A pentameric protein ring with novel architecture is required for herpesviral packaging
Figures
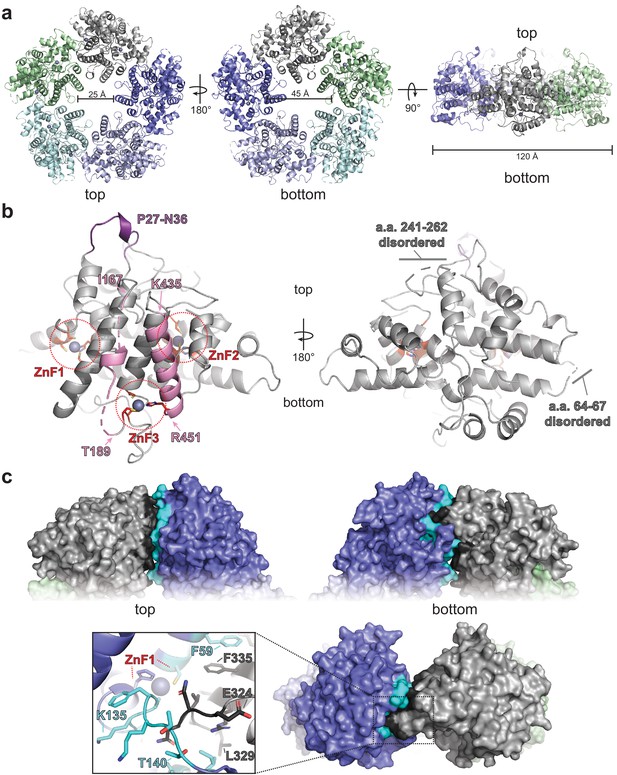
ORF68 forms a homopentameric ring.
(a) View from the top, bottom, and side of the ORF68 crystal structure. The central channel size and overall diameter are highlighted. (b) Each monomer of ORF68 contains three zinc fingers (ZnF; coordinating residues shown in red sticks); Zn+2 shown in gray spheres. Residues 167–188 and 435–451 (pink) span the central channel. Residues 435–451 form an α-helix, whereas residues 177–188 are largely disordered (highlighted in pink). Both regions are anchored by ZnF3. The ‘top’ of the ring has a semi-structured loop consisting of residues P27-N36 (purple). Residues 241–262 and 64–67 are disordered. (c) Subunit interface within the ORF68 pentamer, with the monomer–monomer interface of the gray monomer highlighted in black and the interface on the blue monomer highlighted in cyan. ZnF1 is near the interface; a loop consisting of residues E324-L329 extends into the adjacent monomer. Residues F59 and F335 from adjacent monomers stack.
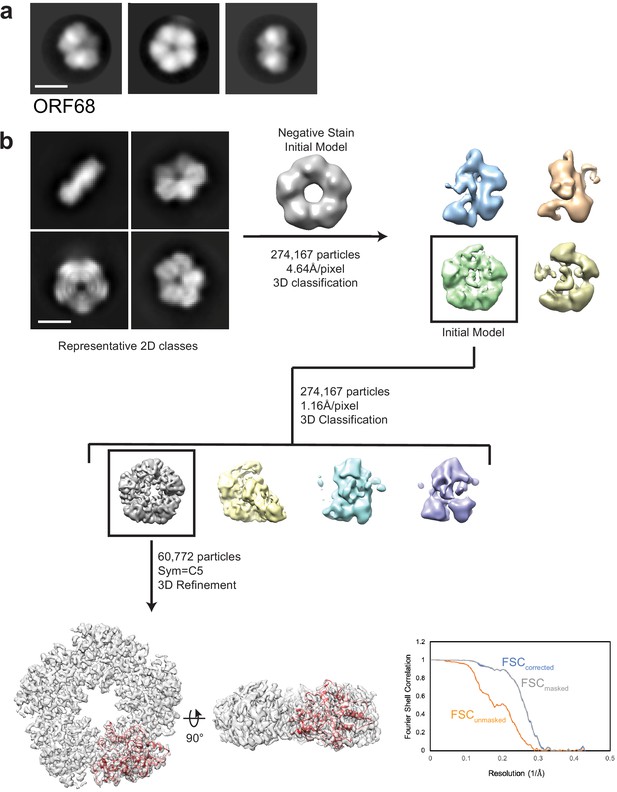
EM data analysis for ORF68.
(a) Representative negative stain 2D class averages of ORF68. Scale bar = 100 Å. (b) Representative 2D class averages and data processing workflow for single-particle cryo-EM analyses of ORF68. The best class was refined to an estimated 3.37 Å resolution, as indicated by the depicted Fourier Shell Correlation (FSC) plot. Scale bar = 100 Å.
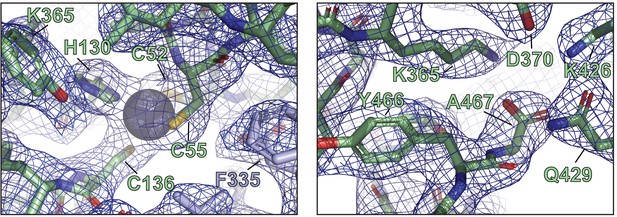
Representative electron densities and X-ray structural details of ORF68.
Representative local 2mFo-DFc maps are contoured at 1σ.
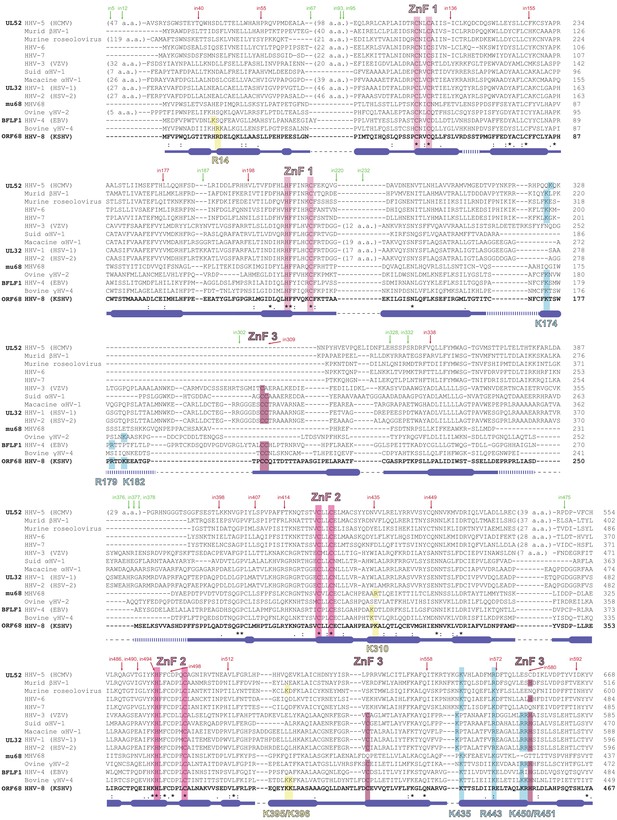
Sequence alignment of homologs of ORF68.
Homologs of ORF68 were identified using BLAST and aligned using Clustal Omega, then manually edited to condense long insertions relative to ORF68. * indicates perfect conservation, : indicates strong conservation, and . indicates weak conservation. Secondary structure as observed in the ORF68 structure is indicated under the alignment, with thick lines for α-helices, thin lines for unstructured regions or loops, and dotted lines for disordered regions of the model. Thin dotted black lines indicate an insertion in the alignment relative to ORF68. The residues involved in the three zinc finger (ZnF) motifs are highlighted in shades of pink. Positively charged residues selected for mutation located in the central channel of the ring are highlighted in blue, while residues selected for mutation outside of the central channel are highlighted in yellow. A transposon screen of UL32 (the ORF68 homolog from HSV-1) was previously performed (Palmer, 2010) and revealed insertion sites (‘in#') within the protein that were either tolerant (labeled in green) or intolerant (labeled in red) of insertion of a 5-amino acid transposon, as assessed by complementation of a UL32-null virus.
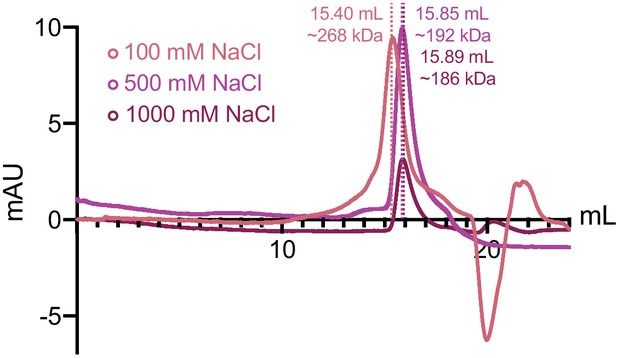
ORF68 remains oligomeric in the presence of high monovalent salt.
Size exclusion chromatography of ORF68 in buffer containing 100 mM (salmon), 500 mM (purple), or 1 M NaCl (maroon).
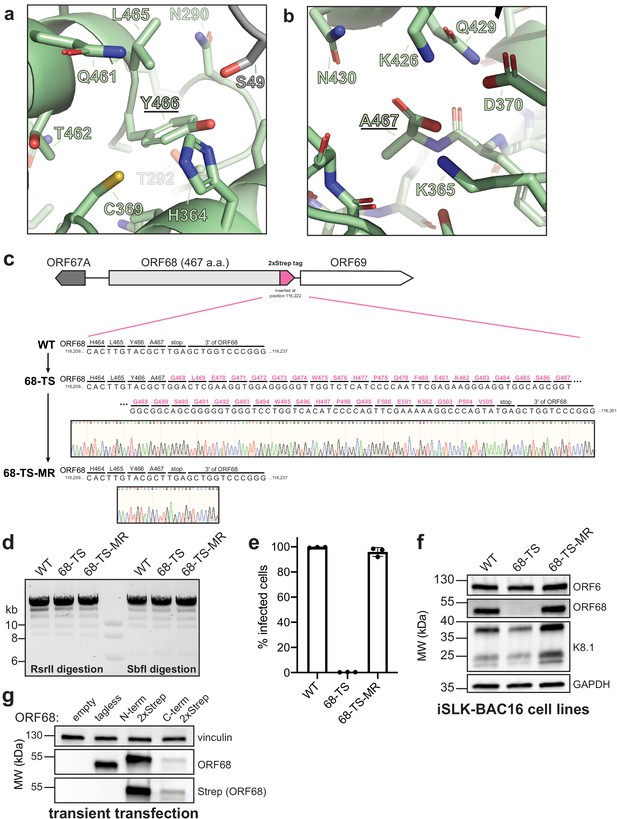
Extension of the C-terminal tail of ORF68 reduces protein levels and prevents infectious virion production.
(a) The penultimate residue of ORF68, Y466, is buried near conserved residues C369 and H364, along with S49 from a neighboring monomer. (b) The last residue of ORF68, A467, is surrounded by conserved residues, including K365, D370, K426, and Q429. (c) Schematic of the genomic locus of ORF68, with the location and sequence of the C-terminal 2xStrep (TS) tag indicated. Sanger sequencing traces for the tag (68-TS) and mutant rescue (68-TS-MR) are shown below. (d) Digestion of recombinant BACs with RsrII and SbfI was used to assess whether large-scale recombination had occurred during mutagenesis. (e) Wild-type (WT), ORF68-TS, and ORF68-TS-MR iSLK cell lines were established using the KSHV BAC16 system. Progeny virion production by these cell lines was assayed by supernatant transfer and flow cytometry of target cells. (f) Western blot of whole cell lysate (20 μg) from WT, ORF68-TS, and ORF68-TS-MR iSLK cell lines. GAPDH was used as a loading control. ORF6 and K8.1 are representative early and late genes, respectively. (g) Western blot of whole cell lysate (20 μg) from HEK293T cells that were transfected with plasmids encoding tagless, N-terminal 2xStrep-tagged, or C-terminal 2xStrep-tagged ORF68. Vinculin serves as a loading control. MR: mutant rescue.
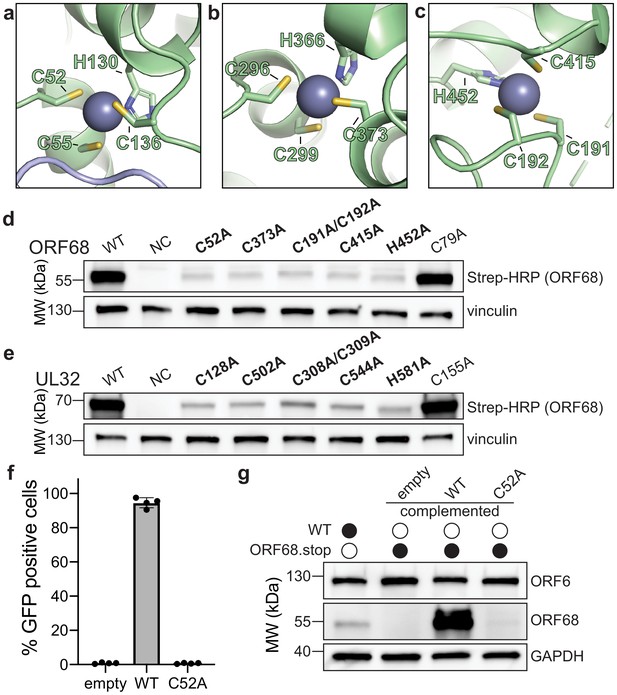
ORF68 and homologs are zinc finger-containing proteins.
(a–c) The Zn2+ ion within the three zinc finger motifs is shown as a blue-gray sphere, while coordinating cysteines and histidines are shown in sticks. (d, e) Western blot of whole cell lysate (33 μg) from HEK293T cells that were transfected with plasmids encoding wild-type or mutant variants of ORF68 (e) or UL32 (f). Vinculin serves as a loading control. (f) ORF68.stop iSLK cells were lentivirally transcomplemented with empty vector or with plasmids encoding wild-type or C52A ORF68. Progeny virion production by these cell lines was assayed by supernatant transfer and flow cytometry of target cells. (g) Western blot of transcomplemented ORF68.stop iSLK cells used in (e).
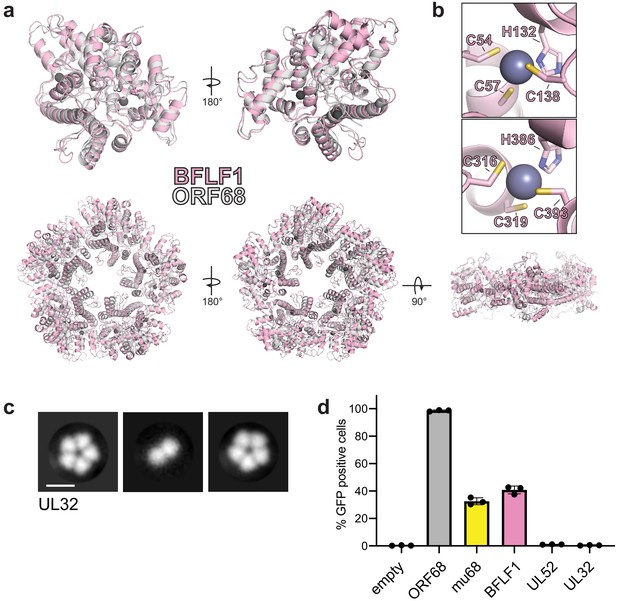
Homologs of ORF68 possess similar structures.
(a) Overlay of ORF68 (gray) and BFLF1 (pink) monomers (top) and their homopentameric complexes (bottom). (b) BFLF1 contains at least two zinc fingers, with Zn2+ ions shown as a blue-gray sphere. (c) Representative 2D class averages from negative stain EM of UL32. Scale bar = 100 Å. (d) ORF68.stop iSLK cells were lentivirally transcomplemented with plasmids encoding N-terminally Strep-tagged ORF68 or homologs from EBV (BFLF1), MHV68 (mu68), HCMV (UL52), or untagged HSV-1 (UL32). Progeny virion production by these cell lines was assayed by supernatant transfer and flow cytometry of target cells.
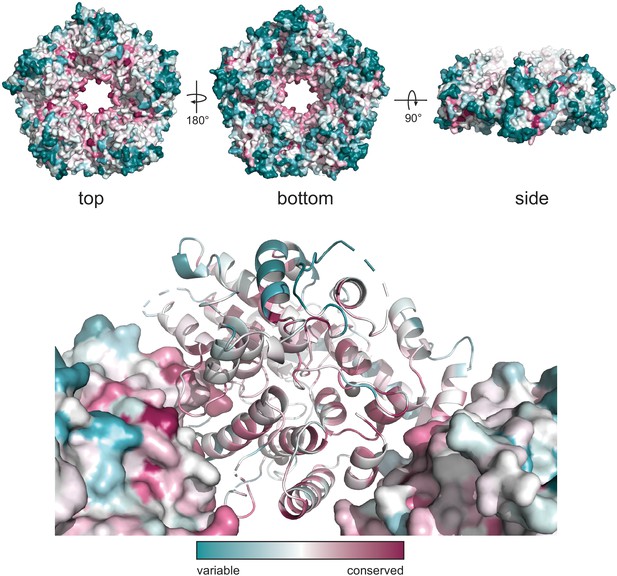
ConSurf model of ORF68.
Conservation of residues within ORF68 as calculated by the ConSurf server (Ashkenazy et al., 2016), where blue residues are variable (nonconserved) and red are conserved.
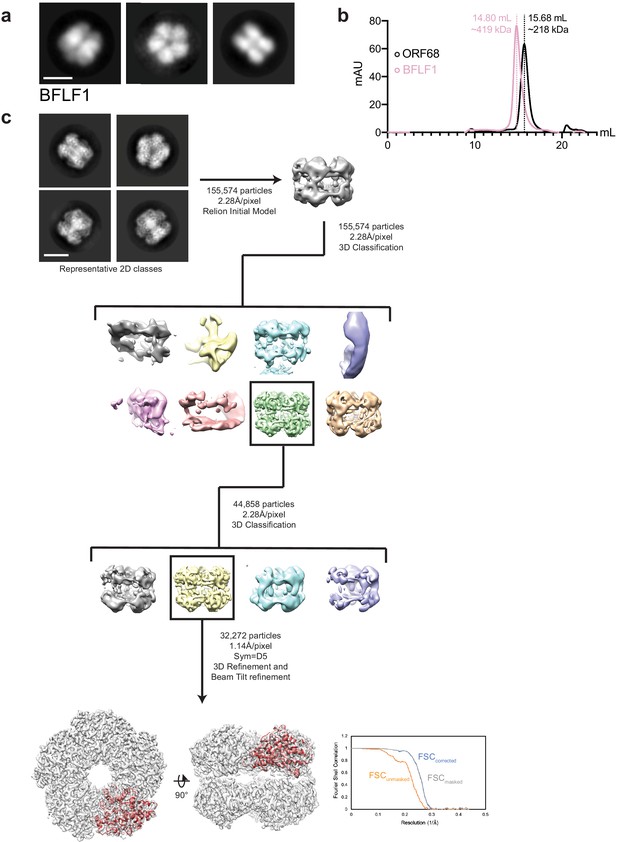
EM data analysis for BFLF1.
(a) Representative negative stain 2D class averages of BFLF1. Scale bar = 100 Å. (b) Size exclusion chromatography of BFLF1 reveals that it is likely a decamer in solution, rather than a pentamer like ORF68. (c) Representative 2D class averages and data processing workflow for single-particle cryo-EM analyses of BFLF1. The best class was refined to an estimated 3.60 Å resolution, as indicated by the Fourier Shell Correlation (FSC) plot. Scale bar = 100 Å.
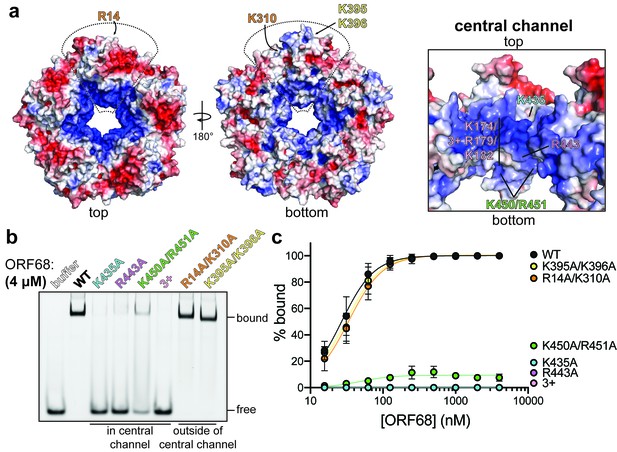
ORF68 binds nucleic acid in vitro via its central channel.
(a) Electrostatic surface of the ORF68 pentamer, contoured from +5 kT/e (blue) to −5 kT/e (red) and shown from the top (left), bottom (middle), and through the central channel (right). The electrostatic surface lacks regions that were disordered in the structure, including residues 169–172 and 179–188, which face the central channel. The locations of residues selected for mutation are indicated on one monomer of the pentamer. (b) Electrophoretic mobility shift assay using fluorescein-labeled 20 bp dsDNA probe (10 nM) and wild-type or mutant ORF68 (4 μM). (c) Binding curves for wild-type and mutant ORF68 interacting with the 20 bp dsDNA probe were determined by electrophoretic mobility shift assays as in (b). Data represent the mean ± s.d. of three independent experiments. Data were fit with a nonlinear regression to the Hill equation.
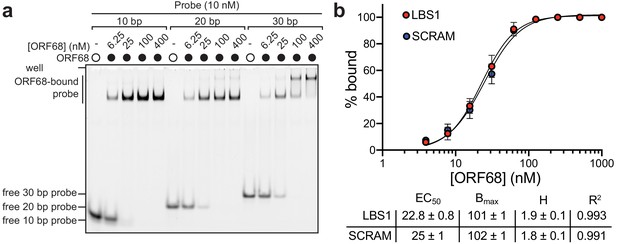
ORF68 nonspecifically binds nucleic acid.
(a) Native gel for the electrophoretic mobility shift assay of ORF68 binding to 5′-fluorescein labeled dsDNA probe with 10, 20, or 30 bp. (b) Binding curves for a 85% GC-rich probe (LBS1) and a 50% GC-rich probe (SCRAM) (top). Data represent the mean ± s.d. of three independent experiments. Data were fit with a nonlinear regression to the Hill equation, with best fit derived binding parameters within the 95% CI (bottom): EC50 (effective binding affinity), Bmax (maximum specific binding), H (Hill coefficient), and R2 (goodness of fit).
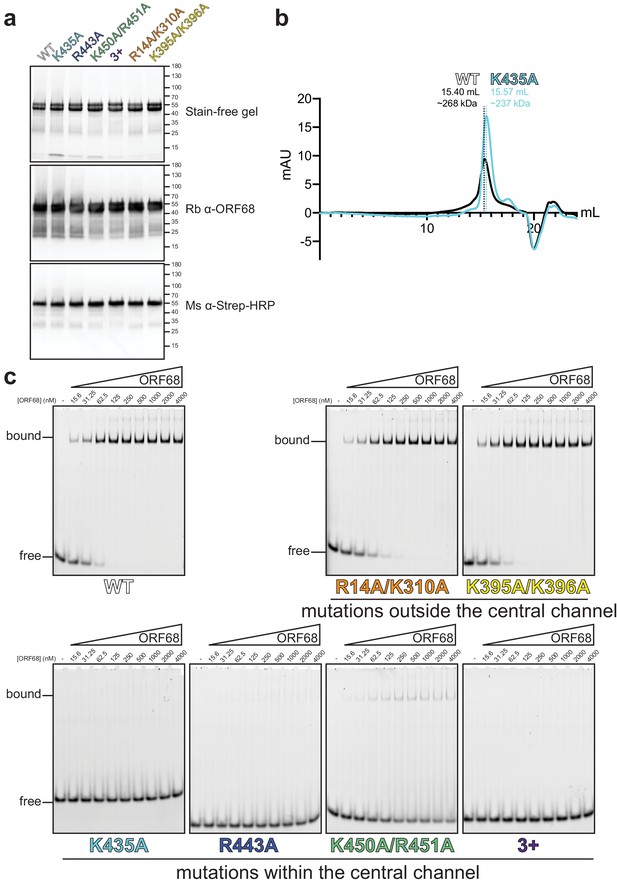
ORF68 mutants can be purified, but mutations in the central channel prevent dsDNA binding.
(a) SDS-PAGE gel of purified recombinant wild-type and mutant ORF68 variants (7.5 μg) used for dsDNA-binding assays. Proteins were visualized by stain-free imaging (top), followed by western blotting for ORF68 (middle), and the Strep tag (bottom). (b) Size exclusion chromatography of wild-type ORF68 and ORF68-K435A reveals that mutation in the central channel does not affect oligomerization. (c) Representative native gels for electrophoretic mobility shift assays with wild-type or mutant ORF68 and a 20 bp dsDNA probe.
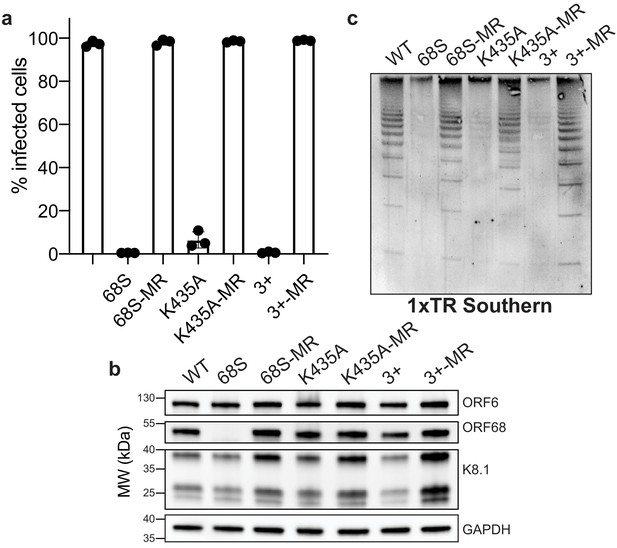
Residues in ORF68 that ablate dsDNA binding in vitro are required for genome cleavage and packaging in vivo.
(a) iSLK cell lines containing ORF68 mutants (68S, K435A, and 3+) and their corresponding mutant rescues (MR) were established using the KSHV BAC16 system. Progeny virion production by these cell lines was assayed by supernatant transfer and flow cytometry of target cells. (b) Western blot of whole cell lysate (25 μg) from ORF68.stop iSLK cell lines. GAPDH was used as a loading control. ORF6 is an early gene and K8.1 is a late gene. (c) Southern blot of DNA isolated from iSLK cell lines using a probe for the terminal repeats. DNA was digested with PstI, which cuts within the genome but not within the terminal repeats and generates a ladder of terminal repeat-containing DNA when successful cleavage and packaging occurs.
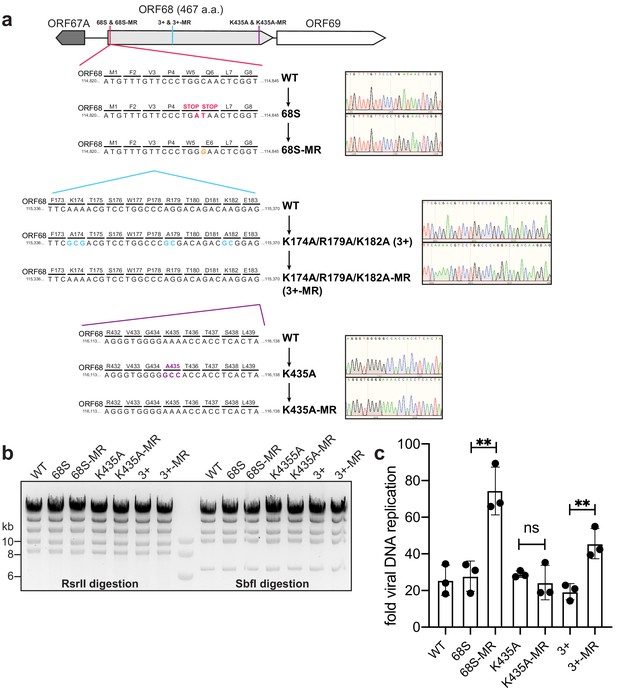
Construction and validation of mutant viruses.
(a) Schematic of the genomic locus of ORF68, with the location of introduced mutations depicted in detail below. Sanger sequencing traces for the mutants and corresponding mutant rescues are shown to the right. (b) Digestion of recombinant BACs with RsrII and SbfI was used to assess whether large-scale recombination had occurred during mutagenesis. (c) Viral DNA replication was measured by qPCR before and after reactivation. Data are from three independent biological replicates, with statistics being calculated using an unpaired t test. **p<0.01. MR, mutant rescue.
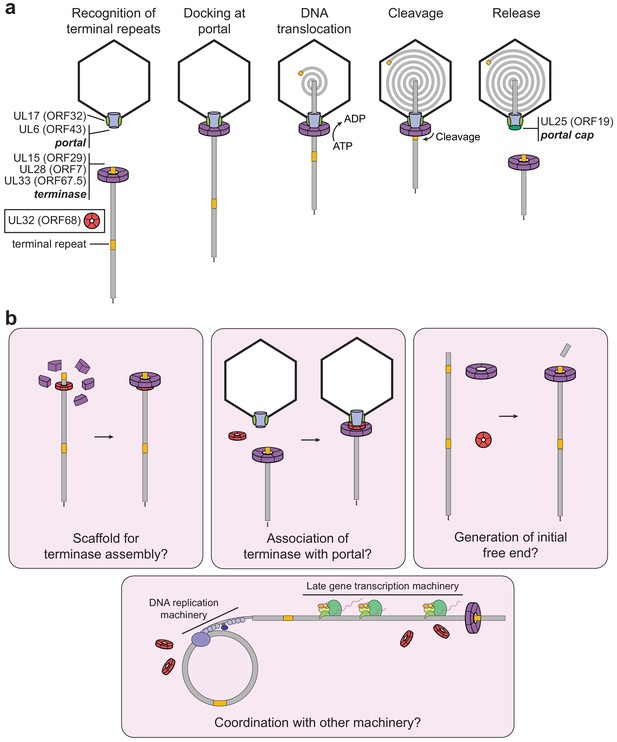
Model of herpesviral cleavage and packaging.
(a) Genes required for cleavage and packaging in HSV-1 and their homologs in KSHV (listed in brackets) are listed. UL15, UL28, and UL33 form the terminase complex that must dock with the portal protein (UL6), capsid, and capsid-associated proteins (including UL17). The terminase translocates the dsDNA genome into the capsid and cleaves within the terminal repeats once a full unit-length genome has been packaged. After release of the remaining genome, the UL25 portal cap binds to stabilize the packaged genome. The precise role of UL32 (KSHV ORF68) has not been determined. (b) Possible roles of ORF68 during packaging could include acting as a scaffold for the terminase to bind the nascent genome (left), acting as an adaptor terminase association with the portal (middle), or promoting formation of the initial free end on nascent genomes (right). Further potential roles include interfacing with the DNA replication machinery or late gene transcription machinery (bottom).
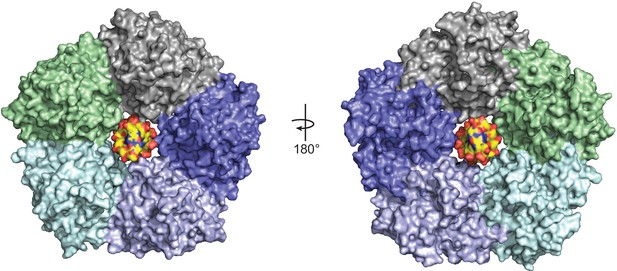
Model of DNA binding in the central channel of ORF68.
Idealized B-form DNA (PDB 1BNA) (yellow) manually docked into the central channel of ORF68. Minimal clashes with residues in the two helices facing the central channel, particularly K174, and K435, T436, D440, and R443, suggest that some conformational changes may be required to accommodate DNA spanning the central channel.
Tables
| Reagent type (species) or resource | Designation | Source or reference | Identifiers | Additional information |
|---|---|---|---|---|
| Gene (Kaposi’s sarcoma-associated herpesvirus) | ORF68 | Human gammaherpesvirus 8 clone BAC16, complete genome | GenBank: MK733609.1; Uniprot: D2XQF2 | |
| Gene (Epstein–Barr virus) | BFLF1 | Human gammaherpesvirus 4 isolate NPCT090, complete genome | GenBank: MK540447.1 | |
| Gene (MHV68) | mu68 | Murid herpesvirus 4 strain g2.4, complete genome | GenBank: AF105037.1 Uniprot: O41969 | |
| Gene (human cytomegalovirus) | UL52 | Human herpesvirus 5 strain Towne, complete genome | GenBank: FJ616285.1; Uniprot: O56765 | |
| Gene (Herpes simplex virus 1) | UL32 | Human herpesvirus 1 isolate KOS, complete genome | GenBank: KT899744.1; Uniprot: H9E939 | |
| Strain, strain background (Escherichia coli) | GS1783 | Brulois et al., 2012 | PMID: 22740391 | WT BAC16-containing E. coli used for construction of BAC16 mutants |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 WT | Brulois et al., 2012 | PMID: 22740391 | WT BAC16 containing KSHV genome |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 ORF68.stop (68S) | Gardner and Glaunsinger, 2018 | PMID: 29875246 | BAC16 mutant containing a premature stop codon in ORF68 |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 ORF68.stop-MR (68S-MR) | Gardner and Glaunsinger, 2018 | PMID: 29875246 | Mutant rescue of BAC16 ORF68.stop |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 ORF68-K435A | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | BAC16 mutant containing ORF68-K435A | |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 ORF68-K435A-MR | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Mutant rescue of BAC16 ORF68-K435A | |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 ORF68-K174A/R179A/K182A (3+) | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | BAC16 mutant containing ORF68-K174A/R179A/K182A | |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 K174A/R179A/K182A-MR (3+-MR) | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Mutant rescue of BAC16 ORF68-K174A/R179A/K182A | |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 ORF68-TS | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | BAC16 mutant containing ORF68-TS | |
| Genetic reagent (Kaposi’s sarcoma-associated herpesvirus) | KSHV BAC16 ORF68-TS-MR | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | BAC16 mutant containing ORF68-TS-MR | |
| Cell line (Homo sapiens) | HEK-293T | ATCC | CRL-3216 | This cell line is commercially available from ATCC |
| Cell line (Homo sapiens) | iSLK-puro | Myoung and Ganem, 2011; Stürzl et al., 2013 | PMID: 21419799; PMID: 22987579 | Renal-cell carcinoma cell line containing doxycycline-inducible KSHV RTA |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 WT | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 WT | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 ORF68.stop | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop-MR | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 ORF68.stop-MR | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68-K435A | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 ORF68-K435A | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68-K435A-MR | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 ORF68-K435A-MR | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68-K174A/R179A/K182A | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 ORF68- K174A/R179A/K182A | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68- K174A/R179A/K182A -MR | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 ORF68- K174A/R179A/K182A -MR | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68-TS | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 ORF68-TS | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68-TS-MR | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | iSLK-puro latently infected with KSHV derived from BAC16 ORF68-TS-MR | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop + pLJM1-zeo-empty | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Lentivirally transduced iSLK-BAC16 ORF68.stop | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop + pLJM1-zeo-2xStrep-ORF68 | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Lentivirally transduced iSLK-BAC16 ORF68.stop | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop + pLJM1-zeo-2xStrep-ORF68-C52A | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Lentivirally transduced iSLK-BAC16 ORF68.stop | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop + pLJM1-zeo-2xStrep-mu68 | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Lentivirally transduced iSLK-BAC16 ORF68.stop | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop + pLJM1-zeo-2xStrep-BFLF1 | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Lentivirally transduced iSLK-BAC16 ORF68.stop | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop + pLJM1-zeo-2xStrep-UL52 | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Lentivirally transduced iSLK-BAC16 ORF68.stop | |
| Cell line (Homo sapiens, Kaposi’s sarcoma-associated herpesvirus) | iSLK-BAC16 ORF68.stop + pLJM1-zeo-UL32 | This paper; 'Materials and methods' (Cell line establishment and viral mutagenesis) | Lentivirally transduced iSLK-BAC16 ORF68.stop | |
| Antibody | Anti-vinculin (rabbit polyclonal) | Abcam | Cat. no. ab91459 | WB primary (1:1000) |
| Antibody | Strep-Tag II HRP conjugate | Novagen | Cat. no. 71591-3 | WB primary (1:2500) |
| Antibody | Anti-K8.1 (rabbit polyclonal) | Gardner and Glaunsinger, 2018 | PMID: 29875246 | WB primary (1:10,000) |
| Antibody | Anti-ORF68 (rabbit polyclonal) | Gardner and Glaunsinger, 2018 | PMID: 29875246 | WB primary (1:5000) |
| Antibody | Anti-ORF6 (Rabbit polyclonal) | Gardner and Glaunsinger, 2018 | PMID: 29875246 | WB primary (1:10,000) |
| Antibody | Anti-GAPDH (mouse monoclonal) | Abcam | Cat. no. ab8245 | WB primary (1:5000) |
| Antibody | Anti-rabbit IgG-HRP (goat polyclonal) | Southern Biotech | Cat. no. 4030-05 | WB secondary (1:5000) |
| Antibody | Anti-mouse IgG-HRP (goat polyclonal) | Southern Biotech | Cat. no. 1031-05 | WB secondary (1:5000) |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162625 | For transient expression of ORF68 |
| Recombinant DNA reagent | pcDNA4/TO-ORF68-2xStrep | Davis et al., 2015 | Addgene: 136229 | For transient expression of ORF68 |
| Recombinant DNA reagent | pcDNA4/TO-ORF68 (tagless) | This paper; 'Materials and methods' (Plasmids) | Addgene: 166025 | For transient expression of ORF68 |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-BFLF1 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162626 | For transient expression of BFLF1 |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-mu68 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162627 | For transient expression of mu68 |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-UL52 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162628 | For transient expression of UL52 |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-UL32 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162629 | For transient expression of UL32 |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 C52A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162630 | For transient expression of ORF68 C52A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 C373A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162631 | For transient expression of ORF68 C373A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 C191A/C192A | This paper; 'Materials and methods' (Plasmids) | Addgene:162632 | For transient expression of ORF68 C191A/C192A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 C415A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162633 | For transient expression of ORF68 C415A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 H452A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162634 | For transient expression of ORF68 H452A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 C79A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162635 | For transient expression of ORF68 C79A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 K435A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162636 | For transient expression of ORF68 K435A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 R443A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162637 | For transient expression of ORF68 R443A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 K450A/R451A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162638 | For transient expression of ORF68 K450A/R451A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 K174A/R179A/K182A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162639 | For transient expression of ORF68 K174A/R179A/K182A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 R14A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162640 | For transient expression of ORF68 R14A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 K310A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162641 | For transient expression of ORF68 K310A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 R14A/K310A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162642 | For transient expression of ORF68 R14A/K310A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-ORF68 K395A/K396A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162643 | For transient expression of ORF68 K395A/K396A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-UL32 C128A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162644 | For transient expression of UL32 C128A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-UL32 C502A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162645 | For transient expression of UL32 C502A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-UL32 C308A/C309A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162646 | For transient expression of UL32 C308A/C309A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-UL32 C544A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162647 | For transient expression of UL32 C544A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-UL32 H581A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162648 | For transient expression of UL32 H581A |
| Recombinant DNA reagent | pcDNA4/TO-2xStrep-UL32 C155A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162649 | For transient expression of UL32 C155A |
| Recombinant DNA reagent | pUE1-TSP-ORF68 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162650 | For transient overexpression of ORF68 |
| Recombinant DNA reagent | pUE1-TSP-ORF68 K435A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162651 | For transient overexpression of ORF68 K435A |
| Recombinant DNA reagent | pUE1-TSP-ORF68 R443A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162652 | For transient overexpression of ORF68 R443A |
| Recombinant DNA reagent | pUE1-TSP-ORF68 K450A/R451A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162653 | For transient overexpression of ORF68 K450A/R451A |
| Recombinant DNA reagent | pUE1-TSP-ORF68 K174A/R179A/K182A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162654 | For transient overexpression of ORF68 K174A/R179A/K182A |
| Recombinant DNA reagent | pUE1-TSP-ORF68 R14A/K310A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162655 | For transient overexpression of ORF68 R14A/K310A |
| Recombinant DNA reagent | pUE1-TSP-ORF68 K395A/K396A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162656 | For transient overexpression of ORF68 K395A/K396A |
| Recombinant DNA reagent | pUE1-TSP-BFLF1 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162657 | For transient overexpression of BFLF1 |
| Recombinant DNA reagent | pUE1-TSP-UL32 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162658 | For transient overexpression of UL32 |
| Recombinant DNA reagent | pLJM1-zeo-empty | Gardner and Glaunsinger, 2018 | PMID: 29875246 | Empty lentiviral vector |
| Recombinant DNA reagent | pLJM1-zeo-2xStrep-ORF68 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162659 | Lentiviral vector for stable expression of ORF68 |
| Recombinant DNA reagent | pLJM1-zeo-2xStrep-ORF68 C52A | This paper; 'Materials and methods' (Plasmids) | Addgene: 162660 | Lentiviral vector for stable expression of ORF68 C52A |
| Recombinant DNA reagent | pLJM1-zeo-2xStrep-BFLF1 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162661 | Lentiviral vector for stable expression of BFLF1 |
| Recombinant DNA reagent | pLJM1-zeo-2xStrep-mu68 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162662 | Lentiviral vector for stable expression of mu68 |
| Recombinant DNA reagent | pLJM1-zeo-2xStrep-UL52 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162663 | Lentiviral vector for stable expression of UL52 |
| Recombinant DNA reagent | pLJM1-zeo-UL32 | This paper; 'Materials and methods' (Plasmids) | Addgene: 162664 | Lentiviral vector for stable expression of UL32 |
| Recombinant DNA reagent | psPAX2 | A gift from Didier Trono | Addgene: 12260 | Lentiviral packaging plasmid |
| Recombinant DNA reagent | pMD2.G | A gift from Didier Trono | Addgene: 12259 | Lentiviral packaging plasmid |
| Peptide, recombinant protein | HRV 3C protease | Millipore Sigma | Cat. no: 71493 | For removal of Twin-Strep tag during protein purification |
| Commercial assay or kit | In-Fusion HD Cloning kit | Clontech | Cat. no. 639650 | For cloning |
| Commercial assay or kit | NucleoBond BAC 100 kit | Macherey Nagel | Cat. no. 740579 | For preparation of BAC DNA |
| Commercial assay or kit | NucleoSpin Blood kit | Macherey Nagel | Cat. no. 740951.50 | For isolation of DNA from iSLK BAC16 cell lines |
| Commercial assay or kit | iTaq Universal SYBR Green Supermix | Bio-Rad | Cat. no. 1725122 | For qPCR assays |
| Commercial assay or kit | DIG High Prime DNA Labeling and Detection Starter Kit II | Roche | Cat. no. 11585614910 | For probe labeling and detection of Southern blots |
| Software, algorithm | CTTFFIND4 | PMID: 26278980 | RRID: SCR_016732 | |
| Software, algorithm | Gautomatch | K. Zhang (MRC-LMB (https://www2.mrc-lmb.cam.ac.uk/research/locally-developed-software/zhang-software/)) | ||
| Software, algorithm | Relion | PMID: 23000701 | RRID: SCR_016274 | |
| Software, algorithm | SerialEM | PMID: 16182563 | RRID: SCR_017293 | |
| Software, algorithm | Motioncor2 | PMID: 28250466 | RRID: SCR_016499 | |
| Software, algorithm | Coot | PMID: 20383002 | RRID: SCR_014222 | |
| Software, algorithm | phenix.refine | PMID: 20124702, 22505256, 31588918 | RRID: SCR_016736 | |
| Software, algorithm | XDS | PMID: 20124692 | RRID: SCR_015652 | |
| Software, algorithm | POINTLESS | PMID: 21460446 | RRID: SCR_014218 | |
| Software, algorithm | STARANISO server | STARANISO (staraniso.globalphasing.org) | RRID: SCR_018362 | |
| Software, algorithm | PHASER | PMID: 19461840 | RRID: SCR_014219 | |
| Software, algorithm | PyMol | PyMol (pymol.org) | RRID: SCR_000305 | |
| Software, algorithm | APBS | PMID: 11517324 | RRID: SCR_008387 | |
| Software, algorithm | ConSurf Server | PMID: 27166375 | RRID: SCR_002320 | |
| Software, algorithm | SWISS-MODEL | PMID: 29788355 | RRID: SCR_018123 | |
| Software, algorithm | Clustal Omega | PMID: 30976793 | RRID: SCR_001591 | |
| Software, algorithm | GraphPad Prism | GraphPad | RRID: SCR_002798 | Version 8 |
| Other | Strep-Tactin Superflow high-capacity 50% suspension | IBA Lifesciences | Cat. no. 2-4030-025 |
Additional files
-
Supplementary file 1
Cryo-EM data collection statistics.
The cryo-EM maps for ORF68 and BFLF1 and the coordinate set for BFLF1 are available in Supplementary file 4.
- https://cdn.elifesciences.org/articles/62261/elife-62261-supp1-v2.docx
-
Supplementary file 2
X-ray data collection and refinement statistics for ORF68.
Statistics for the highest-resolution shell are shown in parentheses. The STARANISO server was used for ellipsoidal truncation (Tickle et al., 2018). The worst diffraction limit after cutoff was 2.99 Å. The ellipsoidally truncated data set was deposited in the Protein Data Bank and is available in Supplementary file 4. Merged diffraction data that has not been ellipsoidally truncated is also available in Supplementary file 4. The coordinate set deposited in the Protein Data Bank is available as Supplementary file 4.
- https://cdn.elifesciences.org/articles/62261/elife-62261-supp2-v2.docx
-
Supplementary file 3
Oligonucleotides used for cloning, qPCR, and EMSAs.
- https://cdn.elifesciences.org/articles/62261/elife-62261-supp3-v2.xlsx
-
Supplementary file 4
Diffraction data sets, cryo-EM maps, and coordinate sets for ORF68 and BFLF1.
- https://cdn.elifesciences.org/articles/62261/elife-62261-supp4-v2.zip
-
Transparent reporting form
- https://cdn.elifesciences.org/articles/62261/elife-62261-transrepform-v2.docx





