig5-data2-v2.csv
Independent 2010.
WHRadjBMI SNPs salvaged via variant-to-gene mapping that go on to reach genome-wide significance by 2018, as well as the set of unconstrained 2010 suggestive SNPs that achieve genome-wide significance by 2018. Positive predictive value is depicted for each bar. Above these percentages, the p-value, as identified through Fisher’s exact test, is posted. These p-values depict the probability that the proportions of salvaged SNPs using variant-to-gene mapping differ from simply salvaging all suggestive SNPs within the same suggestive bin.
-
Figure 6—source data 1
Number of 2010 loci identified by constrained method and the number that achieved GWS by 2018 in each cell type.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig6-data1-v2.csv
-
Figure 6—source data 2
Number of 2010 loci identified with no constraint and the number that achieved GWS by 2018.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig6-data2-v2.csv
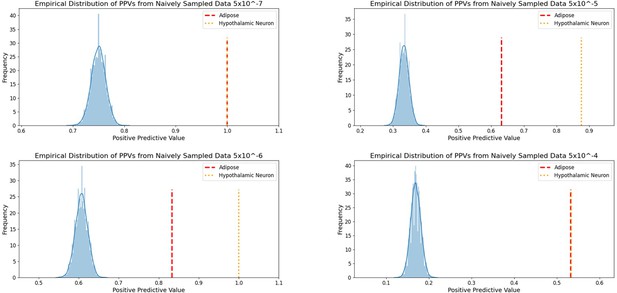
Empirical distribution of positive predictive values of suggestive 2010 WHRadjBMI SNPs achieving GWS by 2018.
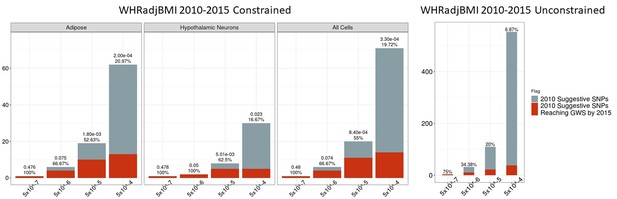
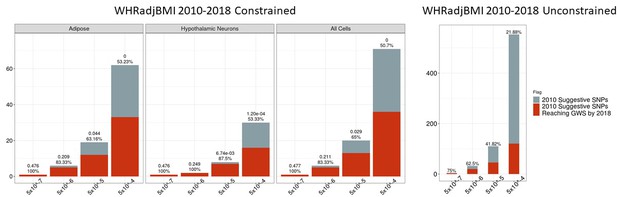
Independent 2010.
WHRadjBMI SNPs salvaged via variant-to-gene mapping that go on to reach genome-wide significance by 2015, as well as the set of unconstrained 2010 suggestive SNPs that achieve genome-wide significance by 2015. Positive predictive value is depicted for each bar. The posterior probability that loci identified by our chromatin-based constraint more often achieve GWS than loci with no constraint is posted above these percentages.
-
Figure 6—figure supplement 2—source data 1
Number of 2010 loci identified by constrained method and the number that achieved GWS by 2015 in each cell type.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig6-figsupp2-data1-v2.csv
-
Figure 6—figure supplement 2—source data 2
Number of 2010 loci identified with no constraint and the number that achieved GWS by 2015.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig6-figsupp2-data2-v2.csv
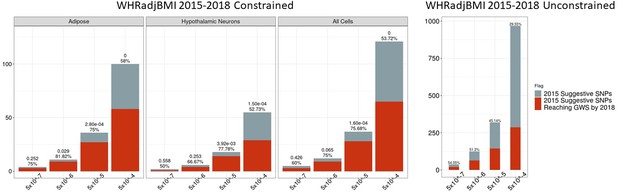
Independent 2015.
WHRadjBMI SNPs salvaged via variant-to-gene mapping that go on to reach genome-wide significance by 2018, as well as the set of unconstrained 2015 suggestive SNPs that achieve genome-wide significance by 2018. Positive predictive value is depicted for each bar. The posterior probability that loci identified by our chromatin-based constraint more often achieve GWS than loci with no constraint is posted above these percentages.
-
Figure 6—figure supplement 3—source data 1
Number of 2015 loci identified by constrained method and the number that achieved GWS by 2018 in each cell type.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig6-figsupp3-data1-v2.csv
-
Figure 6—figure supplement 3—source data 2
Number of 2015 loci identified with no constraint and the number that achieved GWS by 2018.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig6-figsupp3-data2-v2.csv
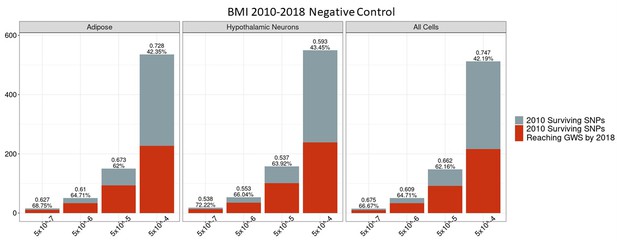
Independent 2010.
BMI SNPs failing the variant-to-gene mapping filter that go on to reach genome-wide significance by 2018. Positive predictive value is depicted for each bar. The posterior probability that loci identified by our chromatin-based constraint more often achieve GWS than loci with no constraint is posted above these percentages. There is no threshold where this data differs significantly from the unconstrained set.
-
Figure 7—source data 1
Number of 2010 loci identified by constrained method and the number that achieved GWS by 2018 in each cell type.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig7-data1-v2.csv
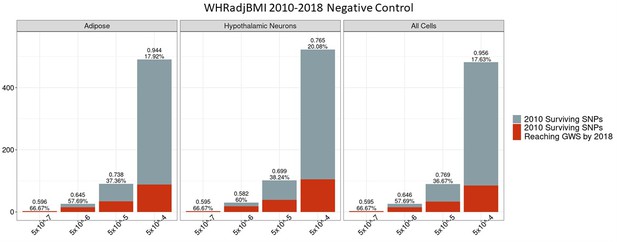
Independent 2010.
WHRadjBMI SNPs failing the variant-to-gene mapping filter that go on to reach genome-wide significance by 2018. Positive predictive value is depicted for each bar. The posterior probability that loci identified by our chromatin-based constraint more often achieve GWS than loci with no constraint is posted above these percentages. There is no threshold where this data differs significantly from the unconstrained set.
-
Figure 8—source data 1
Number of 2010 loci identified by constrained method and the number that achieved GWS by 2018 in each cell type.
- https://cdn.elifesciences.org/articles/62206/elife-62206-fig8-data1-v2.csv
Tables
2015 BMI loci that were implicated with our method in the 2010 data set.
The 2015 genome-wide significant BMI loci whose sentinel SNP was in LD with SNPs implicated from suggestive association in the 2010 BMI GWAS. Key: Notable genes from biological relevance to obesity (B); copy number variation (C); DEPICT analyses (D); GRAIL results (G); BMI-associated variant is in strong LD (r2 ≥ 0.8) with a missense variant in the indicated gene (M); gene nearest to index SNP (N); association and eQTL data converge to affect gene expression (Q) (Locke et al., 2015).
| Novel as of 2015 (Locke et al.) | ||||
|---|---|---|---|---|
| 2015 sentinel SNP | 2010 implicated SNPs | 2015 assigned locus name | Interacting gene | |
| rs4740619 | rs10810462 | C9orf93(C,M,N) | TCONS_00015651 | |
| rs17094222 | rs117597828 | HIF1AN(N) | PAX2 | |
| rs2820292 | rs12086240, rs2820315 | NAV1(N) | TIMM17A | |
| rs758747 | rs2238435 | NLRC3(N) | TCONS_00024950, TCONS_00024564, TCONS_00024949, TCONS_00024562, RP11-462G12.1, TCONS_00024568, TCONS_00024570, RP11-95P2.1, TCONS_00024567, TCONS_00024569, TCONS_00024320, TCONS_00024952 | |
| rs3736485 | rs7183479 | SCG3(B,D); DMXL2(M,N) | LYSMD2, SCG3, CTD-2308G16.1, TMOD2 | |
| Identified between 2010 (Speliotes et al.) and 2015 (Locke et al.) | ||||
| 2015 Sentinel SNP | 2010 Implicated SNPs | 2015 Assigned locus name | Interacting gene | |
| rs17024393 | rs72705210 | GNAT2(N); AMPD2(D) | GSTM3, AHCYL1 | |
| rs4256980 | rs10840079, rs10840087, rs11041999, rs11042023, rs12803166, rs4256980, | TRIM66(D,M,N); TUB(B) | PBLD, RPL27A, TRIM66 | |
| Identified in 2010 (Speliotes et al.), but was not genome-wide significant | ||||
| 2015 Sentinel SNP | 2010 Implicated SNPs | 2015 Assigned Locus Name | Interacting gene | |
| rs10182181 | rs12713419, rs13012304, rs6718510, rs7597332, rs7608976 | ADCY3(B,M,N,Q); POMC(B,G); NCOA1(B); SH2B1(B,M,Q); APOBR(M,Q); | ADCY3, TCONS_00003602 | |
| rs12016871 | rs7988412, | MTIF3(N); GTF3A(Q) *~1 Mb from sentinel | MTIF3 | |
| rs1808579 | rs1788783 | NPC1(B,G,M,Q); C18orf8(N,Q) | NPC1 | |
| rs2287019 | rs11672660, rs34783010 | QPCTL(N); GIPR(B,M) | GIPR | |
2018 BMI loci that were identified using 2010 salvaged.
SNPs 2018 BMI loci identified as genes nearest to genome-wide significant SNPs that could be identified using SNPs salvaged from suggestive regions of the 2010 BMI GWAS.
| Nearest gene to sentinel | Surviving proxy SNPs | Lowest threshold found |
|---|---|---|
| ABHD17A | rs893543, rs893542, rs11671347 | 5 × 10−4 |
| AC007879.5 | rs11677847, rs72951700, rs11689163, rs72966483, rs11694560, rs11692026, rs964621, rs964622 | 5 × 10−4 |
| ADCY3 | rs6718510, rs7597332, rs7608976, rs13012304, rs12713419 | 5 × 10−4 |
| ADCY9 | rs710893, rs2531993, rs2238435 | 5 × 10−5 |
| AK5 | rs12729914 | 5 × 10−5 |
| AP000439.5 | rs11605729 | 5 × 10−4 |
| BCL7A | rs7299842 | 5 × 10−4 |
| C10orf32 | rs7085104 | 5 × 10−4 |
| C18orf8 | rs1788826 | 5 × 10−4 |
| C1orf61 | rs11264483 | 5 × 10−4 |
| CCDC171 | rs10810462 | 5 × 10−4 |
| CNNM2 | rs1926032 | 5 × 10−4 |
| COQ4 | rs1468648 | 5 × 10−4 |
| CRTC1 | rs4808845, rs4808844 | 5 × 10−4 |
| DPYD | rs12077442 | 5 × 10−4 |
| EIF2B5 | rs3914188, rs35637422 | 5 × 10−4 |
| EXOSC10 | rs1884429, rs12041740 | 5 × 10−4 |
| FAIM2 | rs422022 | 5 × 10−4 |
| GAB2 | rs869202 | 5 × 10−4 |
| GIPR | rs34783010, rs11672660 | 5 × 10−7 |
| GPR61 | rs72705210 | 5 × 10−4 |
| HIF1AN | rs117597828 | 5 × 10−4 |
| HOXB1 | rs2326013 | 5 × 10−4 |
| IFNGR1 | rs17258750 | 5 × 10−4 |
| IPO9 | rs2820315 | 5 × 10−5 |
| KCNJ12 | rs9906072 | 5 × 10−4 |
| LMOD1 | rs2047264 | 5 × 10−4 |
| MAP2K3 | rs2001651, rs3785542 | 5 × 10−4 |
| MAP3K7CL | rs928277 | 5 × 10−4 |
| MEF2D | rs2274319, rs1925950, rs12038396, rs3818463, rs2274320, rs2274317 | 5 × 10−4 |
| MLN | rs11752353, rs6921487, rs72880511, rs1887340, rs73746509 | 5 × 10−4 |
| MLXIP | rs28530689, rs10773037, rs28737311, rs36158849, rs2280573 | 5 × 10−4 |
| MST1R | rs3774758, rs2252833, rs6446187 | 5 × 10^−4 |
| MTIF3 | rs7988412 | 5 × 10−7 |
| MTOR | rs11581010, rs10864490 | 5 × 10−4 |
| NAV1 | rs12086240 | 5 × 10−5 |
| NPC1 | rs1788783 | 5 × 10−4 |
| RASA2 | rs2042864 | 5 × 10−4 |
| RCAN2 | rs3934393 | 5 × 10−5 |
| RNU6-543P | rs10761689 | 5 × 10−4 |
| RP11-493K19.3 | rs13100903 | 5 × 10−4 |
| RP11-562L8.1 | rs12887636 | 5 × 10−5 |
| RP11-68I18.10 | rs10788800 | 5 × 10−5 |
| RP11-707P17.1 | rs7183479 | 5 × 10−4 |
| SAE1 | rs466477 | 5 × 10−4 |
| SKAP1 | rs16951519, rs2240121 | 5 × 10−5 |
| STK33 | rs10840087, rs11041999, rs34009921 | 5 × 10−4 |
| TNRC6B | rs6001834, rs4820409 | 5 × 10−4 |
| TRIM66 | rs10840079, rs4256980, rs11042023, rs12803166 | 5 × 10−6 |
| TTC34 | rs6424062 | 5 × 10−5 |
| URM1 | rs7859557, rs2240948 | 5 × 10−4 |
| XXYLT1 | rs58434965 | 5 × 10−4 |
Additional files
-
Source data 1
Classification metrics for each condition.
- https://cdn.elifesciences.org/articles/62206/elife-62206-data1-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62206/elife-62206-transrepform-v2.docx





